Published online Jan 7, 2019. doi: 10.3748/wjg.v25.i1.42
Peer-review started: October 19, 2018
First decision: November 29, 2018
Revised: December 7, 2018
Accepted: December 13, 2018
Article in press: December 13, 2018
Published online: January 7, 2019
Processing time: 80 Days and 15 Hours
Hepatocellular carcinoma (HCC) is the fifth most common cancer, and hepatitis C virus (HCV) infection plays a major role in HCC development. The molecular mechanisms by which HCV infection leads to HCC are varied. HCV core protein is an important risk factor in HCV-associated liver pathogenesis and can modulate several signaling pathways involved in cell cycle regulation, cell growth promotion, cell proliferation, apoptosis, oxidative stress and lipid metabolism. The dysregulation of signaling pathways such as transforming growth factor β (TGF-β), vascular endothelial growth factor (VEGF), Wnt/β-catenin (WNT), cyclooxygenase-2 (COX-2) and peroxisome proliferator-activated receptor α (PPARα) by HCV core protein is implicated in the development of HCC. Therefore, it has been suggested that this protein be considered a favorable target for further studies in the development of HCC. In addition, considering the axial role of these signaling pathways in HCC, they are considered druggable targets for cancer therapy. Therefore, using strategies to limit the dysregulation effects of core protein on these signaling pathways seems necessary to prevent HCV-related HCC.
Core tip: Hepatitis C virus (HCV) core protein can modulate several signaling pathways involved in cell cycle regulation, cell growth promotion, cell proliferation, apoptosis, oxidative stress and lipid metabolism. The dysregulation of these signaling pathways by HCV core protein is implicated in the development of hepatocellular carcinoma (HCC). Considering the axial role of these signaling pathways in HCC, they are considered druggable targets for cancer therapy. Therefore, using strategies to limit the dysregulation effects of core protein on these signaling pathways seems necessary to prevent HCV-related HCC.
- Citation: Mahmoudvand S, Shokri S, Taherkhani R, Farshadpour F. Hepatitis C virus core protein modulates several signaling pathways involved in hepatocellular carcinoma. World J Gastroenterol 2019; 25(1): 42-58
- URL: https://www.wjgnet.com/1007-9327/full/v25/i1/42.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i1.42
At the beginning of the new century, it became clear that infectious agents have an undeniable role in the development of some cancers in humans. It is currently estimated that nearly 16%-18% of all human cancers are attributed to oncogenic viruses[1]. To date, several viruses are linked to cancer in humans, including Epstein-Barr virus (EBV), human papillomavirus (HPV), hepatitis B virus (HBV), human T-cell lymphotropic virus (HTLV), Kaposi’s sarcoma herpesvirus (KSHV), Merkel cell polyomavirus (MCV) and hepatitis C virus (HCV)[2]. HCV is classified as a member of the Flaviviridae family and the Hepacivirus genus. HCV primarily affects the liver and causes chronic HCV infection. Chronic HCV infection inevitably causes additional liver damage, such as hepatitis, cirrhosis and hepatocellular carcinoma (HCC)[3]. Globally, an estimated 185 million people, equating to about 2.8% of the world population, have been infected with HCV[4]. Although the prevalence of HCV is declining, the burden of HCV-related mortality due to advanced liver disease is on the rise[4,5]. Two major forms of HCV infection are acute and chronic infection. Acute HCV infection can be seen in nearly 20%-25% of infected individuals, and approximately 15% of these acute infections develop recognizable symptomatic disease[6]. Chronic HCV infection develops in 75%-85% of acute HCV infections, and 10%-20% of all cases with chronic HCV infection slowly progress to liver cirrhosis, of which 1%-5% lead to HCC annually[7]. HCC is a significant health burden worldwide, and it is interesting to note that HCC is the fifth common malignant tumor in men (554000 cases) and the ninth common tumor in women (228000 cases). HCC is the second leading cause of cancer deaths worldwide and was responsible for about 746000 deaths in 2012[8]. Interestingly, 27% and 25% of cases with cirrhosis and HCC, respectively, are associated with HCV infection worldwide[9].
Globally, approximately 399000 deaths per year occur due to HCV-related liver diseases. According to the World Health Organization (WHO) treatment guidelines, more than 95% of HCV-infected patients can be cured by antiviral medicines. Therefore, the use of appropriate antiviral therapy can reduce the risk of death from HCC. The current standard of care for patients with HCV infection is therapy with a novel class of direct-acting antivirals (DAAs) in combination with pegylated-interferon α (Peg-IFNα) plus ribavirin. To date, the sustained virologic response (SVR) is the best indicator of successful therapy for chronic HCV infection. SVR is defined as having no detectable HCV RNA at 12-24 wk after completion of antiviral therapy, and increasing the chances of achieving SVR is the main goal of treatment[10]. In the treatment course of HCV infection, the rate of SVR has improved to over 95%[11]. Several studies showed that the risk of HCC is significantly lower in patients who achieved SVR following antiviral therapy compared to untreated patients[12-14]. Overall, more studies are needed to determine whether HCC is reduced among hepatitis C patients after achieving SVR. However, the achievement of SVR is important for HCC prevention. There is currently no prophylactic vaccine for HCV; however, research is ongoing to generate an efficient vaccine[15].
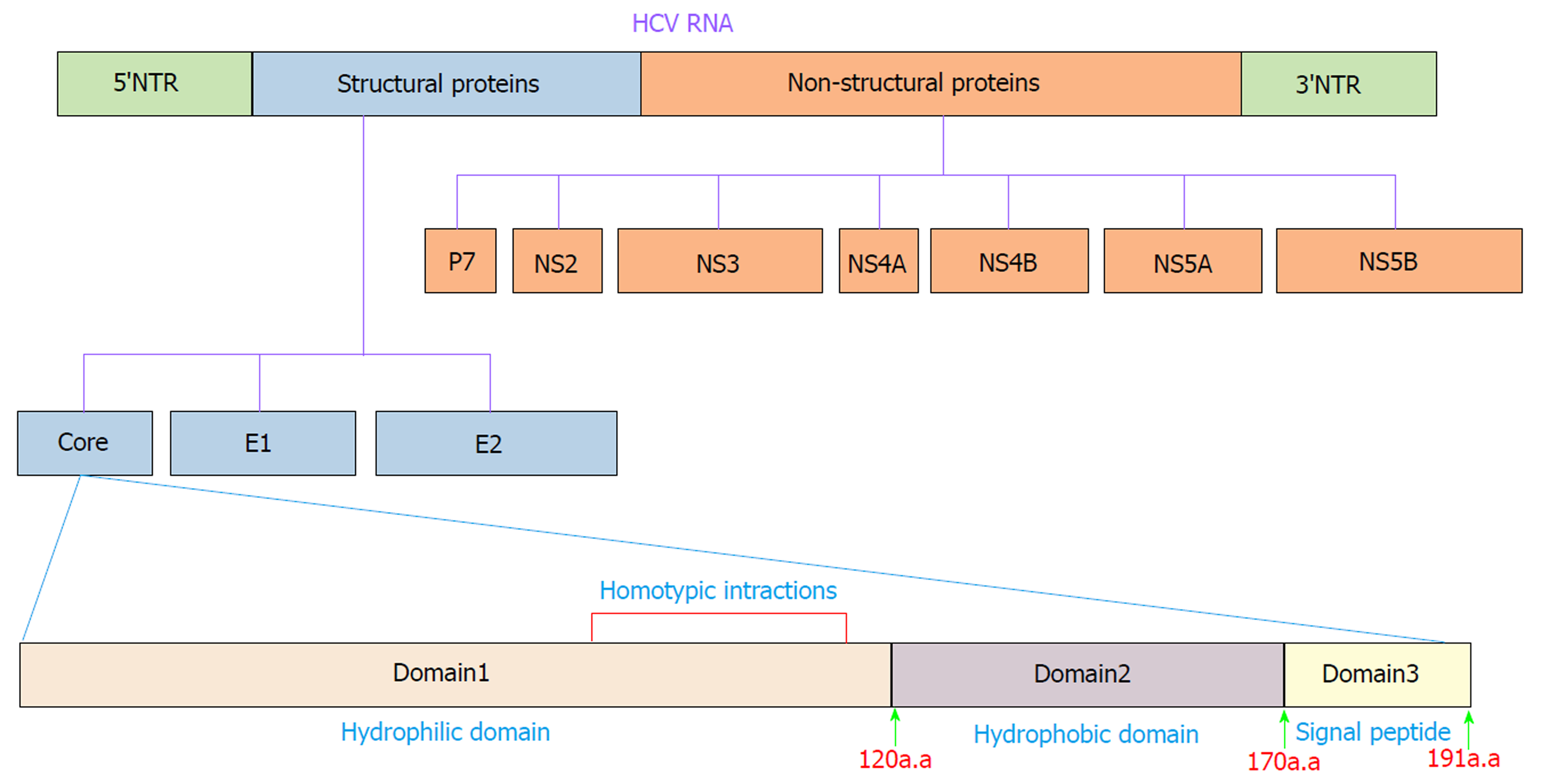
HCV is an enveloped positive-stranded RNA virus that exhibits significant variations across the viral genome. Accordingly, HCV is currently classified into seven genotypes and 67 confirmed subtypes[16]. The HCV genome is approximately 9600 nucleotides in length and encodes a single polyprotein of ~3000 amino acids (aa). The polyprotein is cleaved into ten different structural and nonstructural proteins by viral and cellular proteases. Structural proteins, including core, E1, E2 and p7, are located near the 5′ end of the genome, and nonstructural proteins, including NS1, NS2, NS3, NS4A, NS4B, NS5A and NS5B, are located near the 3′ end of the genome[17] (Figure 1). These proteins make numerous interactions with host cell factors involved in important activities such as cell cycle regulation, cell proliferation, cell growth promotion, transcriptional regulation, apoptosis, oxidative stress and lipid metabolism[18,19]. Many lines of evidence clearly indicate that HCV proteins such as core, NS3, NS5A and NS5B can modulate several potentially oncogenic pathways. These proteins also potentiate oncogenic transformation through direct and indirect interactions with various transcription factors and their induction[20-24]. The core protein is an important HCV protein and is responsible for packaging viral RNA and virion budding. This protein (191 aa) is organized into three main domains that include an N-terminal two-thirds hydrophilic domain (D1, approximately 120 aa), a C-terminal one-third hydrophobic domain (D2, approximately 50 aa), and approximately the last 20 aa that serves as a signal sequence for targeting E1 (D3)[25] (Figure 1). Kunkel et al[26] showed that the residues 76-113 (tryptophan-rich region) are largely solvent exposed, suggesting that it may interact with cellular proteins. It has been shown that core protein has multi-functional activity and can interact with cellular proto-oncogenes and change their expression patterns, thereby leading to hepatocarcinogenesis[27]. Several lines of investigation have demonstrated that core protein plays a pivotal role in the modulation of several key signaling pathways involved in HCC, such as transforming growth factor β (TGF-β), nuclear factor κB (NF-κB), tumor necrosis factor α (TNF-α), cyclooxygenase-2 (COX-2), Wnt/β-catenin (WNT), vascular endothelial growth factor (VEGF), and peroxisome proliferator-activated receptor α (PPARα)[22,28-32]. The mechanisms by which core protein modulates these signaling pathways are extremely complicated. To prevent HCV-related HCC, the molecular events underlying the interactions between HCV core protein and the signaling pathways need to be well understood. In this review, we investigate how the interaction of HCV core protein with several signaling pathways contributes to the development of HCC in HCV-infected patients.
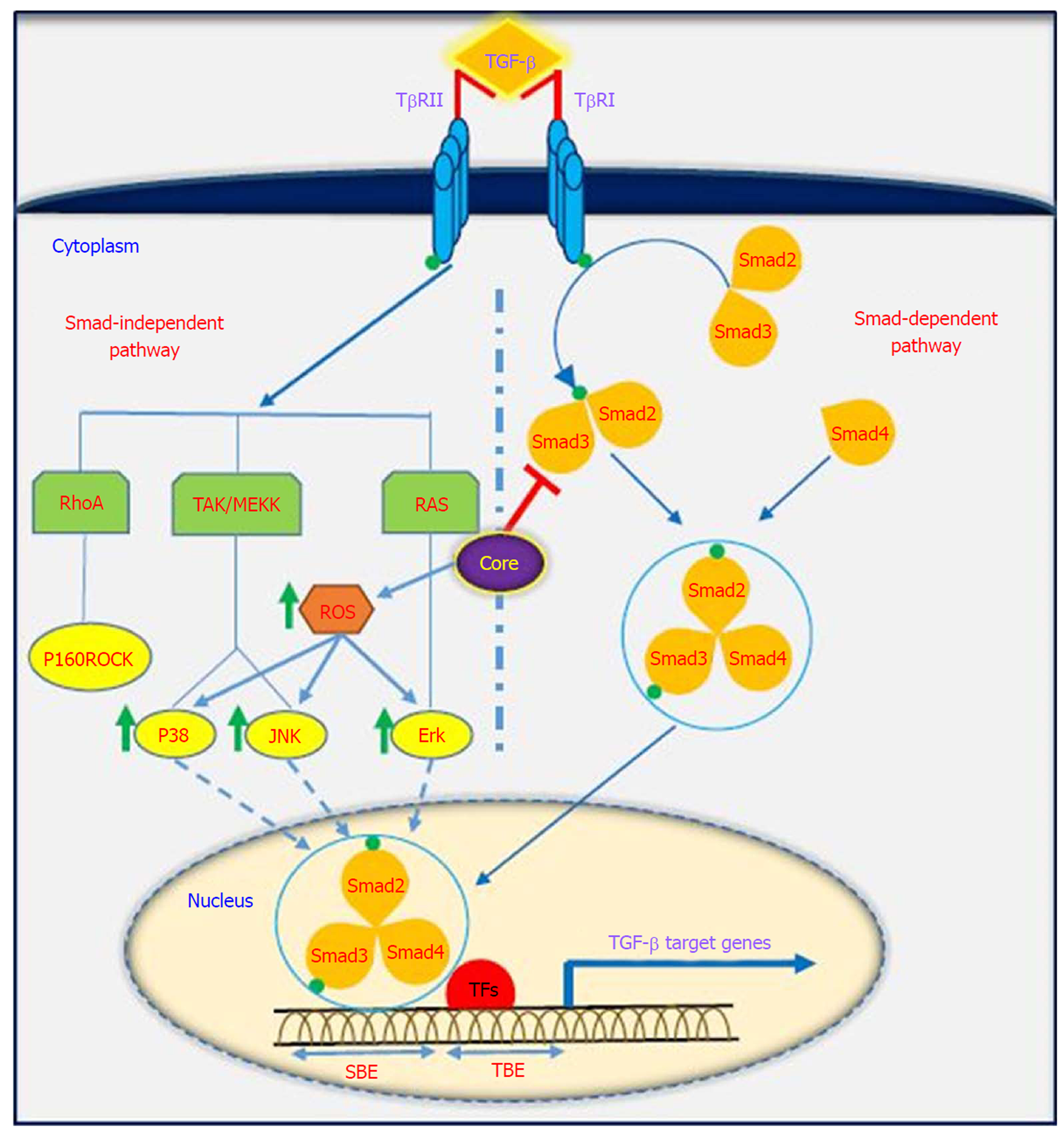
TGF-β is a multifunctional profibrotic cytokine that is found in three isoforms (TGF-β1-3). Of them, TGF-β1 plays a key role in the pathogenesis of liver inflammation, fibrosis, cirrhosis and HCC[33]. It is interesting to note that TGF-β is considered a central mediator of fibrogenesis and plays an important role in the regulation of tumorigenesis, as it controls numerous cellular functions, including apoptosis, differentiation, proliferation, extracellular matrix production, embryonic development, epithelial-mesenchymal transition (commonly known as EMT), and immune response[34,35]. Fibrosis is one of the most important consequences of TGF-β dysregulation, which is characterized by excessive accumulation of extracellular matrix (commonly known as ECM). TGF-β activity is mediated through activation and proliferation of hepatic stellate cells and connective tissue growth factor. Eventually, progressive fibrosis leads to the development of cirrhosis and HCC[36]. TGF-β acts as a double-edged sword depending on the cellular context; in the early stages of cancer development, it exhibits anti-tumor effects, while in the late stages, it has tumor-promoting activities[37]. TGF-β is implicated in several human diseases such as cardiovascular diseases, connective tissue diseases, skeletal and muscular disorders, reproductive disorders, autoimmune disorders, fibrotic disease, atherosclerosis and carcinogenesis[38,39]. Studies have shown that the downstream signaling pathways for TGF-β involve both canonical (Smad-dependent) and non-canonical (Smad-independent) pathways. TGF-β can induce fibrosis via activation of these two pathways, which results in activation of myofibroblasts and excessive production of ECM[40-43]. Smads mediate intracellular responses to TGF-β and have three classes, including receptor-regulated Smads (named R-Smads, including Smad1, 2, 3 ,5 and 8), comediator Smads (named Co-Smad, including Smad4) and inhibitory Smads (named I-Smads, including Smad6 and 7)[44]. Smads are regulated via direct phosphorylation by kinase activities of TGF-β receptors (TβRI and TβRII). The model of TGF-β-induced Smad activation is as follows: TGF-β binds to its receptor TβRII, which further interacts and activates TβRI. Activated TβRI then activates Smad2 and Smad3 via phosphorylation. Subsequently, the activated Smad2/3 forms a heterotrimer with Smad4 that translocates into the cell nucleus. Ultimately, the complex associates with other transcription factors and regulates the expression of target genes by binding to promoters containing the minimal Smad binding element[45,46]. Furthermore, TGF-β can activate non-canonical pathways such as JAK, Erk, JNK, p38 MAPK kinase, Ras and RhoA[40,47] (Figure 2). It has also been shown that HCV-induced transcription factors such as AP-1, Sp1, NF-κB, EGR-1, USF and STAT-3 can active the TGF-β1 promoter[48]. Taken together, it should be noted that the dysregulation of TGF-β signaling pathway could result in cancer development through either direct or indirect effects on other intracellular signaling pathways involved in carcinogenesis.
It has been indicated that oncogenic viruses such as HCV, HBV, HPV, EBV, KSHV and HTLV-1 can modulate TGF-β signaling pathway through various direct or indirect mechanisms, suggesting that this pathway is a desirable target for connecting viral proteins[37]. Several studies support the contention that HCV induces TGF-β1 secretion in HCV patients, as TGF-β1 levels are extremely high in these patients[49-51]. The results of the study performed by Jee et al[52] indicated that TGF-β1 protein in HCV-infected cells and in neighboring cells was more than 20-fold higher than in uninfected cells, and this increased production was observed 2 d after HCV infection. Several studies have demonstrated that HCV core protein can induce TGF-β1 promoter activity and directly and indirectly upregulate its gene expression. These findings suggest that HCV infection is one mechanism by which liver fibrosis can be exacerbated[28,53-56]. Taniguchi et al[28] showed that TGF-β1 mRNA expression is increased by HCV core protein, whereas TGF-β2 and TGF-β3 mRNA levels do not change upon core protein expression. The results of their study suggested that bases 331 to 376 in the TGF-β1 promoter are upregulated by HCV core protein[28]. In studies conducted by Battaglia et al[57] and Pavio et al[58], it was revealed that core protein is able to switch TGF-β from a tumor suppressor to tumor promoter by decreasing hepatocyte apoptosis and increasing EMT by decreasing Smad3 activation. The Smad proteins consist of two principal domains, DNA-binding domain (N-terminal Mad homology 1, MH1 domain) and protein-protein interacting module (C-terminal Mad homology 2, MH2 domain). Pavio et al[58,59] indicated that the central domain (59-126 aa) of core protein binds to the MH1 domain of Smad3, leading to TGF-β inhibition. Cheng et al[60] explained that HCV core protein utilizes various mechanisms to regulate TGF-β, including the following: (1) suppression of TGF-β/Smad3-mediated transcriptional activation through interference with the DNA-binding ability of Smad3; (2) block of TGF-β-induced G1 phase arrest via downregulation of TGF-β-induced p21 promoter activation; and (3) resistance to TGF-β/Smad3-mediated apoptosis. Notably, over-expression of HCV core protein indirectly induces TGF-β production by increased reactive oxygen species production, which in turn increases JNK, Erk and p38 MAP kinase activity in a NF-κB-dependent manner[56,61,62] (Figure 2). In addition, Shin et al[54] revealed that HCV core protein can regulate other factors associated with fibrosis, such as connective tissue growth factor, TβRII and TGF-β1. Nevertheless, considering the pivotal role of TGF-β in the development of fibrosis and tumor progression, this pathway is an important pharmaceutical target to prevent cancer progression. Furthermore, treatment with antiviral drugs such as DAAs in combination with Peg-IFNα and ribavirin increases the chances of achieving SVR. On the other hand, there are some but not conclusive data that TGF-β1 serum levels in chronic hepatitis C patients under antiviral therapy significantly decreases, especially in patients achieving SVR[63-66]. In this condition, HCV proteins are unable to upregulate TGF-β1 expression, which in turn leads to reduced fibrogenesis and prevention of cancer progression.
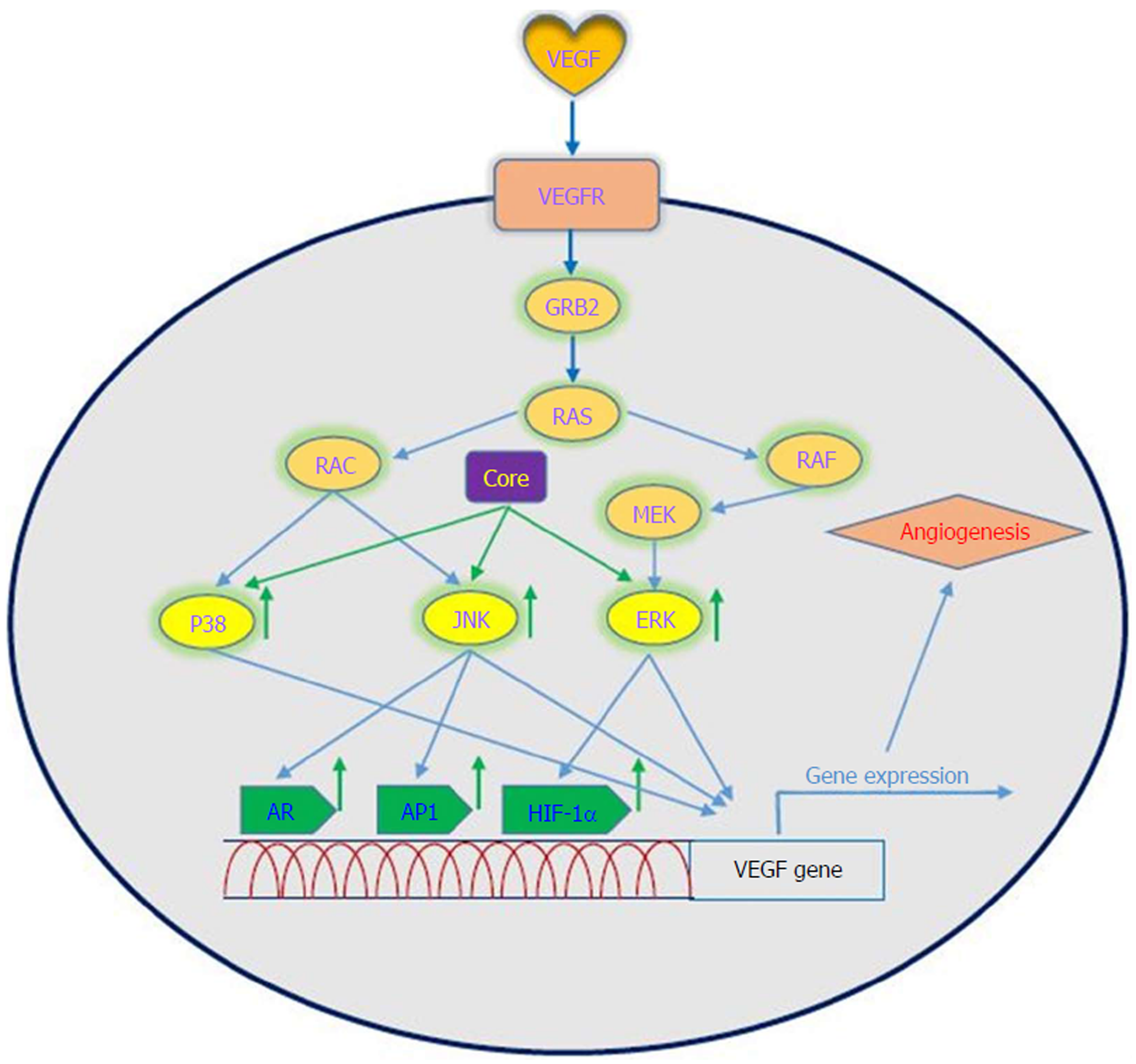
VEGF is a signal protein that is produced by most parenchymal cells and stimulates angiogenesis. In cancer progression, VEGF is the key mediator of angiogenesis and vasculogenesis, which leads to the formation of new blood vessels from pre-existing vessels. In turn, this allows tumors to access oxygen and nutrients[67]. VEGFA, also known as VEGF, is a protein with vascular permeability activity that is a member of a family of growth factors. In addition, VEGFB, VEGFC, VEGFD and placental growth factor are also in this family. These growth factors play an important role in angiogenesis and differ in their biological functions and expression patterns[68]. Several factors play roles in the upregulation of VEGF. It has been demonstrated that a number of growth factors such as PDGF, epidermal growth factor, fibroblast growth factor, TNF, TGF-β and interleukin-1 can induce VEGF gene expression[69]. VEGF applies its effects by binding to VEGF receptors, which are expressed on vascular endothelial cells. The VEGF receptors include VEGF receptor-1 (Flt-1), VEGF receptor-2 (KDR) and VEGF receptor-3 (Flt-4)[70]. VEGF has several roles, including the following: (1) inducing angiogenesis through a direct impact on endothelial cells; (2) inducing cells to invade the underlying matrix and to form capillary-like tubules; (3) elicitation of non-mitogenic responses by vascular endothelial cells; (4) instrumental in maintaining the viability of immature vasculature and inducing hemotaxis; and (5) the expression of plasminogen activators and collagenases in endothelial cells[71]. It should be noted that the induction and activation of hypoxia-inducible factor-1 alpha (HIF-1α) is a major inducer of VEGF expression in tumors. It is one of the first transcription factors in response to hypoxia and is a closely related angiogenesis factor. HIF-1α responds to hypoxia through binding to hypoxic response element, which in turn leads to an increase in VEGF protein expression because it plays a regulatory role in VEGF expression[72,73]. Given the importance of the angiogenic effects of VEGF, dysregulation of its expression is important in disease processes and progression toward cancer. A large body of evidence exists that suggests a role for VEGF in tumorigenesis in human cancers[74].
Oncogenic viruses such as HCV, EBV, HPV, KSHV and HBV can upregulate VEGF with the use of cellular signaling machinery, which leads to angiogenesis[75]. Several reports have shown upregulation of VEGF in patients with HCV-related HCC[76-80]. Mukozu et al[77] demonstrated that VEGF serum levels of patients with HCV-related HCC are significantly higher than those in the control group. HCV core protein can upregulate cellular VEGF expression through HIF-1α transcription factors and activator protein 1 (AP-1). Core protein induces over-expression and stabilization of HIF-1α, which in turn induces VEGF expression[81-84]. Abe et al[82] showed that core protein increases HIF-1α expression level by activating the NF-κB signaling pathway, which leads to an increase in VEGF expression under hypoxia followed by HIF-1α upregulation. They also observed that when cells were incubated with HIF-1α inhibitor, VEGF expression clearly decreased[82]. In a study performed by Zhu et al[83] in Huh7.5.1 cells, it was demonstrated that core protein contributes to VEGF biosynthesis by inducing VEGF expression and secretion. They indicated that using HIF-1α siRNA in Huh7.5.1 cells, which results in reducing expression of HIF-1α, significantly reduces VEGF expression. Shao et al[85] indicated that VEGF expression increased due to activation of AP-1 transcription factor, because the promoter region of VEGF contains binding sites for AP-1 transcription factors, which lead to enhanced VEGF expression via promoter binding. This observation is in line with previous studies of AP-1 activity associated with VEGF expression and HCV core protein-induced AP-1 activity[86-89]. HCV core protein affects androgen receptor (AR) transcriptional activity by activating several signaling pathways such as phosphatidylinositol 3-kinase (PI3K)/AKT and JAK/STAT3. Since VEGF is a target gene for AR in the liver, HCV core protein increases VEGF expression through increased activity of the AR signaling pathway[90]. Hassan et al[31] recently reported that HCV core protein induces VEGF expression mediated by JNK, p38 and ERK signaling pathways (Figure 3). Given the results of various studies, HCV core protein can upregulate the VEGF signaling pathway. These data increase our insights into the molecular mechanisms by which HCV core protein mediates angiogenesis in HCV-infected patients.
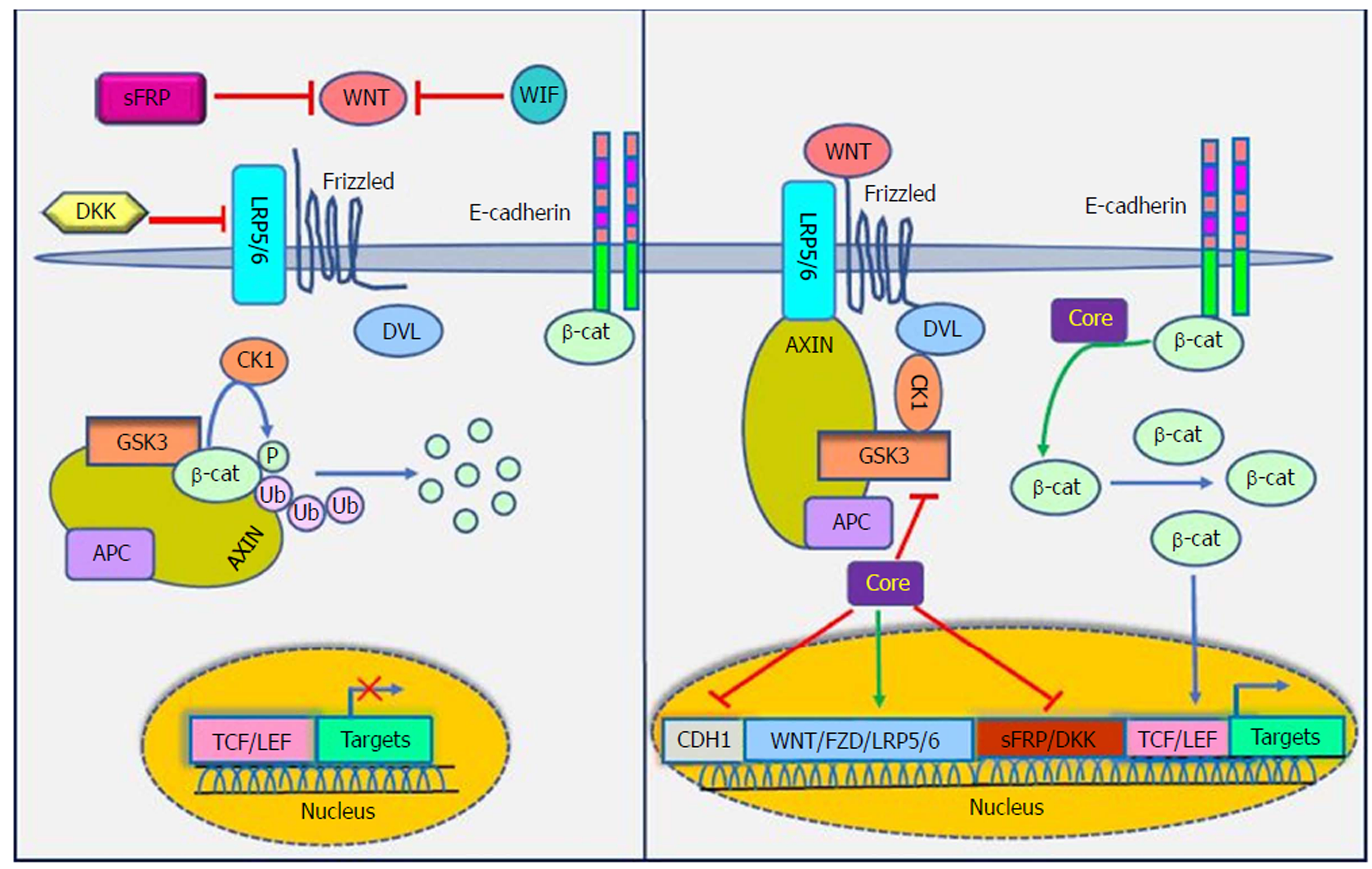
The WNT signaling pathways are a group of signal transduction pathways that regulate different cellular processes such as cell polarity, organogenesis, cell migration and neural patterning during embryonic development[91]. Alteration of WNT activity has been linked to the development of HCC and other liver diseases[92]. β-catenin has a crucial role in Wnt signaling and also tightly binds to the cytoplasmic domain of type I cadherins and is implicated in the structural organization and function of cadherins[93]. Wnts are secreted cysteine-rich lipid-modified glycoproteins[94,95] that bind to the N-terminal extra-cellular cysteine-rich domain of the Frizzled (Fz or Fzd) receptor family and low-density-lipoprotein-related protein 5/6 (LRP5/6) as co-receptors[96,97]. When WNT signaling is inactive, cytoplasmic β-catenin interacts with a multiprotein degradation complex comprised of casein kinase I (CK1), adenomatous polyposis coli gene product (APC), glycogen synthase kinase 3β (GSK3β) and Axin[98]. Axin binds to newly synthesized β-catenin, which is subsequently phosphorylated by CKI and GSK3 on conserved Ser and Thr residues in the amino terminus[99]. Following phosphorylation, β-catenin is targeted for proteasome-dependent degradation, including an interaction with β-transducin repeat-containing protein (β-TrCP), a part of the E3 ubiquitin ligase complex, leading to β-catenin ubiquitination and degradation[100,101]. WNT signaling is regulated by secreted proteins, including secreted Frizzled-related proteins (sFRPs) and Wnt inhibitory protein (WIF) that can bind to Wnts and prevent interactions between Wnt and Wnt receptors. Other Wnt inhibitors include Dickkopf (DKK) proteins, which inhibit Wnt signaling by binding to LRP5/6[95]. If the concentration of Wnts increases, Wnts interact with receptors to activate Dishevelled (Dsh or Dvl) protein. Dvl, a modular phosphoprotein, is phosphorylated by several kinases such as CK1. Activated Dvl recruits the destruction complex to the plasma membrane and binds to Axin and Fz[102,103]. Intriguingly, Axin binds to the cytoplasmic domain of LRP5/6. When Dvl is activated, it leads to the inhibition of GSK3 activity, which activates a complex series of events that decreases β-catenin phosphorylation and degradation[104]. Therefore, β-catenin accumulates in the nucleus and activates the transcription of target genes through interaction with DNA-bound T-cell factor (TCF) and lymphoid enhancer-binding factor 1 (LEF) family members[105]. WNT signaling has been implicated in the modulation of innate immunity by stimulating invariant natural killer T cell (iNKT) responses and production of chemokine-like chemotactic factor leukocyte cell-derived chemotaxin 2 (LECT2) or by decreasing the release of tumor necrosis factor[106-108]. LECT2 is involved in inflammation, chemotaxis, immuno-modulation, cell proliferation and carcinogenesis. In addition, LECT2 signaling induces inflammatory responses by activating the proinflammatory NF-κB pathway[109]. Abnormal Wnt and Fz expression in hepatocytes, stellate and Kupffer cells might play important roles in liver pathobiology[110,111].
Khanizadeh et al[112] indicated that oncogenic viruses can interact with and modulate the Wnt signaling pathway. Because the WNT signaling pathway is a key pathway in HCV-positive HCC, it is important to understand how HCV core protein induces liver carcinogenesis through Wnt signaling[113] (Figure 4). DNA methylation also contributes to the development of HCC. Quan et al[114] showed that HCV core protein can increase Wnt signaling via down-regulation of the sFRP1 promoter in HCC cells through methylation and histone deacetylation. In another study, Umer et al[115] demonstrated that sFRP2 and DKK1 promoters of Wnt inhibitor genes increase DNA methylation in HCC patients compared to normal controls. In addition, HCV core protein is involved in hypermethylation of the CDH1 (E-cadherin) gene promoter. Decreased production of E-cadherin induces Wnt signaling activation[116]. HCV core protein enhances β-catenin expression and nuclear stabilization by inactivating GSK-3β. Nuclear accumulation of β-catenin forms a transcriptional complex with TCF and activates downstream target genes, such as c-Myc, Cyclin D1 and WNT1 inducible signaling pathway (WISP-2), which regulate cell growth and cell cycle progression[22,113]. Additionally, HCV core protein elevates the expression of LRP5/6 co-receptors and FZD receptors and releases β-catenin from the β-catenin-E-cadherin complexes[108]. Taken together, the direct involvement of HCV core protein in the Wnt pathway is an attractive candidate to mediate liver pathogenesis.
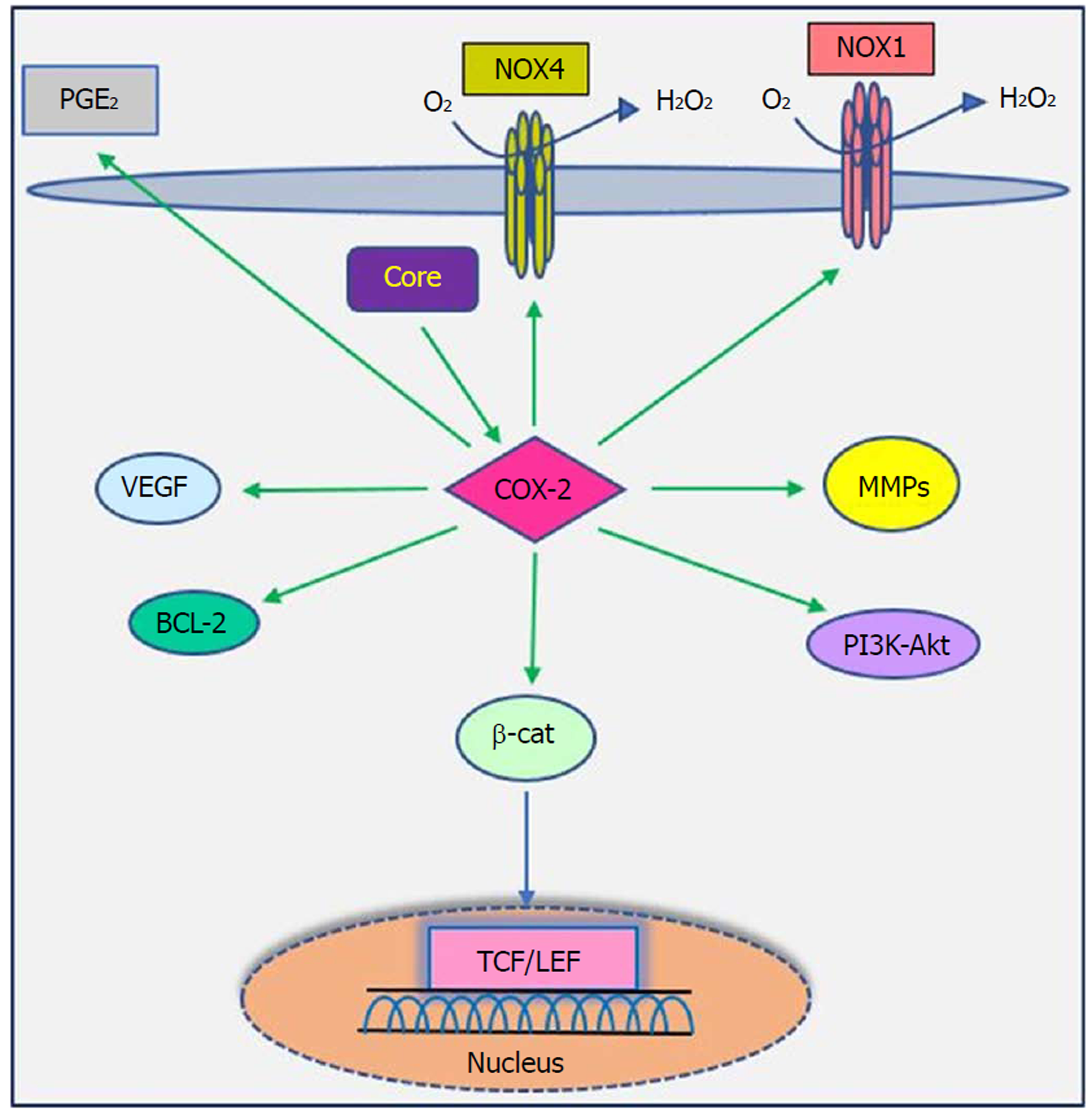
The cyclooxygenase (COX) family consists of constitutive COX-1, inducible COX-2 and COX-3, which are involved in prostaglandin synthesis by conversion of arachidonic acid to prostanoids, including thromboxanes and prostaglandins[117,118]. COX-2 can be induced by tumor promoters, cytokines and growth factors via the cis-acting elements within the 5’ UTR of the COX-2 gene and is known as a pathogenic factor involved in cellular proliferation, anti-apoptosis activity, inflammation, fibrogenesis and tumorigenesis. Moreover, increased levels of prostaglandin E2 and COX-2 contribute to various biological processes, including oxidative stress, liver damage, bacterial and viral infection, acute and chronic inflammation and cancer[117,119,120].
Studies have shown that there is a close relationship between oncogenic viruses such as HCV, HBV, EBV and HPV and COX-2[121-124]. Previous reports demonstrated that increased production of COX-2 is observed in response to HCV infection[125-127]. Overexpression of COX-2 supports HCV replication and has a potential role in hepatocarcinogenesis in HCC and human hepatoma cell lines[121,128,129]. Several studies have previously documented that HCV stimulates COX-2 expression via oxidative stress[130,131]. Oxidative stress is a key contributor in liver fibrosis and carcinogenesis related to HCV infection[132]. HCV core protein upregulates COX-2 levels in hepatocytes and has carcinogenic effects that lead to HCC[62,121,133]. Jahan et al[30] demonstrated that core protein of HCV genotype 3a induces COX-2 expression in Huh-7 cells compared to the core protein of HCV genotype 1a. Conversely, several studies have shown that HCV core protein downregulates COX-2 expression. HCV might avoid the inflammatory responses of host by downregulating this signaling pathway[29,117,134]. COX-2 plays a crucial role in the production of matrix metalloproteinases (MMPs), particularly MMP-2 and MMP-9, by liver cells, which are associated with cell migration via degradation of cellular and extracellular components during organ development, morphogenesis, tissue damage and cancer invasion[135,136]. COX-2 is involved in the regulation of superoxide anion expression, namely NADPH oxidase 1 (NOX1) and NADPH oxidase 4 (NOX4), in hepatocytes[137]. Lan et al[138] demonstrated that NOX1 and NOX4 are increased in cirrhotic patients and play an important role in liver fibrosis via regulating proliferation, inflammation and fibrogenesis in hepatic stellate cells. COX-2 stimulates β-catenin activation, which leads to WNT pathway activation and subsequent cell growth and proliferation through its interaction with TCF in the nucleus[139]. COX-2 induces the production of VEGF and the anti-apoptotic proteins of the Bcl-2 family, which are associated with an increase in angiogenesis, invasiveness, and resistance to apoptosis[140]. Leng et al[141] showed a positive correlation between COX-2 and phosphatidylinositol 3-kinase-Protein Kinase B (PI3K-Akt/PKB) pathway activation in human HCC tissue. PI3K-Akt/PKB plays a major role in cancer development and progression by inhibiting apoptosis and stimulating cell proliferation. A recent study showed that long non-coding RNA (lncRNA) COX-2 inhibits immune evasion, migration and invasion of HCC cells and may provide a novel theoretical basis for HCC treatment[142]. In view of these facts, there is a direct relationship between HCV core protein and COX-2 overexpression, which in turn leads to alterations in cell signaling pathways (Figure 5). Therefore, the COX-2 signaling pathway should be considered a target to prevent HCV replication and HCV-associated diseases.
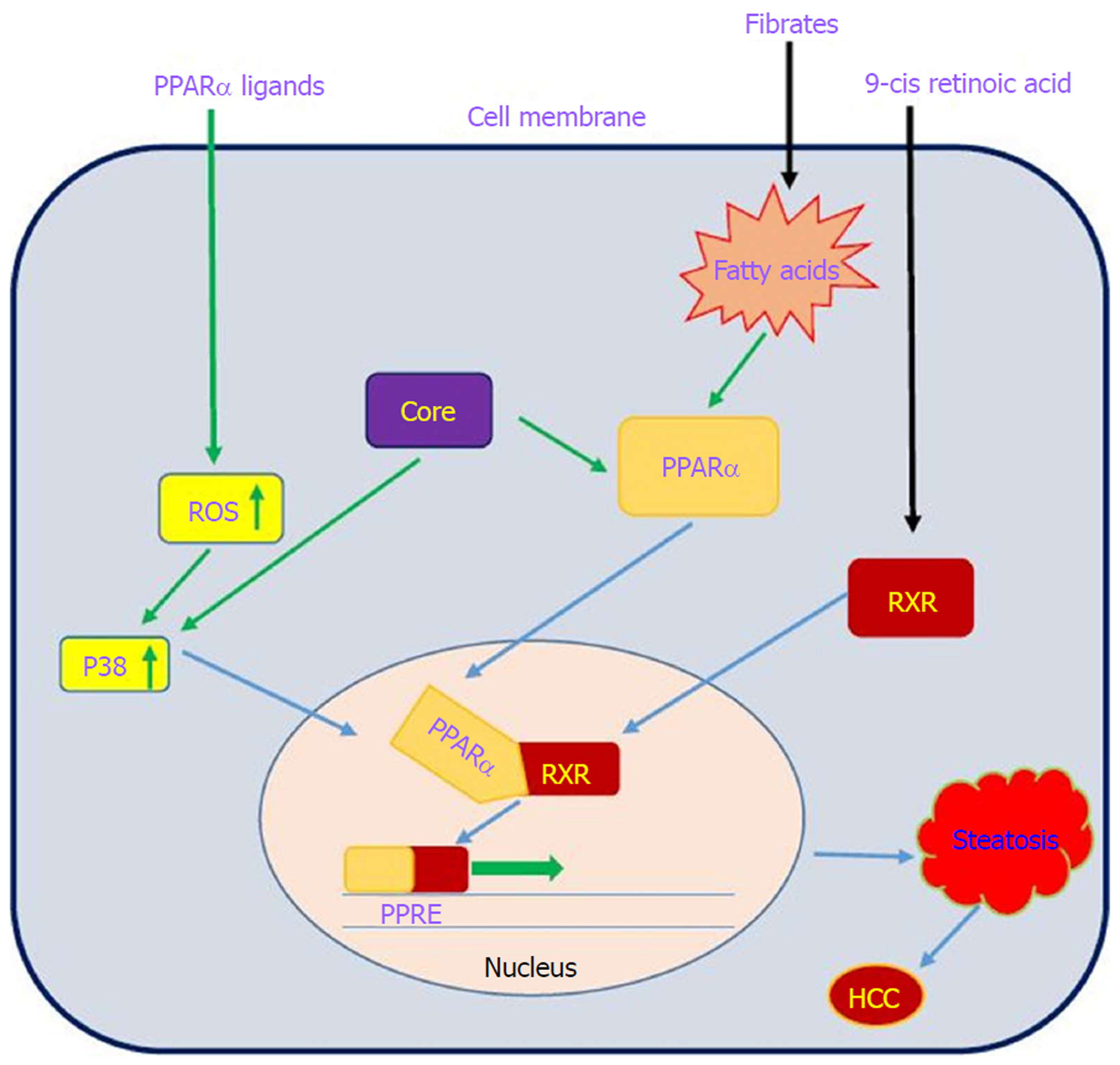
PPARs are a family of nuclear receptor proteins that belong to the steroid/thyroid hormone receptor superfamily and function as transcription factors. This family has three members, including PPARα, PPARγ and PPARβ/δ[143], which play regulatory roles in cellular processes such as differentiation, proliferation, inflammation, oxidative stress, tumorigenesis and metabolism (carbohydrate, lipid, protein)[144]. In order to function, all PPARs initially heterodimerize with the retinoid X receptor (RXR) and then bind to peroxisome proliferator hormone-response elements (PPREs), which are located in the promoter region of PPAR target genes[145]. PPARα is one of the most abundant nuclear receptors expressed in hepatocytes, and it acts as an important and vital regulator in lipid and lipoprotein metabolism. It becomes activated by several molecules such as long-chain unsaturated fatty acids, and once activated, it promotes lipid clearance through β-oxidation upregulation[146]. The relationship between PPARα and cancer development is very complicated. Epidemiological studies have shown that PPARα has carcinogenic consequences in the liver of humans and rodents[147-150]. It is also noted that long-term administration of PPARα ligands can lead to increased ROS generation, accelerated hepatocyte proliferation and development of HCC[151]. In other words, persistent PPARα activation induces hepatic steatosis through increased liver triglyceride synthesis. On the other hand, hepatic steatosis promotes HCC development and acts as an important accelerating factor of HCC in HCV-infected patients[152-154]. Therefore, the hepatic steatosis induced by alteration of fatty acid metabolism in hepatocytes may act as a mediator in causing HCC in HCV-infected patients. Thus, it is important to understand the interaction between HCV infection and PPARα signaling pathway.
On the basis of several lines of evidence, PPARα activity is impaired in patients with chronic hepatitis C infection, which may contribute to hepatocarcinogenesis[32,155]. Dharancy et al[32] showed that PPARα expression is significantly decreased in HCV-infected patients compared to the control group. In this study, HCV core protein expression in HepG2 cells led to disruption of PPARα transcriptional activity, demonstrated by decreased expression of the PPARα target gene CPT1A. Shen et al[156] indicated that PPARα has an inhibitory effect on NF-κB. On the other hand, HCV core protein can disrupt PPARα activity. From this point of view, HCV core protein negatively regulates the repressive effect of PPARα on nuclear NF-κB, which in turn leads to NF-κB activation and HCC development. Studies have shown that HCV core protein can indirectly activate PPARα through activation and phosphorylation of ERK1/2 and P38 MAPK[155,157] (Figure 6). HCV core protein also binds to RXR and enhances RXR transcriptional activity, leading to the upregulation of some lipid metabolism enzymes. This result suggests that the dysregulation of RXR by HCV core protein may contribute to HCC development[158]. These findings demonstrate that HCV core protein disrupts PPARα activity, leading to HCC in HCV-infected patients. Thus, PPARα can be considered a new therapeutic target for preventing HCC.
HCV can cause liver diseases, especially HCC, through the modulation of various signaling pathways. TGF-β, VEGF, WNT, COX-2 and PPARα signaling pathways play important roles in the regulation of fibrogenesis, angiogenesis and tumorigenesis by controlling cell proliferation, apoptosis, transcriptional regulation and cell growth promotion. Therefore, their dysregulation is associated with HCC. As previously mentioned, HCV core protein uses various mechanisms to dysregulate these pathways. Hence, it seems necessary to undertake major research efforts for a better understanding of how HCV core protein leads to the dysregulation of these signaling pathways, which in turn helps in designing effective therapeutic methods. Furthermore, these signaling pathways should be considered therapeutic targets for cancer therapy, and prevention programs should be implemented to prevent their overexpression. Given the commercially available inhibitors against these pathways, HCV-related HCC development can be prevented in the near future. Moreover, the use of antiviral drugs such as DAAs in combination with Peg-IFNα plus ribavirin leads to SVR in chronic hepatitis C patients, which in turn can help reduce some of the factors involved in cancer development, such as TGF-β1. However, these findings require more research.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Iran
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Kai K, Zhao HT S- Editor: Ma RY L- Editor: Filipodia E- Editor: Huang Y
| 1. | Fiorina L, Ricotti M, Vanoli A, Luinetti O, Dallera E, Riboni R, Paolucci S, Brugnatelli S, Paulli M, Pedrazzoli P, Baldanti F, Perfetti V. Systematic analysis of human oncogenic viruses in colon cancer revealed EBV latency in lymphoid infiltrates. Infect Agent Cancer. 2014;9:18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 2. | White MK, Pagano JS, Khalili K. Viruses and human cancers: a long road of discovery of molecular paradigms. Clin Microbiol Rev. 2014;27:463-481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 145] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 3. | Dubuisson J, Cosset FL. Virology and cell biology of the hepatitis C virus life cycle: an update. J Hepatol. 2014;61:S3-S13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 141] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 4. | Taherkhani R, Farshadpour F. Global elimination of hepatitis C virus infection: Progresses and the remaining challenges. World J Hepatol. 2017;9:1239-1252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 50] [Article Influence: 6.3] [Reference Citation Analysis (1)] |
| 5. | Taherkhani R, Farshadpour F. Lurking epidemic of hepatitis C virus infection in Iran: A call to action. World J Hepatol. 2017;9:1040-1042. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 6. | Maheshwari A, Ray S, Thuluvath PJ. Acute hepatitis C. Lancet. 2008;372:321-332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 179] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 7. | Taherkhani R, Farshadpour F. Epidemiology of hepatitis C virus in Iran. World J Gastroenterol. 2015;21:10790-10810. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 65] [Cited by in RCA: 70] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 8. | GLOBOCAN 2012 v1. 0. Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11. Accessed July 25 2016; Available from: http://www.globocan.iarc.fr/. |
| 9. | Webster DP, Klenerman P, Dusheiko GM. Hepatitis C. Lancet. 2015;385:1124-1135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 383] [Cited by in RCA: 361] [Article Influence: 36.1] [Reference Citation Analysis (0)] |
| 10. | Smith-Palmer J, Cerri K, Valentine W. Achieving sustained virologic response in hepatitis C: a systematic review of the clinical, economic and quality of life benefits. BMC Infect Dis. 2015;15:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 142] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 11. | Guarino M, Sessa A, Cossiga V, Morando F, Caporaso N, Morisco F; Special Interest Group on “Hepatocellular carcinoma and new anti-HCV therapies” of the Italian Association for the Study of the Liver. Direct-acting antivirals and hepatocellular carcinoma in chronic hepatitis C: A few lights and many shadows. World J Gastroenterol. 2018;24:2582-2595. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 50] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 12. | Aleman S, Rahbin N, Weiland O, Davidsdottir L, Hedenstierna M, Rose N, Verbaan H, Stål P, Carlsson T, Norrgren H, Ekbom A, Granath F, Hultcrantz R. A risk for hepatocellular carcinoma persists long-term after sustained virologic response in patients with hepatitis C-associated liver cirrhosis. Clin Infect Dis. 2013;57:230-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 195] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 13. | El-Serag HB, Kanwal F, Richardson P, Kramer J. Risk of hepatocellular carcinoma after sustained virological response in Veterans with hepatitis C virus infection. Hepatology. 2016;64:130-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 313] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 14. | van der Meer AJ, Feld JJ, Hofer H, Almasio PL, Calvaruso V, Fernández-Rodríguez CM, Aleman S, Ganne-Carrié N, D’Ambrosio R, Pol S, Trapero-Marugan M, Maan R, Moreno-Otero R, Mallet V, Hultcrantz R, Weiland O, Rutter K, Di Marco V, Alonso S, Bruno S, Colombo M, de Knegt RJ, Veldt BJ, Hansen BE, Janssen HLA. Risk of cirrhosis-related complications in patients with advanced fibrosis following hepatitis C virus eradication. J Hepatol. 2017;66:485-493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 216] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 15. | WHO. Hepatitis C, WHO fact sheet. Available from: http://www.who.int/en/news-room/fact-sheets/detail/hepatitis-c. |
| 16. | Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology. 2014;59:318-327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 965] [Cited by in RCA: 982] [Article Influence: 89.3] [Reference Citation Analysis (1)] |
| 17. | Pirakitikulr N, Kohlway A, Lindenbach BD, Pyle AM. The Coding Region of the HCV Genome Contains a Network of Regulatory RNA Structures. Mol Cell. 2016;62:111-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 90] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 18. | Tellinghuisen TL, Rice CM. Interaction between hepatitis C virus proteins and host cell factors. Curr Opin Microbiol. 2002;5:419-427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 136] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 19. | Banerjee A, Ray RB, Ray R. Oncogenic potential of hepatitis C virus proteins. Viruses. 2010;2:2108-2133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 97] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 20. | Abdallah C, Lejamtel C, Benzoubir N, Battaglia S, Sidahmed-Adrar N, Desterke C, Lemasson M, Rosenberg AR, Samuel D, Bréchot C, Pflieger D, Le Naour F, Bourgeade MF. Hepatitis C virus core protein targets 4E-BP1 expression and phosphorylation and potentiates Myc-induced liver carcinogenesis in transgenic mice. Oncotarget. 2017;8:56228-56242. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 21. | Kasprzak A, Adamek A. Role of hepatitis C virus proteins (C, NS3, NS5A) in hepatic oncogenesis. Hepatol Res. 2008;38:1-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 51] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 22. | Liu J, Ding X, Tang J, Cao Y, Hu P, Zhou F, Shan X, Cai X, Chen Q, Ling N, Zhang B, Bi Y, Chen K, Ren H, Huang A, He TC, Tang N. Enhancement of canonical Wnt/β-catenin signaling activity by HCV core protein promotes cell growth of hepatocellular carcinoma cells. PLoS One. 2011;6:e27496. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 100] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 23. | Jiang YF, He B, Li NP, Ma J, Gong GZ, Zhang M. The oncogenic role of NS5A of hepatitis C virus is mediated by up-regulation of survivin gene expression in the hepatocellular cell through p53 and NF-κB pathways. Cell Biol Int. 2011;35:1225-1232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 21] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 24. | Qadri I, Iwahashi M, Capasso JM, Hopken MW, Flores S, Schaack J, Simon FR. Induced oxidative stress and activated expression of manganese superoxide dismutase during hepatitis C virus replication: role of JNK, p38 MAPK and AP-1. Biochem J. 2004;378:919-928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 119] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 25. | Boulant S, Vanbelle C, Ebel C, Penin F, Lavergne JP. Hepatitis C virus core protein is a dimeric alpha-helical protein exhibiting membrane protein features. J Virol. 2005;79:11353-11365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 119] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 26. | Kunkel M, Watowich SJ. Biophysical characterization of hepatitis C virus core protein: implications for interactions within the virus and host. FEBS Lett. 2004;557:174-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 27. | Bartosch B, Thimme R, Blum HE, Zoulim F. Hepatitis C virus-induced hepatocarcinogenesis. J Hepatol. 2009;51:810-820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 115] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 28. | Taniguchi H, Kato N, Otsuka M, Goto T, Yoshida H, Shiratori Y, Omata M. Hepatitis C virus core protein upregulates transforming growth factor-beta 1 transcription. J Med Virol. 2004;72:52-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 105] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 29. | Joo M, Hahn YS, Kwon M, Sadikot RT, Blackwell TS, Christman JW. Hepatitis C virus core protein suppresses NF-kappaB activation and cyclooxygenase-2 expression by direct interaction with IkappaB kinase beta. J Virol. 2005;79:7648-7657. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 77] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 30. | Jahan S, Khaliq S, Ijaz B, Ahmad W, Hassan S. Role of HCV Core gene of genotype 1a and 3a and host gene Cox-2 in HCV-induced pathogenesis. Virol J. 2011;8:155. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 31. | Hassan M, Selimovic D, Ghozlan H, Abdel-kader O. Hepatitis C virus core protein triggers hepatic angiogenesis by a mechanism including multiple pathways. Hepatology. 2009;49:1469-1482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 85] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 32. | Dharancy S, Malapel M, Perlemuter G, Roskams T, Cheng Y, Dubuquoy L, Podevin P, Conti F, Canva V, Philippe D, Gambiez L, Mathurin P, Paris JC, Schoonjans K, Calmus Y, Pol S, Auwerx J, Desreumaux P. Impaired expression of the peroxisome proliferator-activated receptor alpha during hepatitis C virus infection. Gastroenterology. 2005;128:334-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 157] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 33. | Massagué J. TGFbeta in Cancer. Cell. 2008;134:215-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2725] [Cited by in RCA: 3135] [Article Influence: 184.4] [Reference Citation Analysis (0)] |
| 34. | Syed V. TGF-β Signaling in Cancer. J Cell Biochem. 2016;117:1279-1287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 355] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 35. | Xu F, Liu C, Zhou D, Zhang L. TGF-β/SMAD Pathway and Its Regulation in Hepatic Fibrosis. J Histochem Cytochem. 2016;64:157-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 568] [Article Influence: 63.1] [Reference Citation Analysis (0)] |
| 36. | Biernacka A, Dobaczewski M, Frangogiannis NG. TGF-β signaling in fibrosis. Growth Factors. 2011;29:196-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 746] [Cited by in RCA: 915] [Article Influence: 65.4] [Reference Citation Analysis (0)] |
| 37. | Mirzaei H, Faghihloo E. Viruses as key modulators of the TGF-β pathway; a double-edged sword involved in cancer. Rev Med Virol. 2018;28. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 38. | Gordon KJ, Blobe GC. Role of transforming growth factor-beta superfamily signaling pathways in human disease. Biochim Biophys Acta. 2008;1782:197-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 458] [Cited by in RCA: 510] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 39. | Blobe GC, Schiemann WP, Lodish HF. Role of transforming growth factor beta in human disease. N Engl J Med. 2000;342:1350-1358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1834] [Cited by in RCA: 1884] [Article Influence: 75.4] [Reference Citation Analysis (0)] |
| 40. | Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3797] [Cited by in RCA: 4307] [Article Influence: 195.8] [Reference Citation Analysis (0)] |
| 41. | Moustakas A, Heldin CH. The regulation of TGFbeta signal transduction. Development. 2009;136:3699-3714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 626] [Cited by in RCA: 654] [Article Influence: 40.9] [Reference Citation Analysis (0)] |
| 42. | Ikushima H, Miyazono K. TGFbeta signalling: a complex web in cancer progression. Nat Rev Cancer. 2010;10:415-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 843] [Cited by in RCA: 898] [Article Influence: 59.9] [Reference Citation Analysis (0)] |
| 43. | Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128-139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1451] [Cited by in RCA: 1431] [Article Influence: 89.4] [Reference Citation Analysis (0)] |
| 44. | Wu JW, Hu M, Chai J, Seoane J, Huse M, Li C, Rigotti DJ, Kyin S, Muir TW, Fairman R, Massagué J, Shi Y. Crystal structure of a phosphorylated Smad2. Recognition of phosphoserine by the MH2 domain and insights on Smad function in TGF-beta signaling. Mol Cell. 2001;8:1277-1289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 227] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 45. | Budi EH, Duan D, Derynck R. Transforming Growth Factor-β Receptors and Smads: Regulatory Complexity and Functional Versatility. Trends Cell Biol. 2017;27:658-672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 224] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 46. | Imamura T, Oshima Y, Hikita A. Regulation of TGF-β family signalling by ubiquitination and deubiquitination. J Biochem. 2013;154:481-489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 81] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 47. | Yu L, Hébert MC, Zhang YE. TGF-beta receptor-activated p38 MAP kinase mediates Smad-independent TGF-beta responses. EMBO J. 2002;21:3749-3759. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 546] [Cited by in RCA: 571] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 48. | Presser LD, McRae S, Waris G. Activation of TGF-β1 promoter by hepatitis C virus-induced AP-1 and Sp1: role of TGF-β1 in hepatic stellate cell activation and invasion. PLoS One. 2013;8:e56367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 63] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 49. | Bedossa P, Poynard T, Mathurin P, Lemaigre G, Chaput JC. Transforming growth factor beta 1: in situ expression in the liver of patients with chronic hepatitis C treated with alpha interferon. Gut. 1993;34:S146-S147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 50. | Paradis V, Mathurin P, Laurent A, Charlotte F, Vidaud M, Poynard T, Hoang C, Opolon P, Bedossa P. Histological features predictive of liver fibrosis in chronic hepatitis C infection. J Clin Pathol. 1996;49:998-1004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 28] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 51. | Ray S, Broor SL, Vaishnav Y, Sarkar C, Girish R, Dar L, Seth P, Broor S. Transforming growth factor beta in hepatitis C virus infection: in vivo and in vitro findings. J Gastroenterol Hepatol. 2003;18:393-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 52. | Jee MH, Hong KY, Park JH, Lee JS, Kim HS, Lee SH, Jang SK. New Mechanism of Hepatic Fibrogenesis: Hepatitis C Virus Infection Induces Transforming Growth Factor β1 Production through Glucose-Regulated Protein 94. J Virol. 2015;90:3044-3055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 53. | Benzoubir N, Lejamtel C, Battaglia S, Testoni B, Benassi B, Gondeau C, Perrin-Cocon L, Desterke C, Thiers V, Samuel D, Levrero M, Bréchot C, Bourgeade MF. HCV core-mediated activation of latent TGF-β via thrombospondin drives the crosstalk between hepatocytes and stromal environment. J Hepatol. 2013;59:1160-1168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 48] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 54. | Shin JY, Hur W, Wang JS, Jang JW, Kim CW, Bae SH, Jang SK, Yang SH, Sung YC, Kwon OJ, Yoon SK. HCV core protein promotes liver fibrogenesis via up-regulation of CTGF with TGF-beta1. Exp Mol Med. 2005;37:138-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 74] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 55. | Chusri P, Kumthip K, Hong J, Zhu C, Duan X, Jilg N, Fusco DN, Brisac C, Schaefer EA, Cai D, Peng LF, Maneekarn N, Lin W, Chung RT. HCV induces transforming growth factor β1 through activation of endoplasmic reticulum stress and the unfolded protein response. Sci Rep. 2016;6:22487. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 60] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 56. | Lin W, Tsai WL, Shao RX, Wu G, Peng LF, Barlow LL, Chung WJ, Zhang L, Zhao H, Jang JY, Chung RT. Hepatitis C virus regulates transforming growth factor beta1 production through the generation of reactive oxygen species in a nuclear factor kappaB-dependent manner. Gastroenterology. 2010;138:2509-2518, 2518.e1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 163] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 57. | Battaglia S, Benzoubir N, Nobilet S, Charneau P, Samuel D, Zignego AL, Atfi A, Bréchot C, Bourgeade MF. Liver cancer-derived hepatitis C virus core proteins shift TGF-beta responses from tumor suppression to epithelial-mesenchymal transition. PLoS One. 2009;4:e4355. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 98] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 58. | Pavio N, Battaglia S, Boucreux D, Arnulf B, Sobesky R, Hermine O, Brechot C. Hepatitis C virus core variants isolated from liver tumor but not from adjacent non-tumor tissue interact with Smad3 and inhibit the TGF-beta pathway. Oncogene. 2005;24:6119-6132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 67] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 59. | Macias MJ, Martin-Malpartida P, Massagué J. Structural determinants of Smad function in TGF-β signaling. Trends Biochem Sci. 2015;40:296-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 294] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 60. | Cheng PL, Chang MH, Chao CH, Lee YH. Hepatitis C viral proteins interact with Smad3 and differentially regulate TGF-beta/Smad3-mediated transcriptional activation. Oncogene. 2004;23:7821-7838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 59] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 61. | Erhardt A, Hassan M, Heintges T, Häussinger D. Hepatitis C virus core protein induces cell proliferation and activates ERK, JNK, and p38 MAP kinases together with the MAP kinase phosphatase MKP-1 in a HepG2 Tet-Off cell line. Virology. 2002;292:272-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 105] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 62. | Okuda M, Li K, Beard MR, Showalter LA, Scholle F, Lemon SM, Weinman SA. Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitis C virus core protein. Gastroenterology. 2002;122:366-375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 690] [Cited by in RCA: 679] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 63. | Kotsiri I, Hadziyannis E, Georgiou A, Papageorgiou MV, Vlachogiannakos I, Papatheodoridis G. Changes in serum transforming growth factor-β1 levels in chronic hepatitis C patients under antiviral therapy. Ann Gastroenterol. 2016;29:79-84. [PubMed] |
| 64. | Tsushima H, Kawata S, Tamura S, Ito N, Shirai Y, Kiso S, Doi Y, Yamada A, Oshikawa O, Matsuzawa Y. Reduced plasma transforming growth factor-beta1 levels in patients with chronic hepatitis C after interferon-alpha therapy: association with regression of hepatic fibrosis. J Hepatol. 1999;30:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 70] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 65. | Flisiak R, Jaroszewicz J, Lapinski TW, Flisiak I, Prokopowiczi D. Effect of pegylated interferon alpha 2b plus ribavirin treatment on plasma transforming growth factor-beta1, metalloproteinase-1, and tissue metalloproteinase inhibitor-1 in patients with chronic hepatitis C. World J Gastroenterol. 2005;11:6833-6838. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 6] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 66. | Janczewska-Kazek E, Marek B, Kajdaniuk D, Borgiel-Marek H. Effect of interferon alpha and ribavirin treatment on serum levels of transforming growth factor-beta1, vascular endothelial growth factor, and basic fibroblast growth factor in patients with chronic hepatitis C. World J Gastroenterol. 2006;12:961-965. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 12] [Cited by in RCA: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 67. | Holmes DI, Zachary I. The vascular endothelial growth factor (VEGF) family: angiogenic factors in health and disease. Genome Biol. 2005;6:209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 359] [Cited by in RCA: 476] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 68. | Koch S, Claesson-Welsh L. Signal transduction by vascular endothelial growth factor receptors. Cold Spring Harb Perspect Med. 2012;2:a006502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 535] [Cited by in RCA: 634] [Article Influence: 52.8] [Reference Citation Analysis (0)] |
| 69. | Ferrara N. Vascular endothelial growth factor as a target for anticancer therapy. Oncologist. 2004;9 Suppl 1:2-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 403] [Cited by in RCA: 407] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 70. | Goel HL, Mercurio AM. VEGF targets the tumour cell. Nat Rev Cancer. 2013;13:871-882. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 799] [Cited by in RCA: 962] [Article Influence: 80.2] [Reference Citation Analysis (0)] |
| 71. | Carmeliet P. VEGF as a key mediator of angiogenesis in cancer. Oncology. 2005;69 Suppl 3:4-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 953] [Cited by in RCA: 1212] [Article Influence: 60.6] [Reference Citation Analysis (0)] |
| 72. | Lemus-Varela ML, Flores-Soto ME, Cervantes-Munguía R, Torres-Mendoza BM, Gudiño-Cabrera G, Chaparro-Huerta V, Ortuño-Sahagún D, Beas-Zárate C. Expression of HIF-1 alpha, VEGF and EPO in peripheral blood from patients with two cardiac abnormalities associated with hypoxia. Clin Biochem. 2010;43:234-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 50] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 73. | Wong C, Wellman TL, Lounsbury KM. VEGF and HIF-1alpha expression are increased in advanced stages of epithelial ovarian cancer. Gynecol Oncol. 2003;91:513-517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 87] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 74. | Poon RT, Fan ST, Wong J. Clinical implications of circulating angiogenic factors in cancer patients. J Clin Oncol. 2001;19:1207-1225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 428] [Cited by in RCA: 426] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 75. | Alkharsah KR. VEGF Upregulation in Viral Infections and Its Possible Therapeutic Implications. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 63] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 76. | Llovet JM, Peña CE, Lathia CD, Shan M, Meinhardt G, Bruix J; SHARP Investigators Study Group. Plasma biomarkers as predictors of outcome in patients with advanced hepatocellular carcinoma. Clin Cancer Res. 2012;18:2290-2300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 464] [Cited by in RCA: 455] [Article Influence: 35.0] [Reference Citation Analysis (2)] |
| 77. | Mukozu T, Nagai H, Matsui D, Kanekawa T, Sumino Y. Serum VEGF as a tumor marker in patients with HCV-related liver cirrhosis and hepatocellular carcinoma. Anticancer Res. 2013;33:1013-1021. [PubMed] |
| 78. | Yvamoto EY, Ferreira RF, Nogueira V, Pinhe MA, Tenani GD, Andrade JG, Baitello ME, Gregório ML, Fucuta PS, Silva RF, Souza DR, Silva RC. Influence of vascular endothelial growth factor and alpha-fetoprotein on hepatocellular carcinoma. Genet Mol Res. 2015;14:17453-17462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 79. | Nasimuzzaman M, Waris G, Mikolon D, Stupack DG, Siddiqui A. Hepatitis C virus stabilizes hypoxia-inducible factor 1alpha and stimulates the synthesis of vascular endothelial growth factor. J Virol. 2007;81:10249-10257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 109] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 80. | Mee CJ, Farquhar MJ, Harris HJ, Hu K, Ramma W, Ahmed A, Maurel P, Bicknell R, Balfe P, McKeating JA. Hepatitis C virus infection reduces hepatocellular polarity in a vascular endothelial growth factor-dependent manner. Gastroenterology. 2010;138:1134-1142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 65] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 81. | Shimoda K, Mori M, Shibuta K, Banner BF, Barnard GF. Vascular endothelial growth factor/vascular permeability factor mRNA expression in patients with chronic hepatitis C and hepatocellular carcinoma. Int J Oncol. 1999;14:353-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 13] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 82. | Abe M, Koga H, Yoshida T, Masuda H, Iwamoto H, Sakata M, Hanada S, Nakamura T, Taniguchi E, Kawaguchi T, Yano H, Torimura T, Ueno T, Sata M. Hepatitis C virus core protein upregulates the expression of vascular endothelial growth factor via the nuclear factor-κB/hypoxia-inducible factor-1α axis under hypoxic conditions. Hepatol Res. 2012;42:591-600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 83. | Zhu C, Liu X, Wang S, Yan X, Tang Z, Wu K, Li Y, Liu F. Hepatitis C virus core protein induces hypoxia-inducible factor 1α-mediated vascular endothelial growth factor expression in Huh7.5.1 cells. Mol Med Rep. 2014;9:2010-2014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 84. | Liu XH, Zhou X, Zhu CL, Song H, Liu F. [Effects of HCV core protein on the expression of hypoxia-inducible factor 1 alpha and vascular endothelial growth factor]. Zhonghua Gan Zang Bing Za Zhi. 2011;19:751-754. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 85. | Shao YY, Hsieh MS, Wang HY, Li YS, Lin H, Hsu HW, Huang CY, Hsu CH, Cheng AL. Hepatitis C virus core protein potentiates proangiogenic activity of hepatocellular carcinoma cells. Oncotarget. 2017;8:86681-86692. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 86. | Tzeng HE, Tsai CH, Chang ZL, Su CM, Wang SW, Hwang WL, Tang CH. Interleukin-6 induces vascular endothelial growth factor expression and promotes angiogenesis through apoptosis signal-regulating kinase 1 in human osteosarcoma. Biochem Pharmacol. 2013;85:531-540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 100] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 87. | Dong W, Li Y, Gao M, Hu M, Li X, Mai S, Guo N, Yuan S, Song L. IKKα contributes to UVB-induced VEGF expression by regulating AP-1 transactivation. Nucleic Acids Res. 2012;40:2940-2955. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 88. | Tsutsumi T, Suzuki T, Moriya K, Yotsuyanagi H, Shintani Y, Fujie H, Matsuura Y, Kimura S, Koike K, Miyamura T. Alteration of intrahepatic cytokine expression and AP-1 activation in transgenic mice expressing hepatitis C virus core protein. Virology. 2002;304:415-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 63] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 89. | Shrivastava A, Manna SK, Ray R, Aggarwal BB. Ectopic expression of hepatitis C virus core protein differentially regulates nuclear transcription factors. J Virol. 1998;72:9722-9728. [PubMed] |
| 90. | Kanda T, Steele R, Ray R, Ray RB. Hepatitis C virus core protein augments androgen receptor-mediated signaling. J Virol. 2008;82:11066-11072. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 75] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 91. | Komiya Y, Habas R. Wnt signal transduction pathways. Organogenesis. 2008;4:68-75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1164] [Cited by in RCA: 1072] [Article Influence: 63.1] [Reference Citation Analysis (0)] |
| 92. | Monga SP. β-Catenin Signaling and Roles in Liver Homeostasis, Injury, and Tumorigenesis. Gastroenterology. 2015;148:1294-1310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 363] [Cited by in RCA: 398] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 93. | Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483-1487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1944] [Cited by in RCA: 2018] [Article Influence: 96.1] [Reference Citation Analysis (0)] |
| 94. | Kikuchi A, Yamamoto H, Sato A, Matsumoto S. New insights into the mechanism of Wnt signaling pathway activation. Int Rev Cell Mol Biol. 2011;291:21-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 214] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 95. | Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192-1205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3740] [Cited by in RCA: 4398] [Article Influence: 338.3] [Reference Citation Analysis (0)] |
| 96. | MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9-26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4613] [Cited by in RCA: 4509] [Article Influence: 281.8] [Reference Citation Analysis (0)] |
| 97. | King TD, Zhang W, Suto MJ, Li Y. Frizzled7 as an emerging target for cancer therapy. Cell Signal. 2012;24:846-851. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 111] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 98. | Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781-810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3831] [Cited by in RCA: 4054] [Article Influence: 202.7] [Reference Citation Analysis (0)] |
| 99. | Nusse R, Clevers H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell. 2017;169:985-999. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2031] [Cited by in RCA: 3107] [Article Influence: 388.4] [Reference Citation Analysis (0)] |
| 100. | Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843-850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2659] [Cited by in RCA: 2765] [Article Influence: 138.3] [Reference Citation Analysis (0)] |
| 101. | Heuberger J, Birchmeier W. Interplay of cadherin-mediated cell adhesion and canonical Wnt signaling. Cold Spring Harb Perspect Biol. 2010;2:a002915. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 424] [Cited by in RCA: 482] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 102. | Flanagan DJ, Austin CR, Vincan E, Phesse TJ. Wnt Signalling in Gastrointestinal Epithelial Stem Cells. Genes (Basel). 2018;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 103. | Wallingford JB, Habas R. The developmental biology of Dishevelled: an enigmatic protein governing cell fate and cell polarity. Development. 2005;132:4421-4436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 377] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 104. | Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387-398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1156] [Cited by in RCA: 1213] [Article Influence: 71.4] [Reference Citation Analysis (0)] |
| 105. | Jeong WJ, Ro EJ, Choi KY. Interaction between Wnt/β-catenin and RAS-ERK pathways and an anti-cancer strategy via degradations of β-catenin and RAS by targeting the Wnt/β-catenin pathway. NPJ Precis Oncol. 2018;2:5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 156] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 106. | Trischler J, Shiomi T, Turner DL, Sklepkiewicz PL, Goldklang MP, Tanaka KF, Xu M, Farber DL, D’Armiento JM. Immune Modulation of the T Cell Response in Asthma through Wnt10b. Am J Respir Cell Mol Biol. 2016;54:584-593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 107. | Reuter S, Martin H, Beckert H, Bros M, Montermann E, Belz C, Heinz A, Ohngemach S, Sahin U, Stassen M, Buhl R, Eshkind L, Taube C. The Wnt/β-catenin pathway attenuates experimental allergic airway disease. J Immunol. 2014;193:485-495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 45] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 108. | Wang W, Pan Q, Fuhler GM, Smits R, Peppelenbosch MP. Action and function of Wnt/β-catenin signaling in the progression from chronic hepatitis C to hepatocellular carcinoma. J Gastroenterol. 2017;52:419-431. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 64] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 109. | Anson M, Crain-Denoyelle AM, Baud V, Chereau F, Gougelet A, Terris B, Yamagoe S, Colnot S, Viguier M, Perret C, Couty JP. Oncogenic β-catenin triggers an inflammatory response that determines the aggressiveness of hepatocellular carcinoma in mice. J Clin Invest. 2012;122:586-599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 153] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 110. | Zeng G, Awan F, Otruba W, Muller P, Apte U, Tan X, Gandhi C, Demetris AJ, Monga SP. Wnt’er in liver: expression of Wnt and frizzled genes in mouse. Hepatology. 2007;45:195-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 113] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 111. | Wang Q, Chou X, Guan F, Fang Z, Lu S, Lei J, Li Y, Liu W. Enhanced Wnt Signalling in Hepatocytes is Associated with Schistosoma japonicum Infection and Contributes to Liver Fibrosis. Sci Rep. 2017;7:230. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 112. | Khanizadeh S, Hasanvand B, Esmaeil Lashgarian H, Almasian M, Goudarzi G. Interaction of viral oncogenic proteins with the Wnt signaling pathway. Iran J Basic Med Sci. 2018;21:651-659. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 113. | Liu J, Wang Z, Tang J, Tang R, Shan X, Zhang W, Chen Q, Zhou F, Chen K, Huang A, Tang N. Hepatitis C virus core protein activates Wnt/β-catenin signaling through multiple regulation of upstream molecules in the SMMC-7721 cell line. Arch Virol. 2011;156:1013-1023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 114. | Quan H, Zhou F, Nie D, Chen Q, Cai X, Shan X, Zhou Z, Chen K, Huang A, Li S, Tang N. Hepatitis C virus core protein epigenetically silences SFRP1 and enhances HCC aggressiveness by inducing epithelial-mesenchymal transition. Oncogene. 2014;33:2826-2835. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 88] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 115. | Umer M, Qureshi SA, Hashmi ZY, Raza A, Ahmad J, Rahman M, Iqbal M. Promoter hypermethylation of Wnt pathway inhibitors in hepatitis C virus - induced multistep hepatocarcinogenesis. Virol J. 2014;11:117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 116. | Ripoli M, Barbano R, Balsamo T, Piccoli C, Brunetti V, Coco M, Mazzoccoli G, Vinciguerra M, Pazienza V. Hypermethylated levels of E-cadherin promoter in Huh-7 cells expressing the HCV core protein. Virus Res. 2011;160:74-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 48] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 117. | Jhaveri R, Kundu P, Shapiro AM, Venkatesan A, Dasgupta A. Effect of heptitis C virus core protein on cellular gene expression: specific inhibition of cyclooxygenase 2. J Infect Dis. 2005;191:1498-1506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 14] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 118. | Simmons DL, Botting RM, Hla T. Cyclooxygenase isozymes: the biology of prostaglandin synthesis and inhibition. Pharmacol Rev. 2004;56:387-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1308] [Cited by in RCA: 1214] [Article Influence: 57.8] [Reference Citation Analysis (0)] |
| 119. | Chen WC, Tseng CK, Chen YH, Lin CK, Hsu SH, Wang SN, Lee JC. HCV NS5A Up-Regulates COX-2 Expression via IL-8-Mediated Activation of the ERK/JNK MAPK Pathway. PLoS One. 2015;10:e0133264. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 120. | Greenhough A, Smartt HJ, Moore AE, Roberts HR, Williams AC, Paraskeva C, Kaidi A. The COX-2/PGE2 pathway: key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis. 2009;30:377-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 848] [Cited by in RCA: 937] [Article Influence: 58.6] [Reference Citation Analysis (0)] |
| 121. | Núñez O, Fernández-Martínez A, Majano PL, Apolinario A, Gómez-Gonzalo M, Benedicto I, López-Cabrera M, Boscá L, Clemente G, García-Monzón C, Martín-Sanz P. Increased intrahepatic cyclooxygenase 2, matrix metalloproteinase 2, and matrix metalloproteinase 9 expression is associated with progressive liver disease in chronic hepatitis C virus infection: role of viral core and NS5A proteins. Gut. 2004;53:1665-1672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 123] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 122. | Cheng AS, Chan HL, Leung WK, To KF, Go MY, Chan JY, Liew CT, Sung JJ. Expression of HBx and COX-2 in chronic hepatitis B, cirrhosis and hepatocellular carcinoma: implication of HBx in upregulation of COX-2. Mod Pathol. 2004;17:1169-1179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 78] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 123. | Kaul R, Verma SC, Murakami M, Lan K, Choudhuri T, Robertson ES. Epstein-Barr virus protein can upregulate cyclo-oxygenase-2 expression through association with the suppressor of metastasis Nm23-H1. J Virol. 2006;80:1321-1331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 37] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 124. | Subbaramaiah K, Dannenberg AJ. Cyclooxygenase-2 transcription is regulated by human papillomavirus 16 E6 and E7 oncoproteins: evidence of a corepressor/coactivator exchange. Cancer Res. 2007;67:3976-3985. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 99] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 125. | Moss SF, Blaser MJ. Mechanisms of disease: Inflammation and the origins of cancer. Nat Clin Pract Oncol. 2005;2:90-7; quiz 1 p following 113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 214] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 126. | Aggarwal BB, Vijayalekshmi RV, Sung B. Targeting inflammatory pathways for prevention and therapy of cancer: short-term friend, long-term foe. Clin Cancer Res. 2009;15:425-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 502] [Cited by in RCA: 560] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 127. | Gomaa WM, Ibrahim MA, Shatat ME. Overexpression of cyclooxygenase-2 and transforming growth factor-beta 1 is an independent predictor of poor virological response to interferon therapy in chronic HCV genotype 4 patients. Saudi J Gastroenterol. 2014;20:59-65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 128. | Trujillo-Murillo K, Alvarez-Martínez O, Garza-Rodríguez L, Martínez-Rodríguez H, Bosques-Padilla F, Ramos-Jiménez J, Barrera-Saldaña H, Rincón-Sánchez AR, Rivas-Estilla AM. Additive effect of ethanol and HCV subgenomic replicon expression on COX-2 protein levels and activity. J Viral Hepat. 2007;14:608-617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 129. | Trujillo-Murillo K, Rincón-Sánchez AR, Martínez-Rodríguez H, Bosques-Padilla F, Ramos-Jiménez J, Barrera-Saldaña HA, Rojkind M, Rivas-Estilla AM. Acetylsalicylic acid inhibits hepatitis C virus RNA and protein expression through cyclooxygenase 2 signaling pathways. Hepatology. 2008;47:1462-1472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 61] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 130. | Ivanov AV, Bartosch B, Smirnova OA, Isaguliants MG, Kochetkov SN. HCV and oxidative stress in the liver. Viruses. 2013;5:439-469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 174] [Cited by in RCA: 160] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 131. | Waris G, Siddiqui A. Hepatitis C virus stimulates the expression of cyclooxygenase-2 via oxidative stress: role of prostaglandin E2 in RNA replication. J Virol. 2005;79:9725-9734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 107] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 132. | Simula MP, De Re V. Hepatitis C virus-induced oxidative stress and mitochondrial dysfunction: a focus on recent advances in proteomics. Proteomics Clin Appl. 2010;4:782-793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 33] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 133. | Jahan S, Ashfaq UA, Qasim M, Khaliq S, Saleem MJ, Afzal N. Hepatitis C virus to hepatocellular carcinoma. Infect Agent Cancer. 2012;7:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 134. | Madejón A, Sheldon J, Francisco-Recuero I, Perales C, Domínguez-Beato M, Lasa M, Sánchez-Perez I, Muntané J, Domingo E, García-Samaniego J, Sánchez-Pacheco A. Hepatitis C virus-mediated Aurora B kinase inhibition modulates inflammatory pathway and viral infectivity. J Hepatol. 2015;63:312-319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 135. | Friedman SL. Liver fibrosis -- from bench to bedside. J Hepatol. 2003;38 Suppl 1:S38-S53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1199] [Cited by in RCA: 1293] [Article Influence: 58.8] [Reference Citation Analysis (0)] |
| 136. | Callejas NA, Casado M, Díaz-Guerra MJ, Boscá L, Martín-Sanz P. Expression of cyclooxygenase-2 promotes the release of matrix metalloproteinase-2 and -9 in fetal rat hepatocytes. Hepatology. 2001;33:860-867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 80] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 137. | Ivanov AV, Smirnova OA, Petrushanko IY, Ivanova ON, Karpenko IL, Alekseeva E, Sominskaya I, Makarov AA, Bartosch B, Kochetkov SN, Isaguliants MG. HCV core protein uses multiple mechanisms to induce oxidative stress in human hepatoma Huh7 cells. Viruses. 2015;7:2745-2770. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 69] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 138. | Lan T, Kisseleva T, Brenner DA. Deficiency of NOX1 or NOX4 Prevents Liver Inflammation and Fibrosis in Mice through Inhibition of Hepatic Stellate Cell Activation. PLoS One. 2015;10:e0129743. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 128] [Cited by in RCA: 172] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 139. | Castellone MD, Teramoto H, Williams BO, Druey KM, Gutkind JS. Prostaglandin E2 promotes colon cancer cell growth through a Gs-axin-beta-catenin signaling axis. Science. 2005;310:1504-1510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 695] [Cited by in RCA: 720] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 140. | Yamanaka Y, Shiraki K, Inoue T, Miyashita K, Fuke H, Yamaguchi Y, Yamamoto N, Ito K, Sugimoto K, Nakano T. COX-2 inhibitors sensitize human hepatocellular carcinoma cells to TRAIL-induced apoptosis. Int J Mol Med. 2006;18:41-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 141. | Leng J, Han C, Demetris AJ, Michalopoulos GK, Wu T. Cyclooxygenase-2 promotes hepatocellular carcinoma cell growth through Akt activation: evidence for Akt inhibition in celecoxib-induced apoptosis. Hepatology. 2003;38:756-768. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 214] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 142. | Ye Y, Xu Y, Lai Y, He W, Li Y, Wang R, Luo X, Chen R, Chen T. Long non-coding RNA cox-2 prevents immune evasion and metastasis of hepatocellular carcinoma by altering M1/M2 macrophage polarization. J Cell Biochem. 2018;119:2951-2963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 175] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 143. | Tyagi S, Gupta P, Saini AS, Kaushal C, Sharma S. The peroxisome proliferator-activated receptor: A family of nuclear receptors role in various diseases. J Adv Pharm Technol Res. 2011;2:236-240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 509] [Cited by in RCA: 713] [Article Influence: 54.8] [Reference Citation Analysis (0)] |
| 144. | Michalik L, Wahli W. PPARs Mediate Lipid Signaling in Inflammation and Cancer. PPAR Res. 2008;2008:134059. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 76] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 145. | Ament Z, Masoodi M, Griffin JL. Applications of metabolomics for understanding the action of peroxisome proliferator-activated receptors (PPARs) in diabetes, obesity and cancer. Genome Med. 2012;4:32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 146. | Agriesti F, Tataranni T, Ruggieri V, Capitanio N, Piccoli C. PPARs and HCV-Related Hepatocarcinoma: A Mitochondrial Point of View. PPAR Res. 2012;2012:605302. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 147. | Ashby J, Brady A, Elcombe CR, Elliott BM, Ishmael J, Odum J, Tugwood JD, Kettle S, Purchase IF. Mechanistically-based human hazard assessment of peroxisome proliferator-induced hepatocarcinogenesis. Hum Exp Toxicol. 1994;13 Suppl 2:S1-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 197] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 148. | Saha SA, Kizhakepunnur LG, Bahekar A, Arora RR. The role of fibrates in the prevention of cardiovascular disease--a pooled meta-analysis of long-term randomized placebo-controlled clinical trials. Am Heart J. 2007;154:943-953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 81] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 149. | Issemann I, Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990;347:645-650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2546] [Cited by in RCA: 2520] [Article Influence: 72.0] [Reference Citation Analysis (0)] |
| 150. | Peters JM, Cattley RC, Gonzalez FJ. Role of PPAR alpha in the mechanism of action of the nongenotoxic carcinogen and peroxisome proliferator Wy-14,643. Carcinogenesis. 1997;18:2029-2033. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 375] [Cited by in RCA: 353] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 151. | Tanaka N, Moriya K, Kiyosawa K, Koike K, Aoyama T. Hepatitis C virus core protein induces spontaneous and persistent activation of peroxisome proliferator-activated receptor alpha in transgenic mice: implications for HCV-associated hepatocarcinogenesis. Int J Cancer. 2008;122:124-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 63] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 152. | Yan F, Wang Q, Xu C, Cao M, Zhou X, Wang T, Yu C, Jing F, Chen W, Gao L, Zhao J. Peroxisome proliferator-activated receptor α activation induces hepatic steatosis, suggesting an adverse effect. PLoS One. 2014;9:e99245. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 153. | Ohata K, Hamasaki K, Toriyama K, Matsumoto K, Saeki A, Yanagi K, Abiru S, Nakagawa Y, Shigeno M, Miyazoe S, Ichikawa T, Ishikawa H, Nakao K, Eguchi K. Hepatic steatosis is a risk factor for hepatocellular carcinoma in patients with chronic hepatitis C virus infection. Cancer. 2003;97:3036-3043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 245] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 154. | Koike K. Steatosis, liver injury, and hepatocarcinogenesis in hepatitis C viral infection. J Gastroenterol. 2009;44 Suppl 19:82-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 155. | Tanaka N, Moriya K, Kiyosawa K, Koike K, Gonzalez FJ, Aoyama T. PPARalpha activation is essential for HCV core protein-induced hepatic steatosis and hepatocellular carcinoma in mice. J Clin Invest. 2008;118:683-694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 121] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 156. | Shen W, Gao Y, Lu B, Zhang Q, Hu Y, Chen Y. Negatively regulating TLR4/NF-κB signaling via PPARα in endotoxin-induced uveitis. Biochim Biophys Acta. 2014;1842:1109-1120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 157. | Barger PM, Browning AC, Garner AN, Kelly DP. p38 mitogen-activated protein kinase activates peroxisome proliferator-activated receptor alpha: a potential role in the cardiac metabolic stress response. J Biol Chem. 2001;276:44495-44501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 202] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 158. | Tsutsumi T, Suzuki T, Shimoike T, Suzuki R, Moriya K, Shintani Y, Fujie H, Matsuura Y, Koike K, Miyamura T. Interaction of hepatitis C virus core protein with retinoid X receptor alpha modulates its transcriptional activity. Hepatology. 2002;35:937-946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 125] [Article Influence: 5.4] [Reference Citation Analysis (0)] |