Published online Apr 14, 2019. doi: 10.3748/wjg.v25.i14.1666
Peer-review started: February 12, 2019
First decision: February 26, 2019
Revised: March 4, 2019
Accepted: March 16, 2019
Article in press: March 16, 2019
Published online: April 14, 2019
Processing time: 63 Days and 13.4 Hours
Artificial intelligence (AI) using deep-learning (DL) has emerged as a breakthrough computer technology. By the era of big data, the accumulation of an enormous number of digital images and medical records drove the need for the utilization of AI to efficiently deal with these data, which have become fundamental resources for a machine to learn by itself. Among several DL models, the convolutional neural network showed outstanding performance in image analysis. In the field of gastroenterology, physicians handle large amounts of clinical data and various kinds of image devices such as endoscopy and ultrasound. AI has been applied in gastroenterology in terms of diagnosis, prognosis, and image analysis. However, potential inherent selection bias cannot be excluded in the form of retrospective study. Because overfitting and spectrum bias (class imbalance) have the possibility of overestimating the accuracy, external validation using unused datasets for model development, collected in a way that minimizes the spectrum bias, is mandatory. For robust verification, prospective studies with adequate inclusion/exclusion criteria, which represent the target populations, are needed. DL has its own lack of interpretability. Because interpretability is important in that it can provide safety measures, help to detect bias, and create social acceptance, further investigations should be performed.
Core tip: Artificial intelligence (AI) using deep-learning (DL) has emerged as a breakthrough computer technology. The convolutional neural network exhibited outstanding performance in image analysis. AI has been applied in the field of gastroenterology in terms of diagnosis, prognosis, and image analysis. However, potential inherent pitfalls of selection bias, overfitting, and spectrum bias (class imbalance) have the possibility of overestimating the accuracy and generalizing the result. Therefore, external validation using unused datasets for model development, collected in a way that minimizes the spectrum bias, is mandatory. DL has its own lack of interpretability, and further investigations should be performed on this issue.
- Citation: Yang YJ, Bang CS. Application of artificial intelligence in gastroenterology. World J Gastroenterol 2019; 25(14): 1666-1683
- URL: https://www.wjgnet.com/1007-9327/full/v25/i14/1666.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i14.1666
Recently, artificial intelligence (AI) using deep-learning (DL) has emerged as a breakthrough computer technology, and numerous research studies, using AI applications to identify or differentiate images in various medical fields including radiology, neurology, orthopedics, pathology, ophthalmology, and gastroenterology, have been published[1]. However, AI, the display of intelligent behavior indistinguishable from that of a human being, was already mentioned in the 1950s[2]. Although AI has waxed and waned over the past six decades with seemingly little improvement, it was constantly applied to the medical field using various models of machine learning (ML) including Bayesian inferences, decision trees, linear discriminants, support vector machines (SVM), logistic regression, and artificial neural networks (ANNs).
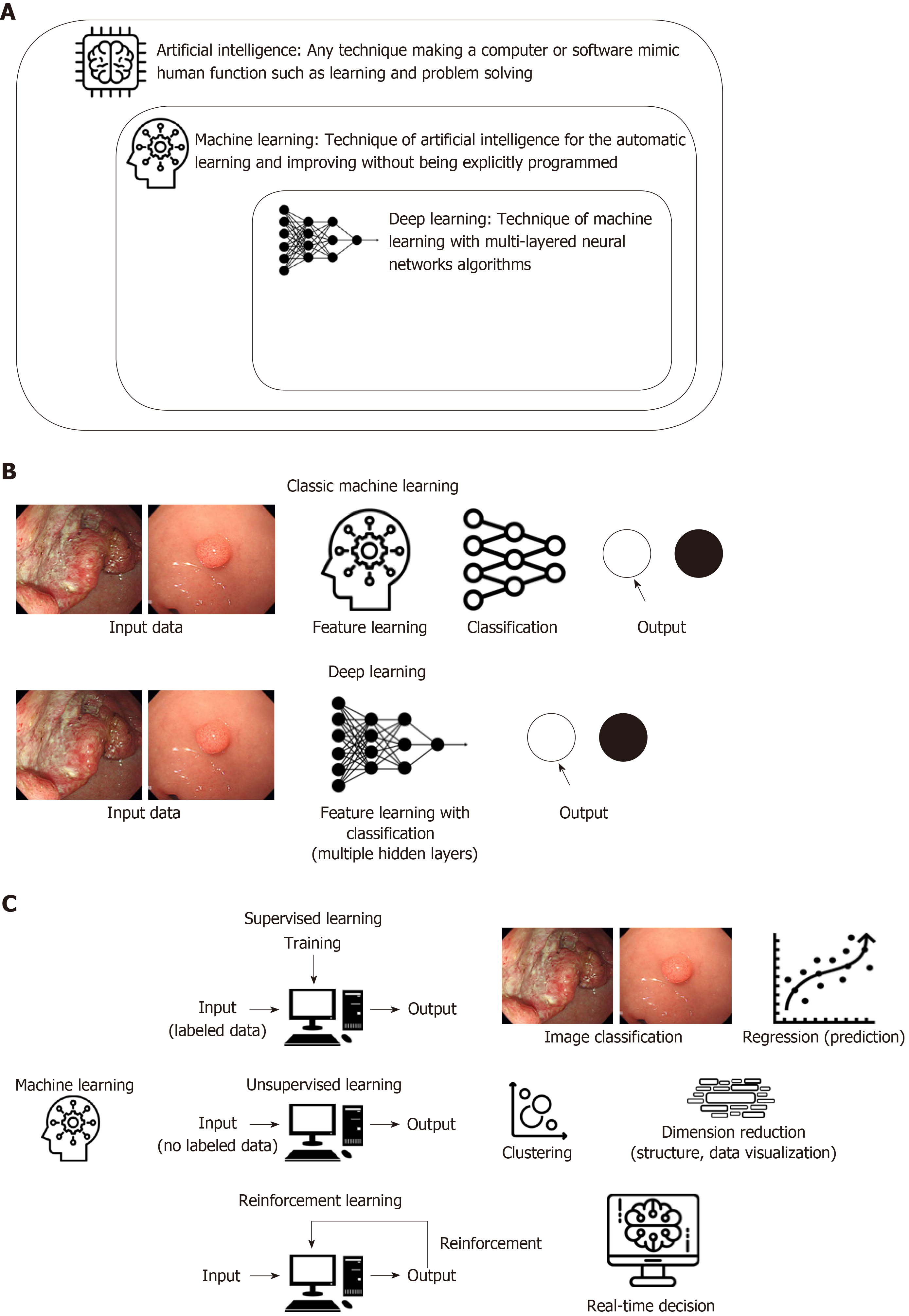
By the era of big data, the accumulation of enormous digital images and medical records drove a need for the utilization of AI to efficiently deal with these data, which also become fundamental resources for the machine to learn by itself. Furthermore, the evolution of computing power with graphic processing units can overcome the limitations of traditional ML, particularly overtraining for input data (overfitting). This led to a revival of AI, especially when using DL technology, a new form of ML. Among several DL methods, the convolutional neural network (CNN), which consists of multilayers of ANN with step-by-step minimal processing, showed outstanding performance in image analysis and has received attention in AI (Figure 1 and Table 1).
| Artificial intelligence | Machine intelligence that has cognitive functions similar to those of humans such as “learning” and “problem solving.” |
| Machine learning | Mathematical algorithms which is automatically built from given data (known as input training data) and predicts or makes decisions in uncertain conditions without being explicitly programmed |
| Support vector machines | Discriminative classifier formally defined by an optimizing hyperplane with the largest functional margin |
| Artificial neural networks | Multilayered interconnected network which consists of an input, hidden connection (between the input and output layer), and output layer |
| Deep learning | Subset of machine learning technique that composed of multiple-layered neural network algorithms |
| Convolutional neural networks | Specific class of artificial neural networks that consists of (1) convolutional and pooling layers, which are the two main components to extract distinct features; and (2) fully connected layers to make an overall classification |
| Overfitting | Modelling error which occurs when a certain learning model tailors itself too much on the training dataset and predictions are not well generalized to new datasets |
| Spectrum bias | Systematic error occurs when the dataset used for model development does not adequately represent or reflect the range of patients who will be applied in clinical practice (target population) |
In the field of gastroenterology, physicians handle large amounts of clinical data and various kinds of image devices such as esophagogastroduodenoscopy (EGD), colonoscopy, capsule endoscopy (CE), and ultrasound equipment. AI has been applied in the field of gastroenterology when making a diagnosis, predicting a prognosis, and analyzing images. Previous studies reported remarkable results of AI in gastroenterology. The rapid progression of AI demands that gastroenterologists learn the utility, strengths, and pitfalls of AI. In addition, physicians should prepare for the changes and effects of AI on real clinical practice in the near future. Hence, in this review, we aim to (1) briefly introduce an ML technology; (2) summarize an AI application in the field of gastroenterology, which is divided into two categories (statistical analysis for recognition of diagnosis or prediction of prognosis, and analyze images for patient applications excluding animal studies); and (3) discuss the challenges for the application and future directions of AI.
Generally, AI is considered as a machine intelligence that has cognitive functions similar to those of humans including “learning” and “problem solving[3]”. Currently, ML is the most common approach of AI. It automatically builds mathematical algorithms from given data (known as input training data) and predicts or makes decisions in uncertain conditions without human instructions (Figure 1A)[4]. In the medical field, ML methods such as Bayesian networks, linear discriminants, SVMs, and ANNs have been used[5]. A naïve Bayes classifier that represents the probabilistic relationship between input and output data is a typical classification model[6]. The SVM, which was invented by Vladimir N Vapnik and Alexey Ya Chervonenkis in 1963[7], is a discriminative model that uses a dividing hyperplane. Before DL development, SVM showed the best performance for classification and regression, which were achieved by optimizing a hyperplane with the largest functional margin (distance from the hyperplane in a high- or infinite-dimensional space to the nearest training data point of any class)[8].
An ANN is a multilayered interconnected network inspired by the neuronal connections of the human brain. Although the ANN was introduced by McCulloch and Walter in 1943[9], it was studied in 1957 by Frank Rosenblatt using the concept of the perceptron[10]. The ANN as a hierarchical structure consists of an input, hidden connection (between the input and output layer), and output layer. The connection in the hidden layer has a strength (known as weight) that is used for the learning process of the network (Figure 1B). Through an appropriate training process (learning process), the network can adjust the value of the connection weight to optimize the best result (Figure 1C).
In the 1980s, an ANN with several hidden layers between the input and output layer was introduced. This was known as a DL (or a deep neural network). Although the ANN showed remarkable performance in managing nonlinear datasets regarding diagnosis and prognostic prediction in the medical field, the ANN revealed several weaknesses as well: a vanishing gradient, overfitting, insufficient computing capacity, and lack of training data. These weaknesses hampered the advancement of the ANN. Finally, the recent availability of big data provided sufficient input data for training, and the rapid progression of computing power allowed researchers to overcome prior limitations. Among several AI methods, DL received the attention of the public and has shown excellent performance in the computer vision area using CNNs.
A CNN consists of (1) convolutional and pooling layers, which are the two main components to extract distinct features; and (2) fully connected layers to make an overall classification. The input images were filtered to extract specialized features using numerous specific filters, and to create multiple feature maps. This preprocessing operation for filtering is called convolution. A learning process for the convolution filter to make the best feature maps is essential for success in a CNN. These feature maps are compressed to smaller sizes by pooling the pixels to capture a larger field of the image, and these convolutional and pooling layers are iterated many times. Finally, fully connected layers combine all features and produce the final outcomes (Figure 1B).
The rapid growth of the CNN was demonstrated at the ImageNet Large Scale Visual Recognition Competition (ILSVRC) in 2012 by Geoffrey Hinton, and several CNNs such as Inception from google and ResNet from Microsoft have shown excellent performance. A graphical summary of AI, ML, and DL development is shown in Figure 1.
Although AI in the field of gastroenterology recently focused on image analysis, several ML models have shown promising results in the recognition of diagnosis and prediction of prognosis. The ANN is appropriate for dealing with complex datasets to overcome the drawbacks of traditional linear statistics. In addition, the ANN can stand for the sophisticated interactions between demographic, environmental, and clinical characteristics.
In terms of diagnosis, Pace et al[11] demonstrated an ANN model in 2005 that made a diagnosis of gastroesophageal reflux disease using only 45 clinical variables in 159 cases with an accuracy of 100%. Lahner et al[12] performed a similar pilot study to recognize atrophic gastritis solely by using clinical and biochemical variables from 350 outpatients by using ANNs and linear discriminant analysis. This study showed great accuracy.
Regarding the prediction of prognosis, in 1998, Pofahl et al[13] compared an ANN model to the Ranson criteria and the Acute Physiologic and Chronic Health Evaluation (APACHE II) scoring system to predict the length of stay for patients with acute pancreatitis. The authors used a backpropagation neural network that was trained using 156 patients. Although the highest specificity (94%) was observed in the Ranson criteria, the ANN model showed the highest sensitivity (75%) when predicting a length of stay more than 7 d. Similar accuracy was observed for the Ranson criteria and APACHE II scoring system[13]. In 2003, Das et al[14] used an ANN to predict the outcomes of acute lower gastrointestinal bleeding using 190 patients. The authors compared the performance of ANNs to a previously validated scoring system (BLEED), which revealed a significantly better predictive accuracy of mortality (87% vs 21%), recurrent bleeding (89% vs 41%), and the need for therapeutic intervention (96% vs 46%) in the ANN model.
Sato et al[15] presented an ANN model in 2005 to predict 1-year and 5-year survival using 418 esophageal cancer patients. This ANN model showed improved accuracy compared to the conventional linear discriminant analysis model.
Recently, the number of input training data items for ANNs was increased from hundreds to thousands of patients. Rotondano et al[16] compared the Rockall score to a supervised ANN model to predict the mortality of nonvariceal upper gastrointestinal bleeding using 2380 patients. This approach showed superior sensitivity (83.8% vs 71.4%), specificity (97.5% vs 52.0%), accuracy (96.8% vs 52.9%), and area under receiver operating characteristic (AUROC) of the predictive performance (0.95 vs 0.67) in the ANN model to those in the complete Rockall score.
Takayama et al[17] established an ANN model for the prediction of prognosis in patients with ulcerative colitis after cytoapheresis therapy and achieved a sensitivity and specificity for the need of an operation of 96% and 97%, respectively. Hardalaç et al[18] established an ANN model to predict mucosal healing by azathioprine therapy in patients with inflammatory bowel disease (IBD) and achieved 79.1% correct classifications. Peng et al[19] used an ANN model to predict the frequency of the onset, relapse, and severity of IBD. The researchers achieved an average accuracy to predict the frequency of onset and severity of IBD but a high accuracy in predicting the frequency of relapse of IBD (mean square error = 0.009, mean absolute percentage error = 17.1%).
SVMs have been used to analyze data and recognize patterns in classification analyses. Recently, Ichimasa et al[20] analyzed 45 clinicopathological factors in 690 endoscopically resected T1 colorectal cancer patients to predict lymph node metastasis using a SVM. This approach showed superior performance (sensitivity 100%, specificity 66%, accuracy 69%) compared to those of American (sensitivity 100%, specificity 44%, accuracy 49%), European (sensitivity 100%, specificity 0%, accuracy 9%), and Japanese (sensitivity 100%, specificity 0%, accuracy 9%) guidelines. A prediction model using a SVM model reduced the amount of unnecessary additional surgery (77%) when misdiagnosing lymph node metastasis than those of a prediction model using American (85%), European (91%), and Japanese guidelines (91%). Yang et al[21] constructed an SVM-based model using clinicopathological features and 23 immunologic markers from 483 patients who underwent curative surgery for esophageal squamous cell carcinoma. This study revealed reasonable performance in identifying high-risk patients with postoperative distant metastasis [sensitivity 56.6%, specificity 97.7%, positive predictive value (PPV) 95.6%, negative predictive value (NPV) 72.3%, and overall accuracy 78.7%] (Table 2).
| Ref. | Published year | Aim of study | Design of study | Number of subjects | Type of AI | Input variables (number/type) | Outcomes |
| Pace et al[11] | 2005 | Diagnosis of gastroesophageal reflux disease | Retrospective | 159 patients (10 times cross validation) | “backpropagation” ANN | 101/clinical variables | Accuracy: 100% |
| Lahner et al[12] | 2005 | Recognition of atrophic corpus gastritis | Retrospective | 350 patients (subdivided several times into training and test set equally) | ANN | 37 to 3 /clinical and biochemical variables (experiment 1 to 5) | Accuracy: 96.6%, 98.8%, 98.4%, 91.3% and 97.7% (experiment 1-5, respectively) |
| Pofahl et al[13] | 1998 | Prediction of length of stay for patients with acute pancreatitis | Retrospective | 195 patients (training set: 156, test set: 39) | “backpropagation” ANN | 71/clinical variables | Sensitivity: 75 % (for prediction of a length of stay more than 7 d) |
| Das et al[14] | 2003 | Prediction of outcomes in acute lower gastrointestinal bleeding | Prospective | 190 patients (training set: 120, internal validation set: 70, external validation set: 142) | ANN | 26/clinical variables | Accuracy (external validation set): 97% for death, 93% for, recurrent bleeding, 94% for need for intervention |
| Sato et al[15] | 2005 | Prediction of 1-year and 5-year survival of esophageal cancer | Retrospective | 418 patients (training-: validation-: test set = 53%: 27%: 20%) | ANN | 199/ clinicopathologic, biologic, and genetic variables | AUROC for 1 year- and 5 year survival prediction: 0.883 and 0.884, respectively |
| Rotondano et al[16] | 2011 | Prediction of mortality in nonvariceal upper gastrointestinal bleeding | Prospective, multicenter | 2380 patients (5 × 2 cross-validation) | ANN | 68/clinical variables | Accuracy: 96.8%, AUROC: 0.95, sensitivity: 83.8%, specificity: 97.5%, |
| Takayama et al[17] | 2015 | Prediction of prognosis in ulcerative colitis after cytoapheresis therapy | Retrospective | 90 patients (training set: 54, test set: 36) | ANN | 13/clinical variables | Sensitivity: 96.0%, specificity: 97.0% |
| Hardalaç et al[18] | 2015 | Prediction of mucosal healing by azathioprine therapy in IBD | Retrospective | 129 patients (training set: 103, validation set: 13, test set: 13) | “feed-forward back-propagation” and “cascade-forward” ANN | 6/clinical variables | Total correct classification rate: 79.1% |
| Peng et al[19] | 2015 | Prediction of frequency of onset, relapse, and severity of IBD | Retrospective | 569 UC and 332 CD patients (training set: data from 2003-2010, validation set: data in 2011) | ANN | 5/meteorological data | Accuracy in predicting the frequency of relapse of IBD (mean square error = 0.009, mean absolute percentage error = 17.1%) |
| Ichimasa et al[20] | 2018 | Prediction of lymph node metastasis, thus minimizing the need for additional surgery in T1 colorectal cancer | Retrospective | 690 patients (training set: 590, validation set: 100) | SVM | 45/ Clinicopathological variables | Accuracy: 69%, sensitivity: 100%, specificity: 66% |
| Yang et al[21] | 2013 | Prediction of postoperative distant metastasis in esophageal squamous cell carcinoma | Retrospective | 483 patients (training set: 319, validation set: 164) | SVM | 30/7 clinicopathological variables and 23 immunomarkers | Accuracy: 78.7% sensitivity: 56.6%, specificity: 97.7%, PPV: 95.6%, NPV: 72.3% |
Although endoscopic screening programs have reduced the mortality from gastrointestinal malignancies, they are still the leading cause of death worldwide and remain a global economic burden. To enhance the detection rate of gastrointestinal neoplasms and optimize the treatment strategies, a high-quality endoscopic examination for the recognition of gastrointestinal neoplasms and classifications between benign and malignant lesions are essential for the gastroenterologist. Thus, gastroenterologists are interested in the applications of AI, especially when using CNNs and SVMs for image analysis. Furthermore, AI has been increasingly adopted in terms of non-neoplastic gastrointestinal diseases including infection, inflammation, or hemorrhage.
Upper gastrointestinal field: Takiyama et al[22] constructed a CNN model that could recognize the anatomical location of EGD images with AUROCs of 1.00 for the larynx and esophagus, and 0.99 for the stomach and duodenum. This CNN model could also recognize specific anatomical locations within the stomach, with AUROCs of 0.99 for the upper, middle, and lower stomach.
To assist in the discrimination of early neoplastic lesions in Barrett’s esophagus, van der Sommen et al[23] developed an automated algorithm to include specific textures, color filters, and ML from 100 endoscopic images. This algorithm reasonably detected early neoplastic lesions in a per-image analysis with a sensitivity and specificity of 83%. In 2017, the same group investigated a model to improve the detection rate of early neoplastic lesions in Barrett’s esophagus by using 60 ex vivo volumetric laser endomicroscopy images. This novel computer model showed optimal performance compared with a clinical volumetric laser endomicroscopy prediction score with a sensitivity of 90% and specificity of 93%[24].
Several studies evaluated the ML model using specialized endoscopy to differentiate neoplastic/dysplastic and non-neoplastic lesions. Kodashima et al[25] showed that computer-based analysis can easily identify malignant tissue at the cellular level using endocytoscopic images, which enables the microscopic visualization of the mucosal surface. In 2015, Shin et al[26] reported on an image analysis model to detect esophageal squamous dysplasia using high-resolution microendoscopy (HRME). The sensitivity and specificity of this model were 87% and 97%, respectively. During the following year, Quang et al[27] from the same study group evolved this model, which was incorporated in tablet-interfaced HRME with full automation for real-time analysis. As a result, the model reduced the costs compared to previous laptop-interfaced HRME and showed good diagnostic yields of esophageal squamous cell carcinoma with a sensitivity and specificity of 95% and 91%, respectively. However, there was a limitation for the application of this model owing to the unavailability of specialized endoscopy.
Finally, Horie et al[28] demonstrated the utility of AI using CNNs to make a diagnosis of esophageal cancer. This was trained with 8428 conventional endoscopic images including white-light images (WLIs) and narrow-band images (NBIs). This CNN model detected esophageal cancer with a sensitivity of 95% and could identify all small cancers of < 10 mm. This model also distinguished superficial esophageal cancer from advanced cancer with an accuracy of 98%.
Helicobacter pylori (H. pylori) infection is the most important risk factor of peptic ulcers and gastric cancer. Several researchers challenged AI to aid the endoscopic diagnosis of H. pylori infections. In 2004, Huang et al[29] investigated the predictability of H. pylori infection by refined feature selection with a neural network using related gastric histologic features in endoscopic images. This model was trained and analyzed with 84 image parameters from 30 patients. The sensitivity and specificity for the detection of H. pylori infection were 85.4% and 90.9%, respectively. In addition, the accuracy of this model for identifying gastric atrophy, intestinal metaplasia, and predicting the severity of H. pylori-related gastric inflammation was higher than 80%.
Recently, two Japanese researchers reported on the application of a CNN to make a diagnosis of H. pylori infection[30,31]. Itoh et al[31] developed a CNN model to recognize H. pylori infections by using 596 endoscopic images after the data augmentation of a prior set of 149 images. This CNN model showed promising results with a sensitivity and specificity of 86.7% and 86.7%, respectively. Shichijo et al[30] compared the performance of a CNN to that of 23 endoscopists for the diagnosis of H. pylori infection by using endoscopic images. The CNN model showed superior sensitivity (88.9% vs 79.0%), specificity (87.4% vs 83.2%), accuracy (87.7% vs 82.4%), and diagnostic time (194 s vs 230 s).
In 2018, a prospective pilot study was conducted for automated diagnosis of H. pylori infections using image-enhanced endoscopy such as blue laser imaging-bright and linked color imaging. The performance of the developed AI model was significantly higher with blue laser imaging-bright and linked color imaging training (AUROCs of 0.96 and 0.95) than WLI imaging training (0.66)[32].
The utility of AI in the diagnosis of gastrointestinal neoplasms was classified into two main categories: detection and characterization. In 2012, Kubota et al[33] first evaluated a computer-aided pattern recognition system to identify the depth of the wall invasion of gastric cancer using endoscopic images. They used 902 endoscopic images and created a backpropagation model after a 10-time cross validation. As a result, the diagnostic accuracy was 77.2%, 49.1%, 51.0%, and 55.3% for T1-4 staging, respectively. In particular, the accuracy of T1a (mucosal invasion) and T1b staging (submucosal invasion) was 68.9% and 63.6%, respectively. Hirasawa et al[34] reported on the good performance of a CNN-based diagnostic system to detect gastric cancers in endoscopic images. The authors trained the CNN model using 13584 endoscopic images and tested it with 2296 images. The overall sensitivity was 92.2%. In addition, the detection rate with a diameter of 6 mm or more was 98.6%, and all invasive cancers were identified[34]. All missed lesions were superficially depressed and differentiated-type intramucosal cancers that were difficult to distinguish from gastritis even for experienced endoscopists. However, 69.4% of the lesions that the CNN diagnosed as gastric cancer were benign, and the most common reasons for misdiagnosis were gastritis with redness, atrophy, and intestinal metaplasia[34].
Zhu et al[35] further applied a CNN system to discriminate the invasion depth of gastric cancer (M/SM1 vs deeper than SM1) using conventional endoscopic images. They trained a CNN model with 790 images and tested it with another 203 images. The CNN model showed high accuracy (89.2%) and specificity (95.6%) when determining the invasion depth of gastric cancer. This result was significantly superior to that of experienced endoscopists. Kanesaka et al[36] studied a computer-aided diagnosis system using a SVM to facilitate the use of magnifying NBI to distinguish early gastric cancer. The study reported on remarkable potential in terms of diagnostic performance (accuracy 96.3%, PPV 98.3%, sensitivity 96.7%, and specificity 95%) and the performance of area concordance (accuracy 73.8%, PPV 75.3%, sensitivity 65.5%, and specificity 80.8%).
In terms of hepatology, the ultrasound has been challenged for the application of AI. Gatos et al[37] established a SVM diagnostic model of chronic liver disease using ultrasound shear wave elastography (70 patients with chronic liver disease and 56 healthy controls). The performance was promising, with an accuracy of 87.3%, sensitivity of 93.5%, and specificity of 81.2%, although the prospective validation was not conducted. Kuppili et al[38] established a fatty liver detection and characterization model using a single-layer feed-forward neural network, and validated this model with a higher accuracy than the previous SVM-based model. These researchers used ultrasound images of 63 patients, and the gold standard for labeling for each patient was the pathologic results of a liver biopsy.
The determination of liver cirrhosis was also challenged with ML technology. Liu et al[39] developed a CNN model with ultrasound liver capsule images (44 images from controls and 47 images from patients with cirrhosis), and classified these images using a SVM. The AUROC for the classification was 0.951, although the prospective validation was not conducted.
Lower gastrointestinal field: Among various gastrointestinal fields, the development of an AI model using colonoscopy has been the most promising area because polyp detection during colonoscopies is frequent. This provides sufficient sources for AI training, and a missed colorectal polyp is directly associated with interval colorectal cancer development.
In terms of polyp detection, Fernandez-Esparrach et al[40] established an automated computer-vision method using an energy map to detect colonic polyps in 2016. They used 24 videos containing 31 polyps and showed acceptable performance with a sensitivity of 70.4% and a specificity of 72.4% for polyp detection (Table 3). Recently, this performance was improved with a DL application for polyp detection[41,42]. Misawa et al[41] designed a CNN model using 546 short videos from 73 full-length videos, which were divided into two groups of training data (105 polyp-positive videos and 306 polyp-negative videos) and test data (50 polyp-positive videos and 85 polyp-negative videos). The researchers showed the possibility of the automated detection of colonic polyps in real time, and the sensitivity and specificity were 90.0% and 63.3%, respectively. Urban et al[42] also used a CNN system to identify colonic polyps. They used 8641 hand-labeled images and 20 colonoscopy videos in various combinations as training and test data. The CNN model detected polyps in real time with an AUROC of 0.991 and an accuracy of 96.4%. Moreover, it assisted in the identification of an additional nine polyps compared with expert endoscopists in the application of test colonoscopy videos.
| Ref. | Published year | Aim of study | Design of study | Number of subjects | Type of AI | Endoscopic or ultrasoud modality | Outcomes |
| Takiyama et al[22] | 2018 | Recognition of anatomical locations of EGD images | Retrospective | Training set: 27335 images from 1750 patients. Validation set: 17081 images from 435 patients | CNN | White-light endoscopy | AUROCs: 1.00 for the larynx and esophagus, and 0.99 for the stomach and duodenum recognition |
| van der Sommen et al[23] | 2016 | Discrimination of early neoplastic lesions in Barrett’s esophagus | Retrospective | 100 endoscopic images from 44 patients (leave-one-out cross-validation on a per-patient basis) | SVM | White-light endoscopy | Sensitivity: 83%, specificity: 83% (per-image analysis) |
| Swager et al[24] | 2017 | Identification of early Barrett’s esophagus neoplasia on ex vivo volumetric laser endomicroscopy images. | Retrospective | 60 volumetric laser endomicroscopy images | Combination of several methods (SVM, discriminant analysis, AdaBoost, random forest, etc) | Ex vivo volumetric laser endomicroscopy | Sensitivity: 90%, specificity: 93% |
| Kodashima et al[25] | 2007 | Discrimination between normal and malignant tissue at the cellular level in the esophagus | Prospective ex vivo pilot | 10 patients | ImageJ program | Endocytoscopy | Difference in the mean ratio of total nuclei to the entire selected field, 6.4 ± 1.9% in normal tissues and 25.3 ± 3.8% in malignant samples |
| Shin et al[26] | 2015 | Diagnosis of esophageal squamous dysplasia | Prospective, multicenter | 375 sites from 177 patients (training set: 104 sites, test set: 104 sites, validation set: 167 sites) | Linear discriminant analysis | HRME | Sensitivity: 87%, specificity: 97% |
| Quang et al[27] | 2016 | Diagnosis of esophageal squamous cell neoplasia | Retrospective, multicenter | Same data from reference number 26 | Linear discriminant analysis | Tablet-interfaced HRME | Sensitivity: 95%, specificity: 91% |
| Horie et al[28] | 2019 | Diagnosis of esophageal cancer | Retrospective | Training set: 8428 images from 384 patients. Test set: 1118 images from 97 patients | CNN | White-light endoscopy with NBI | Sensitivity 98% |
| Huang et al[29] | 2004 | Diagnosis of H. pylori infection | Prospective | Training set: 30 patients. Test set: 74 patients | Refined feature selection with neural network | White-light endoscopy | Sensitivity: 85.4%, specificity: 90.9% |
| Shichijo et al[30] | 2017 | Diagnosis of H. pylori Infection | Retrospective | Training set: CNN1: 32208 images; CNN2: images classified according to 8 different locations in the stomach. Test set: 11481 images from 397 patients | CNN | White-light endoscopy | Accuracy: 87.7%, sensitivity: 88.9%, specificity: 87.4%, diagnostic time: 194 s. |
| Itoh et al[31] | 2018 | Diagnosis of H. pylori infection | Prospective | Training set: 149 images (596 images through data augmentation. Test set: 30 images | CNN | White-light endoscopy | AUROC: 0.956, sensitivity: 86.7%, specificity: 86.7%, |
| Nakashima et al[32] | 2018 | Diagnosis of H. pylori infection | Prospective pilot | 222 patients (training set: 162, test set: 60) | CNN | White-light endoscopy and image-enhanced endoscopy, such as blue laser imaging-bright and linked color imaging | AUROC: 0.96 (blue laser imaging-bright), 0.95 (linked color imaging) |
| Kubota et al[33] | 2012 | Diagnosis of depth of invasion in gastric cancer | Retrospective | 902 images (10 times cross validation) | “backpropagation” ANN | White-light endoscopy | Accuracy: 77.2%, 49.1%, 51.0%, and 55.3% for T1-4 staging, respectively |
| Hirasawa et al[34] | 2018 | Detection of gastric cancers | Retrospective | Training set: 13584 images. Test set: 2296 images. | CNN | White-light endoscopy, chromoendoscopy, NBI | Sensitivity: 92.2%, detection rate with a diameter of 6 mm or more: 98.6% |
| Zhu et al[35] | 2018 | Diagnosis of depth of invasion in gastric cancer (mucosa/SM1/deeper than SM1) | Retrospective | Training set: 790 images. Test set: 203 images | CNN | White-light endoscopy | Accuracy: 89.2%, AUROC: 0.94, sensitivity: 74.5%, specificity: 95.6% |
| Kanesakaet al[36] | 2018 | Diagnosis of early gastric cancer using magnifying NBI images | Retrospective | Training set: 126 images. Test set: 81 images | SVM | Magnifying NBI | Accuracy: 96.3%, sensitivity: 96.7%, specificity: 95%, PPV: 98.3%, |
| Gatos et al[37] | 2017 | Diagnosis of chronic liver disease | Retrospective | 126 patients (56 healthy controls, 70 with chronic liver disease | SVM | Ultrasound shear wave elastography imaging with a stiffness value-clustering | AUROC: 0.87, highest accuracy: 87.3%, sensitivity: 93.5%, specificity: 81.2% |
| Kuppili et al[38] | 2017 | Detection and characterization of fatty liver | Prospective | 63 patients who underwent liver biopsy (10 times cross validation) | Extreme Learning Machine to train single-layer feed-forward neural network | Ultrasound liver images | Accuracy: 96.75%, AUROC: 0.97 (validation performance) |
| Liu et al[39] | 2017 | Diagnosis of liver cirrhosis | Retrospective | 44 images from controls and 47 images from patients with cirrhosis | SVM | Ultrasound liver capsule images | AUROC: 0.951 |
Although there were many promising performances of the automated polyp detection models, a prospective validation was not conducted[43-45]. However, Klare et al[46] performed a prototype software validation under real-time conditions (55 routine colonoscopies), and the results were comparable between those of endoscopists and the established software. The endoscopists’ polyp detection rates and adenoma detection rates were 56.4% and 30.9%, respectively, and these rates were 50.9% and 29.1% for the software, respectively). Wang et al[47] established a DL algorithm by using data from 1290 patients, and validated this model with 27113 newly collected colonoscopy images from 1138 patients. This model showed remarkable performance with a sensitivity of 94.38%, specificity of 95.2%, and AUROC of 0.984 for at least one polyp detection[47].
For AI applications of polyp characterization, magnifying endoscopic images, which is useful when discriminating pit or vascular patterns, was first adopted to enhance the performance of AI. Tischendort et al[48] developed an automated classification model of colorectal polyps by magnifying NBI images to evaluate vascular patterns in 2010. They reported that the overall accurate classification rates were 91.9% for a consensus decision between the human observers and 90.9% for a safe decision (classifying polyps as neoplastic in cases when there was an interobserver discrepancy)[48]. In 2011, Gross et al[49] compared the performances of a computer-based model for the differentiation of small colonic polyps of < 10 mm using NBI images. The expert endoscopists and computer-based model showed comparable diagnostic performance in sensitivity (93.4% vs 95.0%), specificity (91.8% vs 90.3%), and accuracy (92.7% vs 93.1%)[49].
Takemura et al[50] retrospectively compared the identification of pit patterns of a computer-based model with shape descriptors such as area, perimeter, fit ellipse, or circularity in reference to endoscopic diagnosis by using magnified endoscopic images with crystal violet staining in 2010. The accuracies of the type I, II, IIIL, and IV pit patterns of colorectal lesions were 100%, 100%, 96.6%, and 96.7%, respectively. In 2012, the authors applied an upgraded version of a computer system via SVM to distinguish neoplastic and non-neoplastic lesions by using endoscopic NBI images, which showed a detection accuracy of 97.8%[51]. They further demonstrated the availability of a real-time image recognition system in 2016, and the accuracy between the pathologic results of diminutive polyps and diagnosis by a real-time image recognition model was 93.2%[52].
Byrne et al[53] developed a CNN model for the real-time differentiation of diminutive colorectal polyps by using only NBI video frames in 2017. This model discriminated adenomas from hyperplastic polyps with an accuracy of 94%, and identified the adenoma with a sensitivity of 98% and a specificity of 83%[53]. Likewise, Chen et al[54] made a CNN model trained with 2157 images to identify neoplastic or hyperplastic polyps of < 5 mm with a PPV and NPV of 89.6% and 91.5%, respectively. In 2017, Komeda et al[55] reported on the preliminary data of a CNN model to distinguish adenomas from non-adenomatous polyps. The CNN model was trained with 1800 conventional endoscopic images with WLI, NBI, and chromoendoscopy, and the accuracy of a 10-hold cross-validation was 75.1%.
To enhance the differentiation of polyps, a Japanese study group reported several articles for AI application with endocytoscopy images, which enables the observation of nuclei on site, and showed comparable diagnostic results to those of pathologic examinations. In 2015, these researchers first developed a computer-aided diagnosis system using endocytoscopy for the discrimination of neoplastic changes in small polyps. This approach showed a comparable sensitivity (92.0%) and accuracy (89.2%) with those of expert endoscopists[56]. In 2016, this research team developed a second-generation model that could (1) evaluate both nuclei and ductal lumens, (2) use an SVM instead of multivariate analysis, (3) provide the confidence levels of the decisions, and (4) provide a more rapid process of discriminating neoplastic changes from 0.3 s to 0.2 s. The endocytoscopic microvascular patterns could be effectively evaluated by staining with dye[57]. These researchers also developed endocytoscopy with NBI without staining to evaluate microvascular findings. This approach showed an overall accuracy of 90%[57]. The same group performed a prospective validation of a real-time computer-aided diagnosis system using endocytoscopy with NBI or stained images to identify neoplastic diminutive polyps. The researchers reported a pathologic prediction rate of 98.1%, and the time required to assess one diminutive polyp was about 35 to 47 s[59].
The application of a computer-aided ultrahigh (approximately 400 ×) magnification endocytoscopy system for the diagnosis of invasive colorectal cancers was investigated by Takeda et al[60]. This system was trained with 5543 endocytoscopic images from 238 lesions and reported a sensitivity of 89.4%, specificity of 98.9%, and accuracy of 94.1% using 200 test images[60].
For the application of AI in IBD, Maeda et al[61] developed a diagnosis system using a SVM after refining previous computer-aided endocytoscopy systems[56-58]. They evaluated the diagnostic performance of this model for the prediction of persistent histologic inflammation in ulcerative colitis patients. This model showed good performance with a sensitivity of 74%, specificity of 97%, and an accuracy of 91%[61].
Currently, the resolution of images is relatively low in capsule endoscopy compared to other digestive endoscopies. Moreover, the interpretation and diagnosis of capsule endoscopy images highly depends on the reviewer’s ability and effort. It is also a time-consuming process. Therefore, several conditions were attempted for the automated diagnosis of capsule endoscopy images including angioectasia, celiac disease, or intestinal hookworms, or for small intestinal motility characterization[62-65].
Leenhardt et al[62] developed a gastrointestinal angiectasia detection model using semantic segmentation images with a CNN. They used 600 control images and 600 typical angiectasia images to form 4166 small bowel capsule endoscopy videos, which were divided equally into training and test data sets. The CNN-based model revealed a high diagnostic performance with a sensitivity of 100%, specificity of 96%, PPV of 96%, and NPV of 100%[62] (Table 4). Zhou et al[63] established a CNN model for the classification of celiac disease from control with capsule endoscopy clips from six celiac disease patients and five controls. The researchers achieved 100% sensitivity and specificity for the test data set. Moreover, the evaluation confidence was related to the severity level of small bowel mucosal lesions, reflecting the potential for the quantitative measurement of the existence and degree of pathology throughout the small intestine[63]. Intestinal hookworms are difficult to find with direct visualization because they have small tubular structures with a whitish color and semitransparent features similar to background intestinal mucosa. Moreover, the presence of intestinal secretory materials makes them difficult to detect. He et al[64] established a CNN model for the detection of hookworms in capsule endoscopy images. The CNN-based model showed a reasonable performance with a sensitivity of 84.6%, specificity of 88.6% and only 15% hookworm images and 11% non-hookworm image were falsely detected.
| Ref. | Published year | Aim of study | Design of study | Number of subjects | Type of AI | Endoscopic modality | Outcomes |
| Fernandez-Esparrach et al[40] | 2016 | Detection of colonic polyps | Retrospective | 24 videos containing 31 polyps | Window Median Depth of Valleys Accumulation maps | White-light colonoscopy | Sensitivity: 70.4%. Specificity: 72.4% |
| Misawa et al[41] | 2018 | Detection of colonic polyps | Retrospective | 546 short videos (training set: 105 polyp-positive videos and 306 polyp-negative videos, test set: 50 polyp-positive videos and 85 polyp-negative videos) from 73 full length videos | CNN | White-light colonoscopy | Accuracy: 76.5%. Sensitivity: 90.0%. Specificity: 63.3%. |
| Urban et al[42] | 2018 | Detection of colonic polyps | Retrospective | 8641 images with 20 colonoscopy videos | CNN | White-light colonoscopy with NBI | Accuracy: 96.4%. AUROC: 0.991 |
| Klare et al[46] | 2019 | Detection of colonic polyps | Prospective | 55 patients | Automated polyp detection software | White-light colonoscopy | Polyp detection rate: 50.9%. Adenoma detection rate: 29.1% |
| Wang et al[47] | 2018 | Detection of colonic polyps | Retrospective | Training set: 5545 images from 1290 patients. Validation set A: 27113 images from 1138 patients. Validation set B: 612 images. Validation set C: 138 video clips from 110 patients. Validation set D: 54 videos from 54 patients | CNN | White-light colonoscopy | Dataset A: AUROC: 0.98 for at least one polyp detection, per-image sensitivity: 94.4%, per-image specificity: 95.2%. Dataset B: per-image sensitivity: 88.2%. Dataset C: per-image sensitivity: 91.6%, per-polyp sensitivity: 100%. Dataset D: per-image specificity: 95.4% |
| Tischendort et al[48] | 2010 | Classification of colorectal polyps on the basis of vascularization features. | Prospective pilot | 209 polyps from 128 patients | SVM | Magnifying NBI images | Accurate classification rate: 91.9% |
| Gross et al[49] | 2011 | Differentiation of small colonic polyps of < 10 mm | Prospective | 434 polyps from 214 patients | SVM | Magnifying NBI images | Accuracy: 93.1%. Sensitivity: 95.0%. Specificity: 90.3%. |
| Takemura et al[50] | 2010 | Classification of pit patterns | Retrospective | Training set: 72 images. Validation set: 134 images | HuPAS software version 1.3 | Magnifying endoscopic images with crystal violet staining | Accuracies of the type I, II, IIIL, and IV pit patterns of colorectal lesions: 100%, 100%, 96.6%, and 96.7%, respectively |
| Takemura et al[51] | 2012 | Classification of histology of colorectal tumors | Retrospective | Training set: 1519 images. Validation set: 371 images | HuPAS software version 3.1 using SVM | Magnifying NBI images | Accuracy: 97.8% |
| Kominami et al[52] | 2016 | Classification of histology of colorectal polyps | Prospective | Training set: 2247 images from 1262 colorectal lesion. Validation: 118 colorectal lesions | SVM with logistic regression | Magnifying NBI images | Accuracy: 93.2%, Sensitivity: 93.0%, Specificity: 93.3%, PPV: 93%, NPV: 93.3% |
| Byrne et al[53] | 2017 | Differentiation of histology of diminutive colorectal polyps | Retrospective | Training set: 223 videos, Validation set: 40 videos. Test set: 125 videos | CNN | NBI video frames | Accuracy: 94%, Sensitivity: 98%, Specificity: 83% |
| Chen et al[54] | 2018 | Identification of neoplastic or hyperplastic polyps of < 5 mm | Retrospective | Training set: 2157 images. Test set: 284 images | CNN | Magnifying NBI images | Sensitivity: 96.3%, specificity: 78.1%, PPV: 89.6%, NPV: 91.5% |
| Komeda et al[55] | 2017 | Discrimination adenomas from non-adenomatous polyps | Retrospective | 1200 images from the endoscopic videos (10 times cross validation) | CNN | White-light colonoscopy with NBI and chromoendoscopy | Accuracy in validation: 75.1% |
| Mori et al[56] | 2015 | Discrimination of neoplastic changes in small polyps | Retrospective | Test set: 176 polyps form 152 patients | Multivariate regression analysis | Endocytoscopy | Accuracy: 89.2%, Sensitivity: 92.0% |
| Mori et al[57] | 2016 | Development of 2nd generation model, which was mentioned in reference number 56 | Retrospective | Test set: 205 small colorectal polyps (≤ 10 mm) from 123 patients | SVM | Endocytoscopy | Accuracy: 89% for both diminutive(< 5 mm) and small (< 10 mm) polyps |
| Misawa et al[58] | 2016 | Diagnosis of colorectal lesions using microvascular findings | Retrospective | Training set: 979 images, validation set: 100 images | SVM | Endocytoscopy with NBI | Accuracy: 90% |
| Mori et al[59] | 2018 | Diagnosis of neoplastic diminutive polyp | Prospective | 466 diminutive polyps from 325 patients | SVM | Endocytoscopy with NBI and stained images | Prediction rate: 98.1% |
| Takeda et al[60] | 2017 | Diagnosis of invasive colorectal cancer | Retrospective | Training set: 5543 images from 238 lesions. Test set: 200 images | SVM | Endocytoscopy with NBI and stained images | Accuracy: 94.1% Sensitivity: 89.4%, Specificity: 98.9%, PPV: 98.8%, NPV: 90.1% |
| Maeda et al[61] | 2018 | Prediction of persistent histologic inflammation in ulcerative colitis patients | Retrospective | Training set: 12900 images.Test set: 9935 images | SVM | Endocytoscopy with NBI | Accuracy: 91%, Sensitivity: 74%, Specificity: 97% |
The interpretation of wireless motility capsule endoscopy is a complex task. Seguí et al[65] established a CNN model for small-intestine motility characterization and achieved a mean classification accuracy of 96% for six intestinal motility events (“turbid”, “bubbles”, “clear blob”, “wrinkles”, “wall”, and, “undefined”). This outperformed the other classifiers by a large margin (a 14% relative performance increase).
Although many researchers have investigated the utility of AI and have shown promising results, most studies were designed in retrospective manner: as a case-control study from a single center, or by using endoscopic images that were chosen from specific endoscopic modalities unavailable from many institutions. Potential inherent bias such as selection bias cannot be excluded in this situation. Therefore, it is crucial to meticulously validate the performance of AI before the application of AI in real clinical practice. To properly verify the accuracy of AI, physicians should understand the effects of overfitting and spectrum bias (class imbalance) on the performance of AI, and try to evaluate the performance by avoiding these biases.
Overfitting occurs when a learning model tailors itself too much on the training dataset and predictions are not well generalized to new datasets[66] (Table 5). Although several methods were used to reduce overfitting in the development of DL models, they did not guarantee the resolution of this problem. In addition, datasets that were collected by case-control design are particularly vulnerable to spectrum bias. Spectrum bias occurs when the dataset used for model development does not adequately represent the range of patients who will be applied in clinical practice (target population)[67].
| Ref. | Published year | Aim of study | Design of study | Number of subjects | Type of AI | Outcomes |
| Leenhardt et al[62] | 2019 | Detection of gastrointestinal angiectasia | Retrospective | 600 control images and 600 typical angiectasia images (divided equally into training and test datasets) | CNN | Sensitivity: 100%, specificity: 96%, PPV: 96%, NPV: 100%. |
| Zhou et al[63] | 2017 | Classification of celiac disease | Retrospective | Training set: 6 celiac disease patients, 5 controls. Test set: additional 5 celiac disease patients, 5 controls | CNN | Sensitivity: 100%, specificity: 100% (for test dataset) |
| He et al[64] | 2018 | Detection of intestinal hookworms | Retrospective | 440000 images | CNN | Sensitivity: 84.6%, specificity: 88.6% |
| Seguí et al[65] | 2016 | Characterization of small intestinal motility | Retrospective | 120000 images (training set: 100000, test set: 20000) | CNN | Mean classification accuracy: 96% |
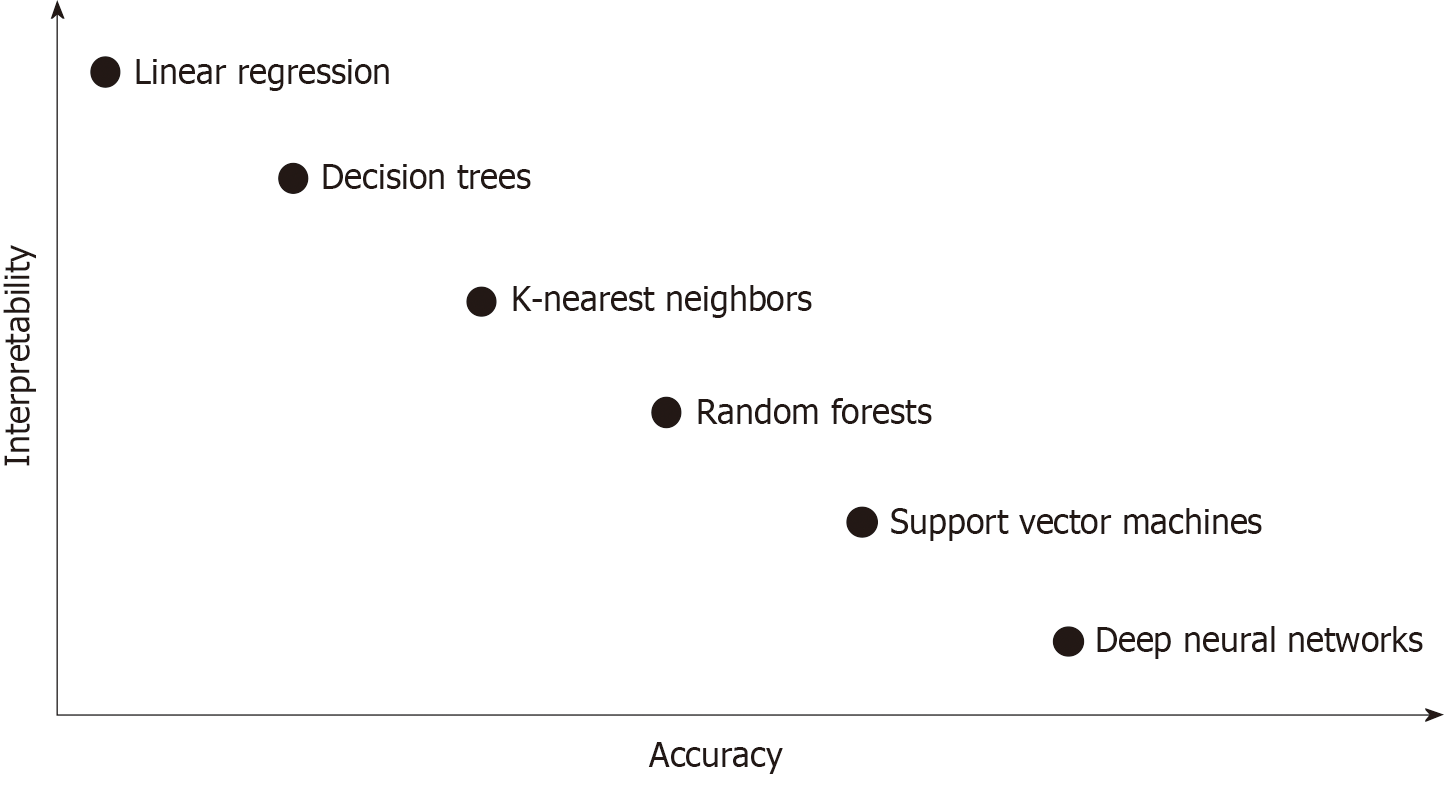
Because overfitting and spectrum bias may lead to overestimation of the accuracy and generalization, external validation using unused datasets for model development, collected in a way that minimizes the spectrum bias, is mandatory. For more robust clinical verification, well-designed multicenter prospective studies with adequate inclusion/exclusion criteria that represent the target population are needed. Furthermore, DL technology has its own “black box” nature (lack of interpretability or explainability), which means the decision mechanism of AI is not clearly demonstrated (Figure 2). Because interpretability is important in that it can provide safety measures, help to detect bias, and establish social acceptance, further investigation to solve this issue should be performed. However, there have been some methods to complement “black box” characteristics, such as the attention map and saliency region[68].
It is obvious that the efficiency and accuracy of ML increases as the amount of data increases; however, it is challenging to develop an efficient ML model owing to the paucity of human labeled data given the issue of privacy with regard to private medical records. To overcome this issue, data augmentation strategies (with synthetically modified data) have been proposed[69]. Spiking neural networks, which more closely mimic the real mechanisms of neurons, can potentially replace current ANN models with more powerful computing ability, although no effective supervised learning method currently exists[70].
The precision of diagnosis or classification using AI does not always mean efficacy in real clinical practice. The actual benefit of the clinical outcome, the satisfaction of physicians, and the cost effectiveness beyond the academic performance must be proven by sophisticated investigation. Finally, the acquisition of reasonable regulations from responsible authorities and a reimbursement policy are essential for integrating AI technology in the current healthcare environment. Moreover, AI is not perfect. That’s why “Augmented Intelligence” emerged emphasizing the fact that AI is designed to improve or enhance human intelligence rather than replace it. Although the aim of applying AI in medical practice is to improve the workflow with enhanced precision and to reduce the number of unintentional errors, established models with inaccuracy or exaggerated performance are likely to cause ethical issues owing to misdiagnosis or misclassification. Moreover, we do not know the impact of AI application on the doctor-patient relationship, which is an essential part of healthcare utilization and the practice of medicine. Therefore, ethical principles relevant to AI model development should be established in the current period when AI research begins to increase.
Since AI was introduced in the 1950s, it has been persistently challenged in terms of statistical or image analyses in the field of gastroenterology. Recent evaluation of big data and computer science enabled the dramatic development of AI technology, particularly DL, which showed promising potential. Now, there is no doubt that the implementation of AI in the gastroenterology field will progress in various healthcare services. To utilize AI wisely, physicians should make great effort to understand its feasibility and ameliorate the drawbacks through further investigation.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: South Korea
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Chiu KW, Triantafyllou K S-Editor: Ma RY L-Editor: A E-Editor: Song H
| 1. | Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. 2019;25:44-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2376] [Cited by in RCA: 2842] [Article Influence: 473.7] [Reference Citation Analysis (0)] |
| 2. | Turing AM. Computing machinery and intelligence. Oxford: Mind 1950; 433-460. |
| 3. | Russell S, Norvig P. Artificial intelligence: a modern approach. 3rd ed. Harlow: Pearson Education 2009; 1-3. |
| 4. | Kevin PM. Machine learning: a probabilistic perspective. 1st ed. Cambridge: The MIT Press 2012; 1-2. |
| 5. | Bishop B. CM: Pattern recognition and machine learning. J Electron Imaging. 2006;16:140-155. |
| 6. | Lee JG, Jun S, Cho YW, Lee H, Kim GB, Seo JB, Kim N. Deep Learning in Medical Imaging: General Overview. Korean J Radiol. 2017;18:570-584. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 555] [Cited by in RCA: 598] [Article Influence: 74.8] [Reference Citation Analysis (2)] |
| 7. | Boser BE, Guyon IM, Vapnik VN. A training algorithm for optimal margin classifiers. Proceedings of the fifth annual workshop on Computational learning theory; 1992 July 1, Pittsburgh, US. New York: ACM 1992; 144-152. |
| 8. | Hastie T, Tibshirani R, Friedman J. The elements of Statistical Learning. New York: Springer 2001; . |
| 9. | McCulloch WS, Pitts W. A logical calculus of the ideas immanent in nervous activity. Bull Math Biophys. 1943;5:115-133. |
| 10. | ROSENBLATT F. The perceptron: a probabilistic model for information storage and organization in the brain. Psychol Rev. 1958;65:386-408. [PubMed] |
| 11. | Pace F, Buscema M, Dominici P, Intraligi M, Baldi F, Cestari R, Passaretti S, Bianchi Porro G, Grossi E. Artificial neural networks are able to recognize gastro-oesophageal reflux disease patients solely on the basis of clinical data. Eur J Gastroenterol Hepatol. 2005;17:605-610. [PubMed] |
| 12. | Lahner E, Grossi E, Intraligi M, Buscema M, Corleto VD, Delle Fave G, Annibale B. Possible contribution of artificial neural networks and linear discriminant analysis in recognition of patients with suspected atrophic body gastritis. World J Gastroenterol. 2005;11:5867-5873. [PubMed] |
| 13. | Pofahl WE, Walczak SM, Rhone E, Izenberg SD. Use of an artificial neural network to predict length of stay in acute pancreatitis. Am Surg. 1998;64:868-872. [PubMed] |
| 14. | Das A, Ben-Menachem T, Cooper GS, Chak A, Sivak MV, Gonet JA, Wong RC. Prediction of outcome in acute lower-gastrointestinal haemorrhage based on an artificial neural network: internal and external validation of a predictive model. Lancet. 2003;362:1261-1266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 150] [Article Influence: 6.8] [Reference Citation Analysis (1)] |
| 15. | Sato F, Shimada Y, Selaru FM, Shibata D, Maeda M, Watanabe G, Mori Y, Stass SA, Imamura M, Meltzer SJ. Prediction of survival in patients with esophageal carcinoma using artificial neural networks. Cancer. 2005;103:1596-1605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 58] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 16. | Rotondano G, Cipolletta L, Grossi E, Koch M, Intraligi M, Buscema M, Marmo R; Italian Registry on Upper Gastrointestinal Bleeding (Progetto Nazionale Emorragie Digestive). Artificial neural networks accurately predict mortality in patients with nonvariceal upper GI bleeding. Gastrointest Endosc. 2011;73:218-226, 226.e1-226.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 48] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 17. | Takayama T, Okamoto S, Hisamatsu T, Naganuma M, Matsuoka K, Mizuno S, Bessho R, Hibi T, Kanai T. Computer-Aided Prediction of Long-Term Prognosis of Patients with Ulcerative Colitis after Cytoapheresis Therapy. PLoS One. 2015;10:e0131197. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 18. | Hardalaç F, Başaranoğlu M, Yüksel M, Kutbay U, Kaplan M, Özderin Özin Y, Kılıç ZM, Demirbağ AE, Coşkun O, Aksoy A, Gangarapu V, Örmeci N, Kayaçetin E. The rate of mucosal healing by azathioprine therapy and prediction by artificial systems. Turk J Gastroenterol. 2015;26:315-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 19. | Peng JC, Ran ZH, Shen J. Seasonal variation in onset and relapse of IBD and a model to predict the frequency of onset, relapse, and severity of IBD based on artificial neural network. Int J Colorectal Dis. 2015;30:1267-1273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 20. | Ichimasa K, Kudo SE, Mori Y, Misawa M, Matsudaira S, Kouyama Y, Baba T, Hidaka E, Wakamura K, Hayashi T, Kudo T, Ishigaki T, Yagawa Y, Nakamura H, Takeda K, Haji A, Hamatani S, Mori K, Ishida F, Miyachi H. Artificial intelligence may help in predicting the need for additional surgery after endoscopic resection of T1 colorectal cancer. Endoscopy. 2018;50:230-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 111] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 21. | Yang HX, Feng W, Wei JC, Zeng TS, Li ZD, Zhang LJ, Lin P, Luo RZ, He JH, Fu JH. Support vector machine-based nomogram predicts postoperative distant metastasis for patients with oesophageal squamous cell carcinoma. Br J Cancer. 2013;109:1109-1116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 22. | Takiyama H, Ozawa T, Ishihara S, Fujishiro M, Shichijo S, Nomura S, Miura M, Tada T. Automatic anatomical classification of esophagogastroduodenoscopy images using deep convolutional neural networks. Sci Rep. 2018;8:7497. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 88] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 23. | van der Sommen F, Zinger S, Curvers WL, Bisschops R, Pech O, Weusten BL, Bergman JJ, de With PH, Schoon EJ. Computer-aided detection of early neoplastic lesions in Barrett's esophagus. Endoscopy. 2016;48:617-624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 127] [Article Influence: 14.1] [Reference Citation Analysis (2)] |
| 24. | Swager AF, van der Sommen F, Klomp SR, Zinger S, Meijer SL, Schoon EJ, Bergman JJGHM, de With PH, Curvers WL. Computer-aided detection of early Barrett's neoplasia using volumetric laser endomicroscopy. Gastrointest Endosc. 2017;86:839-846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 102] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 25. | Kodashima S, Fujishiro M, Takubo K, Kammori M, Nomura S, Kakushima N, Muraki Y, Goto O, Ono S, Kaminishi M, Omata M. Ex vivo pilot study using computed analysis of endo-cytoscopic images to differentiate normal and malignant squamous cell epithelia in the oesophagus. Dig Liver Dis. 2007;39:762-766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 18] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 26. | Shin D, Protano MA, Polydorides AD, Dawsey SM, Pierce MC, Kim MK, Schwarz RA, Quang T, Parikh N, Bhutani MS, Zhang F, Wang G, Xue L, Wang X, Xu H, Anandasabapathy S, Richards-Kortum RR. Quantitative analysis of high-resolution microendoscopic images for diagnosis of esophageal squamous cell carcinoma. Clin Gastroenterol Hepatol. 2015;13:272-279.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 66] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 27. | Quang T, Schwarz RA, Dawsey SM, Tan MC, Patel K, Yu X, Wang G, Zhang F, Xu H, Anandasabapathy S, Richards-Kortum R. A tablet-interfaced high-resolution microendoscope with automated image interpretation for real-time evaluation of esophageal squamous cell neoplasia. Gastrointest Endosc. 2016;84:834-841. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 61] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 28. | Horie Y, Yoshio T, Aoyama K, Yoshimizu S, Horiuchi Y, Ishiyama A, Hirasawa T, Tsuchida T, Ozawa T, Ishihara S, Kumagai Y, Fujishiro M, Maetani I, Fujisaki J, Tada T. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest Endosc. 2019;89:25-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 273] [Article Influence: 45.5] [Reference Citation Analysis (0)] |
| 29. | Huang CR, Sheu BS, Chung PC, Yang HB. Computerized diagnosis of Helicobacter pylori infection and associated gastric inflammation from endoscopic images by refined feature selection using a neural network. Endoscopy. 2004;36:601-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 45] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 30. | Shichijo S, Nomura S, Aoyama K, Nishikawa Y, Miura M, Shinagawa T, Takiyama H, Tanimoto T, Ishihara S, Matsuo K, Tada T. Application of Convolutional Neural Networks in the Diagnosis of Helicobacter pylori Infection Based on Endoscopic Images. EBioMedicine. 2017;25:106-111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 235] [Cited by in RCA: 181] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 31. | Itoh T, Kawahira H, Nakashima H, Yata N. Deep learning analyzes Helicobacter pylori infection by upper gastrointestinal endoscopy images. Endosc Int Open. 2018;6:E139-E144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 130] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 32. | Nakashima H, Kawahira H, Kawachi H, Sakaki N. Artificial intelligence diagnosis of Helicobacter pylori infection using blue laser imaging-bright and linked color imaging: a single-center prospective study. Ann Gastroenterol. 2018;31:462-468. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 63] [Article Influence: 9.0] [Reference Citation Analysis (1)] |
| 33. | Kubota K, Kuroda J, Yoshida M, Ohta K, Kitajima M. Medical image analysis: computer-aided diagnosis of gastric cancer invasion on endoscopic images. Surg Endosc. 2012;26:1485-1489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 58] [Article Influence: 4.1] [Reference Citation Analysis (1)] |
| 34. | Hirasawa T, Aoyama K, Tanimoto T, Ishihara S, Shichijo S, Ozawa T, Ohnishi T, Fujishiro M, Matsuo K, Fujisaki J, Tada T. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer. 2018;21:653-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 568] [Cited by in RCA: 428] [Article Influence: 61.1] [Reference Citation Analysis (0)] |
| 35. | Zhu Y, Wang QC, Xu MD, Zhang Z, Cheng J, Zhong YS, Zhang YQ, Chen WF, Yao LQ, Zhou PH, Li QL. Application of convolutional neural network in the diagnosis of the invasion depth of gastric cancer based on conventional endoscopy. Gastrointest Endosc. 2019;89:806-815.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 231] [Article Influence: 38.5] [Reference Citation Analysis (0)] |
| 36. | Kanesaka T, Lee TC, Uedo N, Lin KP, Chen HZ, Lee JY, Wang HP, Chang HT. Computer-aided diagnosis for identifying and delineating early gastric cancers in magnifying narrow-band imaging. Gastrointest Endosc. 2018;87:1339-1344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 129] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 37. | Gatos I, Tsantis S, Spiliopoulos S, Karnabatidis D, Theotokas I, Zoumpoulis P, Loupas T, Hazle JD, Kagadis GC. A Machine-Learning Algorithm Toward Color Analysis for Chronic Liver Disease Classification, Employing Ultrasound Shear Wave Elastography. Ultrasound Med Biol. 2017;43:1797-1810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 73] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 38. | Kuppili V, Biswas M, Sreekumar A, Suri HS, Saba L, Edla DR, Marinhoe RT, Sanches JM, Suri JS. Extreme Learning Machine Framework for Risk Stratification of Fatty Liver Disease Using Ultrasound Tissue Characterization. J Med Syst. 2017;41:152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 76] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 39. | Liu X, Song JL, Wang SH, Zhao JW, Chen YQ. Learning to Diagnose Cirrhosis with Liver Capsule Guided Ultrasound Image Classification. Sensors (Basel). 2017;17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 64] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 40. | Fernández-Esparrach G, Bernal J, López-Cerón M, Córdova H, Sánchez-Montes C, Rodríguez de Miguel C, Sánchez FJ. Exploring the clinical potential of an automatic colonic polyp detection method based on the creation of energy maps. Endoscopy. 2016;48:837-842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 87] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 41. | Misawa M, Kudo SE, Mori Y, Cho T, Kataoka S, Yamauchi A, Ogawa Y, Maeda Y, Takeda K, Ichimasa K, Nakamura H, Yagawa Y, Toyoshima N, Ogata N, Kudo T, Hisayuki T, Hayashi T, Wakamura K, Baba T, Ishida F, Itoh H, Roth H, Oda M, Mori K. Artificial Intelligence-Assisted Polyp Detection for Colonoscopy: Initial Experience. Gastroenterology. 2018;154:2027-2029.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 269] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 42. | Urban G, Tripathi P, Alkayali T, Mittal M, Jalali F, Karnes W, Baldi P. Deep Learning Localizes and Identifies Polyps in Real Time With 96% Accuracy in Screening Colonoscopy. Gastroenterology. 2018;155:1069-1078.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 433] [Article Influence: 61.9] [Reference Citation Analysis (1)] |
| 43. | Wang H, Liang Z, Li LC, Han H, Song B, Pickhardt PJ, Barish MA, Lascarides CE. An adaptive paradigm for computer-aided detection of colonic polyps. Phys Med Biol. 2015;60:7207-7228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 44. | k G, c R. Automatic Colorectal Polyp Detection in Colonoscopy Video Frames. Asian Pac J Cancer Prev. 2016;17:4869-4873. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 45. | Billah M, Waheed S, Rahman MM. An Automatic Gastrointestinal Polyp Detection System in Video Endoscopy Using Fusion of Color Wavelet and Convolutional Neural Network Features. Int J Biomed Imaging. 2017;2017:9545920. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 62] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 46. | Klare P, Sander C, Prinzen M, Haller B, Nowack S, Abdelhafez M, Poszler A, Brown H, Wilhelm D, Schmid RM, von Delius S, Wittenberg T. Automated polyp detection in the colorectum: a prospective study (with videos). Gastrointest Endosc. 2019;89:576-582.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 74] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 47. | Wang P, Xiao X, Brown JR, Berzin TM, Tu M, Xiong F, Hu X, Liu P, Song Y, Zhang D, Yang X. Development and validation of a deep-learning algorithm for the detection of polyps during colonoscopy. Nat Biomed Eng. 2018;2:741. [RCA] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 270] [Article Influence: 38.6] [Reference Citation Analysis (0)] |
| 48. | Tischendorf JJ, Gross S, Winograd R, Hecker H, Auer R, Behrens A, Trautwein C, Aach T, Stehle T. Computer-aided classification of colorectal polyps based on vascular patterns: a pilot study. Endoscopy. 2010;42:203-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 96] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 49. | Gross S, Trautwein C, Behrens A, Winograd R, Palm S, Lutz HH, Schirin-Sokhan R, Hecker H, Aach T, Tischendorf JJ. Computer-based classification of small colorectal polyps by using narrow-band imaging with optical magnification. Gastrointest Endosc. 2011;74:1354-1359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 97] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 50. | Takemura Y, Yoshida S, Tanaka S, Onji K, Oka S, Tamaki T, Kaneda K, Yoshihara M, Chayama K. Quantitative analysis and development of a computer-aided system for identification of regular pit patterns of colorectal lesions. Gastrointest Endosc. 2010;72:1047-1051. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 47] [Article Influence: 3.1] [Reference Citation Analysis (1)] |
| 51. | Takemura Y, Yoshida S, Tanaka S, Kawase R, Onji K, Oka S, Tamaki T, Raytchev B, Kaneda K, Yoshihara M, Chayama K. Computer-aided system for predicting the histology of colorectal tumors by using narrow-band imaging magnifying colonoscopy (with video). Gastrointest Endosc. 2012;75:179-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 84] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 52. | Kominami Y, Yoshida S, Tanaka S, Sanomura Y, Hirakawa T, Raytchev B, Tamaki T, Koide T, Kaneda K, Chayama K. Computer-aided diagnosis of colorectal polyp histology by using a real-time image recognition system and narrow-band imaging magnifying colonoscopy. Gastrointest Endosc. 2016;83:643-649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 160] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 53. | Byrne MF, Chapados N, Soudan F, Oertel C, Linares Pérez M, Kelly R, Iqbal N, Chandelier F, Rex DK. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut. 2019;68:94-100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 363] [Cited by in RCA: 413] [Article Influence: 68.8] [Reference Citation Analysis (0)] |
| 54. | Chen PJ, Lin MC, Lai MJ, Lin JC, Lu HH, Tseng VS. Accurate Classification of Diminutive Colorectal Polyps Using Computer-Aided Analysis. Gastroenterology. 2018;154:568-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 276] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 55. | Komeda Y, Handa H, Watanabe T, Nomura T, Kitahashi M, Sakurai T, Okamoto A, Minami T, Kono M, Arizumi T, Takenaka M, Hagiwara S, Matsui S, Nishida N, Kashida H, Kudo M. Computer-Aided Diagnosis Based on Convolutional Neural Network System for Colorectal Polyp Classification: Preliminary Experience. Oncology. 2017;93 Suppl 1:30-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 129] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 56. | Mori Y, Kudo SE, Wakamura K, Misawa M, Ogawa Y, Kutsukawa M, Kudo T, Hayashi T, Miyachi H, Ishida F, Inoue H. Novel computer-aided diagnostic system for colorectal lesions by using endocytoscopy (with videos). Gastrointest Endosc. 2015;81:621-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 104] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 57. | Mori Y, Kudo SE, Chiu PW, Singh R, Misawa M, Wakamura K, Kudo T, Hayashi T, Katagiri A, Miyachi H, Ishida F, Maeda Y, Inoue H, Nimura Y, Oda M, Mori K. Impact of an automated system for endocytoscopic diagnosis of small colorectal lesions: an international web-based study. Endoscopy. 2016;48:1110-1118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 74] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 58. | Misawa M, Kudo SE, Mori Y, Nakamura H, Kataoka S, Maeda Y, Kudo T, Hayashi T, Wakamura K, Miyachi H, Katagiri A, Baba T, Ishida F, Inoue H, Nimura Y, Mori K. Characterization of Colorectal Lesions Using a Computer-Aided Diagnostic System for Narrow-Band Imaging Endocytoscopy. Gastroenterology. 2016;150:1531-1532.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 125] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 59. | Mori Y, Kudo SE, Misawa M, Saito Y, Ikematsu H, Hotta K, Ohtsuka K, Urushibara F, Kataoka S, Ogawa Y, Maeda Y, Takeda K, Nakamura H, Ichimasa K, Kudo T, Hayashi T, Wakamura K, Ishida F, Inoue H, Itoh H, Oda M, Mori K. Real-Time Use of Artificial Intelligence in Identification of Diminutive Polyps During Colonoscopy: A Prospective Study. Ann Intern Med. 2018;169:357-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 412] [Cited by in RCA: 355] [Article Influence: 50.7] [Reference Citation Analysis (1)] |
| 60. | Takeda K, Kudo SE, Mori Y, Misawa M, Kudo T, Wakamura K, Katagiri A, Baba T, Hidaka E, Ishida F, Inoue H, Oda M, Mori K. Accuracy of diagnosing invasive colorectal cancer using computer-aided endocytoscopy. Endoscopy. 2017;49:798-802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 101] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 61. | Maeda Y, Kudo SE, Mori Y, Misawa M, Ogata N, Sasanuma S, Wakamura K, Oda M, Mori K, Ohtsuka K. Fully automated diagnostic system with artificial intelligence using endocytoscopy to identify the presence of histologic inflammation associated with ulcerative colitis (with video). Gastrointest Endosc. 2019;89:408-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 165] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 62. | Leenhardt R, Vasseur P, Li C, Saurin JC, Rahmi G, Cholet F, Becq A, Marteau P, Histace A, Dray X; CAD-CAP Database Working Group. A neural network algorithm for detection of GI angiectasia during small-bowel capsule endoscopy. Gastrointest Endosc. 2019;89:189-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 148] [Article Influence: 24.7] [Reference Citation Analysis (1)] |
| 63. | Zhou T, Han G, Li BN, Lin Z, Ciaccio EJ, Green PH, Qin J. Quantitative analysis of patients with celiac disease by video capsule endoscopy: A deep learning method. Comput Biol Med. 2017;85:1-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 104] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 64. | He JY, Wu X, Jiang YG, Peng Q, Jain R. Hookworm Detection in Wireless Capsule Endoscopy Images With Deep Learning. IEEE Trans Image Process. 2018;27:2379-2392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 80] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 65. | Seguí S, Drozdzal M, Pascual G, Radeva P, Malagelada C, Azpiroz F, Vitrià J. Generic feature learning for wireless capsule endoscopy analysis. Comput Biol Med. 2016;79:163-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 56] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 66. | England JR, Cheng PM. Artificial Intelligence for Medical Image Analysis: A Guide for Authors and Reviewers. AJR Am J Roentgenol. 2019;212:513-519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 81] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 67. | Park SH, Han K. Methodologic Guide for Evaluating Clinical Performance and Effect of Artificial Intelligence Technology for Medical Diagnosis and Prediction. Radiology. 2018;286:800-809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 467] [Cited by in RCA: 498] [Article Influence: 71.1] [Reference Citation Analysis (0)] |
| 68. | Park SH. Artificial intelligence in medicine: beginner's guide. J Korean Soc Radiol. 2018;78:301-308. [DOI] [Full Text] |
| 69. | Bae HJ, Kim CW, Kim N, Park B, Kim N, Seo JB, Lee SM. A Perlin Noise-Based Augmentation Strategy for Deep Learning with Small Data Samples of HRCT Images. Sci Rep. 2018;8:17687. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 70. | Tavanaei A, Ghodrati M, Kheradpisheh SR, Masquelier T, Maida A. Deep learning in spiking neural networks. Neural Netw. 2019;111:47-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 338] [Cited by in RCA: 280] [Article Influence: 40.0] [Reference Citation Analysis (0)] |