Published online Mar 7, 2018. doi: 10.3748/wjg.v24.i9.971
Peer-review started: January 10, 2018
First decision: January 18, 2018
Revised: February 5, 2018
Accepted: February 8, 2018
Article in press: February 8, 2018
Published online: March 7, 2018
Processing time: 53 Days and 22.7 Hours
To evaluate the association between virulence factor status and antibiotic resistance in Helicobacter pylori (H. pylori)-infected patients in Ireland.
DNA was extracted from antral and corpus biopsies obtained from 165 H. pylori-infected patients. Genotyping for clarithromycin and fluoroquinolone-mediating mutations was performed using the Genotype HelicoDR assay. cagA and vacA genotypes were investigated using PCR.
Primary, secondary and overall resistance rates for clarithromycin were 50.5% (n = 53/105), 78.3% (n = 47/60) and 60.6% (n = 100/165), respectively. Primary, secondary and overall resistance rates for fluoroquinolones were 15.2% (n = 16/105) and 28.3% (n = 17/60) and 20% (n = 33/165), respectively. Resistance to both antibiotics was 12.4% (n = 13/105) in treatment-naïve patients, 25% (n = 15/60) in those previously treated and 17% (n = 28/165) overall. A cagA-positive genotype was detected in 22.4% (n = 37/165) of patient samples. The dominant vacA genotype was S1/M2 at 44.8% (n = 74/165), followed by S2/M2 at 26.7% (n = 44/165), S1/M1 at 23.6% (n = 39/165) and S2/M1 at 4.8% (n = 8/165). Primary clarithromycin resistance was significantly lower in cagA-positive strains than in cagA-negative strains [32% (n = 8/25) vs 56.3% (n = 45/80) P = 0.03]. Similarly, in patients infected with more virulent H. pylori strains bearing the vacA s1 genotype, primary clarithromycin resistance was significantly lower than in those infected with less virulent strains bearing the vacA s2 genotype, [41% (n = 32/78) vs 77.8% (n = 21/27) P = 0.0001]. No statistically significant association was found between primary fluoroquinolone resistance and virulence factor status.
Genotypic H. pylori clarithromycin resistance is high and cagA-negative strains are dominant in our population. Less virulent (cagA-negative and vacA S2-containing) strains of H. pylori are associated with primary clarithromycin resistance.
Core tip: The management of Helicobacter pylori (H. pylori) infection is challenging, largely due to the emergence of antibiotic resistance. A greater understanding of local antibiotic resistance rates is important in determining the most appropriate treatment regimen in a given population. Furthermore, insight into the virulence of the infecting strains and the association between virulence and antibiotic resistance could potentially be an avenue to explore in the effort to improve eradication rates. This study provides and update on the prevalence of clarithromycin and fluoroquinolone resistance in Ireland and demonstrates that less virulent strains of H. pylori are predictive of primary clarithromycin resistance.
- Citation: Brennan DE, Dowd C, O’Morain C, McNamara D, Smith SM. Can bacterial virulence factors predict antibiotic resistant Helicobacter pylori infection? World J Gastroenterol 2018; 24(9): 971-981
- URL: https://www.wjgnet.com/1007-9327/full/v24/i9/971.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i9.971
Helicobacter pylori (H. pylori) infection causes acute and chronic gastritis, gastric and duodenal ulcers, and in rare cases gastric adenocarcinoma and MALT (mucosa-associated lymphoid tissue) lymphoma[1]. While its prevalence in the developed world has generally decreased, it is still high in indigenous populations and the developing world[2]. The Maastricht consensus recommends that all symptomatic H. pylori-infected adults are treated[1]. There are many different treatment options available, however the most common treatment for first-line eradication of H. pylori is triple therapy, which consists of two antibiotics (clarithromycin and amoxicillin) and a proton pump inhibitor, taken for 7-14 d. An efficacious therapy for H. pylori eradication is one that achieves an eradication rate of over 80%[1]. However, in many countries, the eradication rate for standard triple therapy has fallen below 80%. Indeed in a recent study in Ireland, the eradication rates of standard seven-day triple therapy were just 56.8% and 61% by intention-to-treat and per-protocol analysis, respectively[3]. There are several factors that impact the efficacy of treatment for H. pylori; high bacterial load, high gastric acidity and poor patient compliance. However, undoubtedly the most important is the rapid emergence of antimicrobial resistant strains of H. pylori, particularly to clarithromycin[4-6]. Resistance to clarithromycin can decrease the success rate of clarithromycin-based triple therapy by up to 70%[7]. One study found that the presence of clarithromycin resistant strains in a patient infected with H. pylori predicted treatment failure almost perfectly[8].
H. pylori is a highly heterogeneous bacterium and its virulence varies geographically. Virulence factors not only contribute to the pathogenicity of the bacteria but may play a role in determining treatment outcome[9]. The most commonly studied virulence factors in H. pylori are encoded by the cytotoxin associated gene A (cagA) and the vacuolating associated gene A (vacA). There are at least 4 variable regions in the vacA gene; in the signal (s) region, of which one of two alleles can be present: s1 or s2, and in the middle (m) region, of which one of two alleles can be present; m1 or m2[10]. These variable regions display different levels of toxicity to host cells, with vacA s1/m1 being most cytotoxic, followed by s1/m2. The s2/m2 genotype has been found to induce little or no toxicity[11]. A possible relationship between virulence factors and antimicrobial resistance has been suggested. A study conducted in 2009 in Ireland reported that the absence of cagA may be a risk factor for developing metronidazole resistance[12]. This study aimed to provide an update on the prevalence of virulence factor genotypes and antibiotic resistance in Irish H. pylori strains and assess the relationship between clarithromycin and fluoroquinolone resistance with virulence factor status.
A prospective study was carried out in a tertiary referral teaching hospital (Adelaide and Meath Hospital, Dublin, Ireland) affiliated with Trinity College Dublin. Patients who had been referred to the endoscopy clinic were included from August 2014 until June 2017. The study received ethical approval from the Adelaide and Meath Hospital Research Ethics Committee. Informed consent was obtained from all patients before enrolment.
Inclusion criteria were (1) ability and willingness to participate in the study and to provide informed consent, and (2) confirmed H. pylori infection as indicated by a positive rapid urease test (TRI-MED Distributors, PTY LTD, Washington, United States) at 30 min and by histology.
Exclusion criteria were (1) age less than 18 years, (2) pregnancy or lactation, (3) severe inter-current illness, (4) current PPI use or recent antibiotic use (within 4 wk); and (5) bleeding problems or use of blood thinning drugs.
A single corpus and antrum biopsy from each patient were placed into collection tubes and stored at -20 °C until processed for genomic DNA isolation using the QIAamp DNA Mini Kit (Qiagen GmbH, Hilden, Germany) according to manufacturer’s instructions. All isolated DNA was stored -20 °C until genotyping was performed.
Genotyping for clarithromycin and fluoroquinolone-mediating mutations was performed using the Genotype HelicoDR assay (Hain Lifescience GmbH, Nehren, Germany) according to the manufacturer’s instructions. Briefly, multiplex amplification of DNA regions of interest was performed using biotinylated primers supplied in the GenoType HelicoDR kit and the Hotstart Taq DNA polymerase kit (Qiagen). PCR products were reverse hybridised to DNA strips containing probes for gene regions of interest, developed and interpreted according to the manufacturers’ instructions[13].
To determine virulence factor genotype, PCR was performed as previously described by Taneike et al[12] using the primers described in Table 1. CagA and vacA genotypes were evaluated by performing gel electrophoresis on the PCR products using 1% agarose gel.
| Primer | Primer sequence | Gene | Product size (bp) |
| CAGA-F | 5’-GATAACAGGCAAGCTTTTGATG-3’ | cagA | 349 |
| CAGA-R | 5’-CTGCAAAAGATTGTTTGGCAGA-3’ | ||
| VA1-F | 5’- ATGGAAATACAACAACAAACACAC-3’ | vacA signal region | 259/286 (s1/s2) |
| VA1-R | 5’ – CTGCTTGAATGCGCCAAAC-3’ | ||
| VAG-F | 5’ – CAATCTGTCCAATCAAGCGAG-3’ | vacA middle region | 567/642 (m1/m2) |
| VAG-R | 5’- GCTTCAAAATAATTCCAAGG-3’ |
Statistical analysis was carried out using GraphPad Prism (GraphPad Software Inc., CA, United States). Continuous variables are presented as arithmetic mean and SD. P values for continuous variables were calculated and compared using the two-tailed independent t-test. P values for categorical variables were calculated using the Fisher’s exact test/Pearson χ2-test. In all cases, a P value less than 0.05 was considered significant.
Samples from a total of 165 H. pylori-infected patients were analysed in the study. Patient demographics and clinical characteristics are shown in Table 2. 63.6% (n = 105) of patients had not been treated for H. pylori infection previously, while 36.4% (n = 60) had undergone at least one eradication treatment regimen (Table 2).
| Number of gastric biopsy specimens n (%) | |||
| All patients 165 (100) | Treatment Naïve 105 (63.6) | Previously treated 60 (36.4) | |
| Gender | |||
| Female | 69 (41.8) | 31 (29.5) | 38 (63.3) |
| Male | 96 (58.2) | 74 (70.5) | 22 (36.7) |
| Age | |||
| mean ± SD | 49.2 ± 15.8 | 50.3 ± 16.3 | 47.4 ± 14.7 |
| Histology findings | |||
| Chronic gastritis | 130 (78.8) | 78 (74.3) | 52 (86.7) |
| Intestinal metaplasia | 23 (13.9) | 16 (15.2) | 7 (11.7) |
| No data available | 11 (6.7) | 10 (9.5) | 1 (1.7) |
| Normal mucosa | 1 (0.6) | 1 (1.0) | 0 (0.0) |
| Endoscopic findings | |||
| Gastritis | 92 (55.8) | 57 (54.3) | 35 (58.3) |
| Normal | 32 (19.4) | 19 (18.1) | 13 (21.7) |
| Gastric/duodenal ulcer | 21 (12.7) | 15 (14.3) | 6 (10.0) |
| No data available | 17 (10.3) | 11 (10.5) | 6 (10.0) |
| Atrophic mucosa | 1 (0.6) | 1 (1.0) | 0 (0.0) |
| Other1 | 2 (1.2) | 2 (1.9) | 0 (0.0) |
Primary resistance rates for clarithromycin and fluoroquinolones were 50.5% (n = 53/105; Table 3) and 15.2% (n = 16/105; Table 4), respectively. In those previously treated for H. pylori infection, the resistance rates for both clarithromycin and fluoroquinolones were higher at 78.3% (n = 47/60; Table 3) and 28.3% (n = 17/60; Table 4), respectively. Overall resistance rates, regardless of treatment history, were 60.6% (n = 100/165; Table 3) and 20% (n = 33/165; Table 4) for clarithromycin and fluoroquinolones, respectively. Among patients infected with a clarithromycin-resistant strain, the most common point mutation was A2147G, at 78% (n = 78/100; Table 3). The most common point mutation conferring resistance to fluoroquinolones in resistant patients was gyr91 D91Y, at 54.5% (n = 18/33; Table 4).
| Genotype | Number of gastric biopsy specimens n (%) | P value1 | ||
| All patients 165 (100) | Treatment Naïve 105 (63.6) | Previously treated 60 (36.4) | ||
| ClarithromycinS (WT) | 65 (39.4) | 52 (49.5) | 13 (21.7) | < 0.001 |
| ClarithromycinR | 100 (60.6) | 53 (50.5) | 47 (78.3) | |
| Point mutations | ||||
| A2147G | 78 (78) | 44 (83) | 34 (72.3) | NS |
| A2146G | 8 (8) | 3 (5.7) | 5 (10.6) | NS |
| A2146C | 6 (6) | 3 (5.7) | 3 (6.4) | NS |
| A2146C + A2147G | 5 (5) | 3 (5.7) | 2 (4.3) | NS |
| A2146G + A2147G | 2 (2) | 0 (0) | 2 (4.3) | NS |
| A2146G + A2146C | 1 (1) | 0 (0) | 1 (2.1) | NS |
| Genotype | Number of gastric biopsy specimens n (%) | P value1 | ||
| All patients 165 (100) | Treatment Naïve 105 (63.6) | Previously treated 60 (36.4) | ||
| FluoroquinoloneS (WT) | 132 (80.0) | 89 (84.8) | 43 (71.7) | |
| FluoroquinoloneR | 33 (20.0) | 16 (15.2) | 17 (28.3) | NS |
| Point mutations | ||||
| gyr91 D91Y | 18 (54.5) | 10 (62.5) | 8 (47.1) | NS |
| gyr91 D91N | 6 (18.2) | 2 (12.5) | 4 (23.5) | NS |
| gyr91 D91G | 2 (6.1) | 0 (0.0) | 2 (11.8) | NS |
| gyr91 D91N + gyr91 D91G | 2 (6.1) | 1 (6.3) | 1 (5.9) | NS |
| gyr91 D91N +gyr91 D91Y | 2 (6.1) | 1 (6.3) | 1 (5.9) | NS |
| gyr87 N87K | 1 (3.0) | 1 (6.3) | 0 (0.0) | NS |
| gyr87 N87K + gyr91 D91N + gyr91 D91G | 1 (3.0) | 0 (0.0) | 1 (5.9) | NS |
| gyr87 N87K + gyr91 D91N + gyr91 D91G + gyr91 D91Y | 1 (3.0) | 1 (6.3) | 0 (0.0) | NS |
Dual resistance rates for clarithromycin and fluoroquinolones were 12.4% (n = 13/105) in the treatment naïve, 25% (n = 15/60) in those previously treated and 17% (n = 28/165) in all patients included (Table 5). The overall rate of dual susceptibility among the patients was 36.4% (n = 60/165; Table 5). Dual susceptibility was significantly higher in treatment-naïve patients versus those previously treated (46.6%, n = 49/105 vs 18.3%, n = 11/60; P < 0.05; Fisher’s exact test).
| Genotype | Number of gastric biopsy specimens n (%) | P value1 | ||
| All patients 165 (100) | Treatment Naïve 105 (63.6) | Previously treated 60 (36.4) | ||
| Susceptible (to both) | 60 (36.4) | 49 (46.6) | 11 (18.3) | < 0.05 |
| Resistant (to at least one) | 105 (63.6) | 56 (53.3) | 49 (81.6) | |
| Susceptible/resistant to one | 137 (83.0) | 92 (87.6) | 45 (75.0) | |
| Resistant to both | 28 (17.0) | 13 (12.4) | 15 (25.0) | 0.05 |
Table 6 illustrates the distribution of H. pylori virulence factor genotype in infected patients. Overall, 22.4% (n = 37/165) of patients were infected with strains that were cagA positive and 77.6% (n = 128/165) that were cagA negative. The most prevalent vacA allele was S1/M2 at 44.8% (n = 74/165), followed by S2/M2, S1/M1 and S2/M1 at 26.7% (n = 44/165), 23.6% (n = 39/165) and 4.8% (n = 8/165), respectively (Table 6). Interestingly, the frequency of the vacA S1 genotype (the more virulent S region genotype) was significantly lower in those previously treated than the treatment-naïve group [58.3% (n = 35/60) vs 74.3% (n = 78/105) respectively; P < 0.05; Fisher’s exact test]. Additionally, the frequency of the S2/M2 genotype (the least virulent genotype) was significantly higher in those patients who have been treated previously [36.7% (n = 22/60) vs 21% (n = 22/105) respectively; P < 0.05; Fisher’s exact test; Table 6].
| Genotype | Overall (n = 165) | Treatment naïve (n = 105) | Previous treatment (n = 60) | P value1 |
| cagA status | ||||
| Positive | 37 (22.4) | 25 (23.8) | 12 (20) | NS |
| Negative | 128 (77.6) | 80 (76.2) | 48 (80) | |
| vacA allele | ||||
| S1 | 113 (68.5) | 78 (74.3) | 35 (58.3) | |
| S2 | 52 (31.5) | 27 (25.7) | 25 (41.7) | < 0.05 |
| M1 | 47 (28.5) | 31 (29.5) | 16 (26.7) | |
| M2 | 118 (71.5) | 74 (70.5) | 44 (73.3) | NS |
| S1/M1 | 39 (23.6) | 26 (24.8) | 13 (21.7) | NS |
| S1/M2 | 74 (44.8) | 52 (49.5) | 22 (36.7) | NS |
| S2/M1 | 8 (4.8) | 5 (4.8) | 3 (5.0) | NS |
| S2/M2 | 44 (26.7) | 22 (21.0) | 22 (36.7) | < 0.05 |
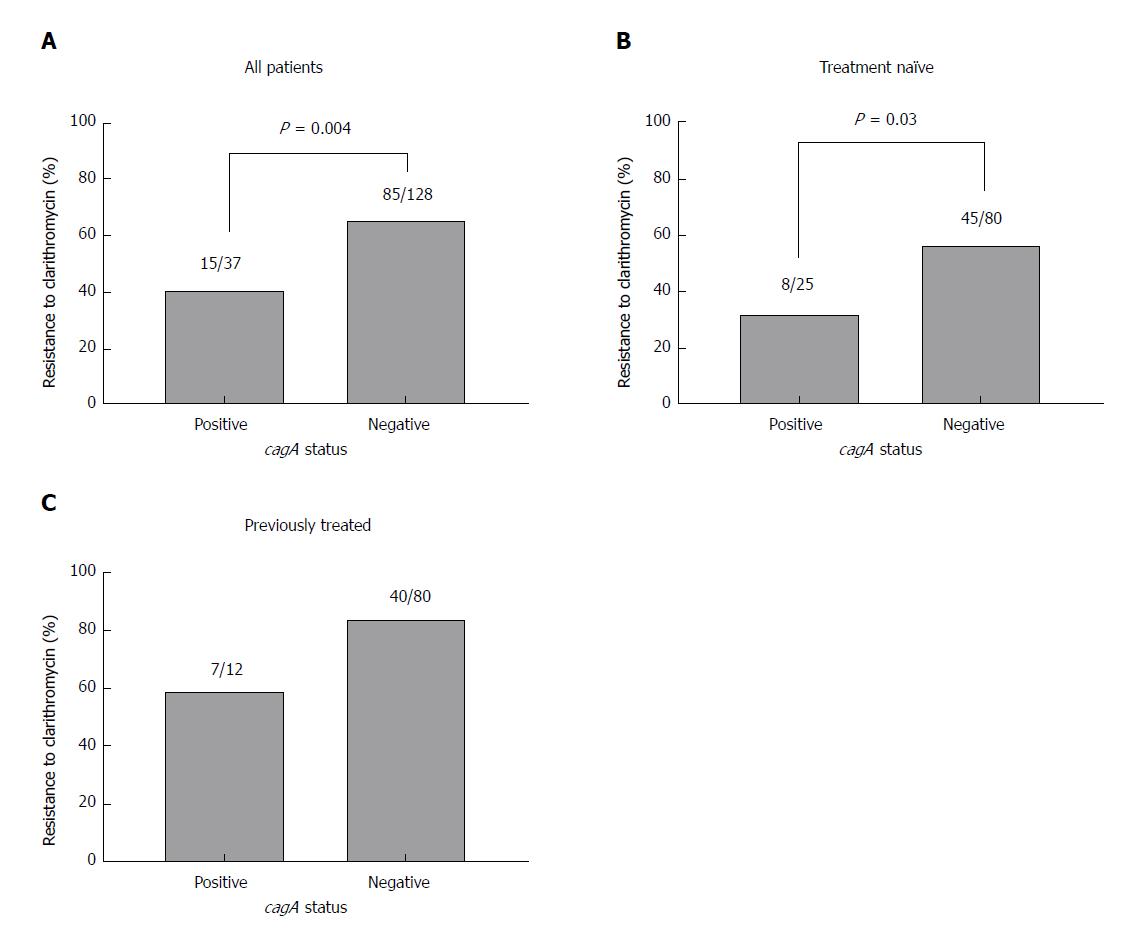
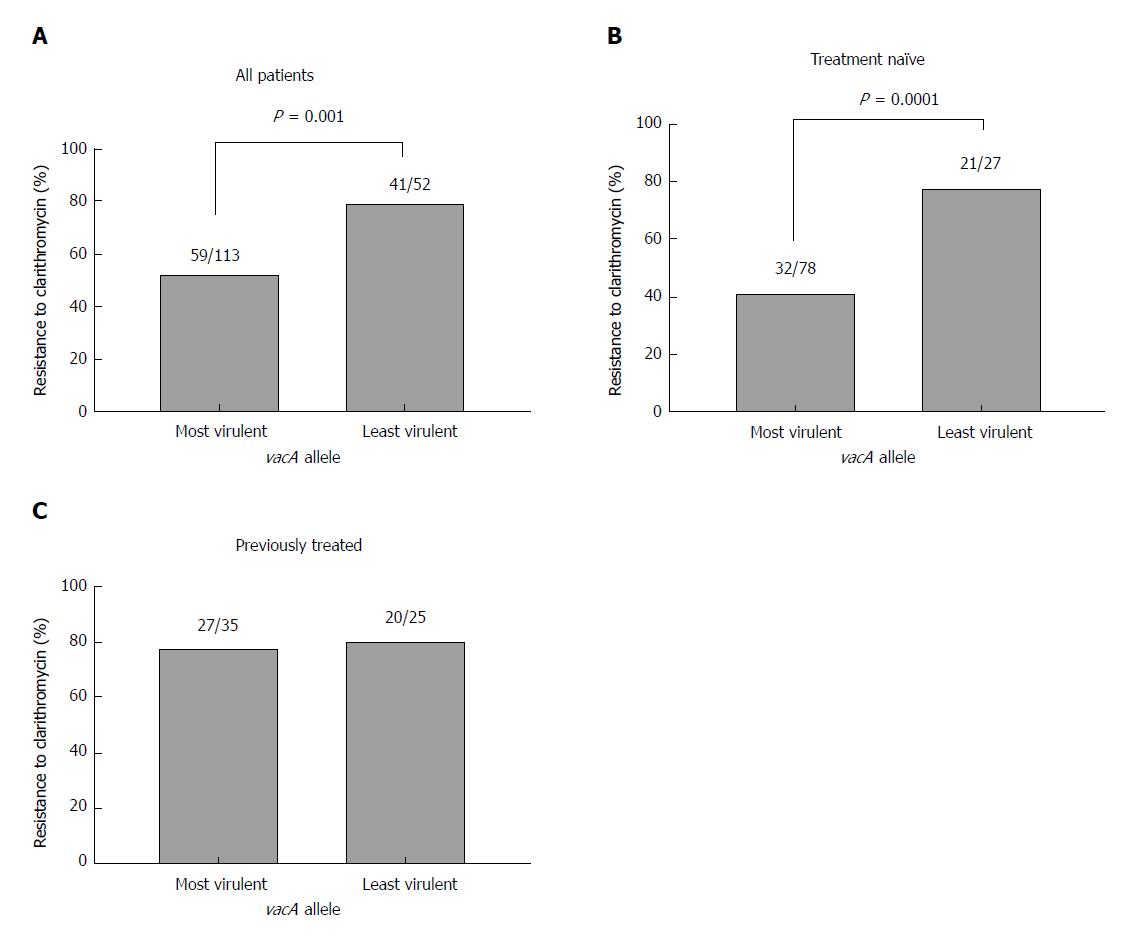
Next, the relationship between antibiotic resistance and virulence factor genotype was assessed. Analysis of all recruited patients revealed that genotypic resistance to clarithromycin was significantly lower in cagA-positive strains than in cagA-negative strains [40.5% (n = 15/37) vs 66.4% (n = 85/128); χ2 = 8.04; P = 0.004; Pearson χ2 test; Figure 1A]. When patients were sub-grouped into treatment-naïve (Figure 1B) and those previously treated (Figure 1C), clarithromycin resistance was also lower in cagA-positive strains compared to cagA-negative strains, although this only reached statistical significance in the treatment-naïve cohort [32% (n = 8/25) vs 56.3% (n = 45/80); χ2 = 4.5; P = 0.03; Pearson χ2 test; Figure 1B]. Similarly, in patients infected with more virulent H. pylori strains bearing the vacA s1 genotype, clarithromycin resistance was significantly lower than in those infected with less virulent strains bearing the vacA s2 genotype, when all patients were included [52.2% (n = 59/113) vs 78.8% (n = 41/52); χ2 = 10.6; P = 0.001; Pearson χ2 test; Figure 2A] and in those that were treatment-naïve [41% (n = 32/78) vs 77.8% (n = 21/27); χ2 = 10.8; P = 0.0001; Pearson χ2 test; Figure 2B], but not in patients that were previously treated (Figure 2C).
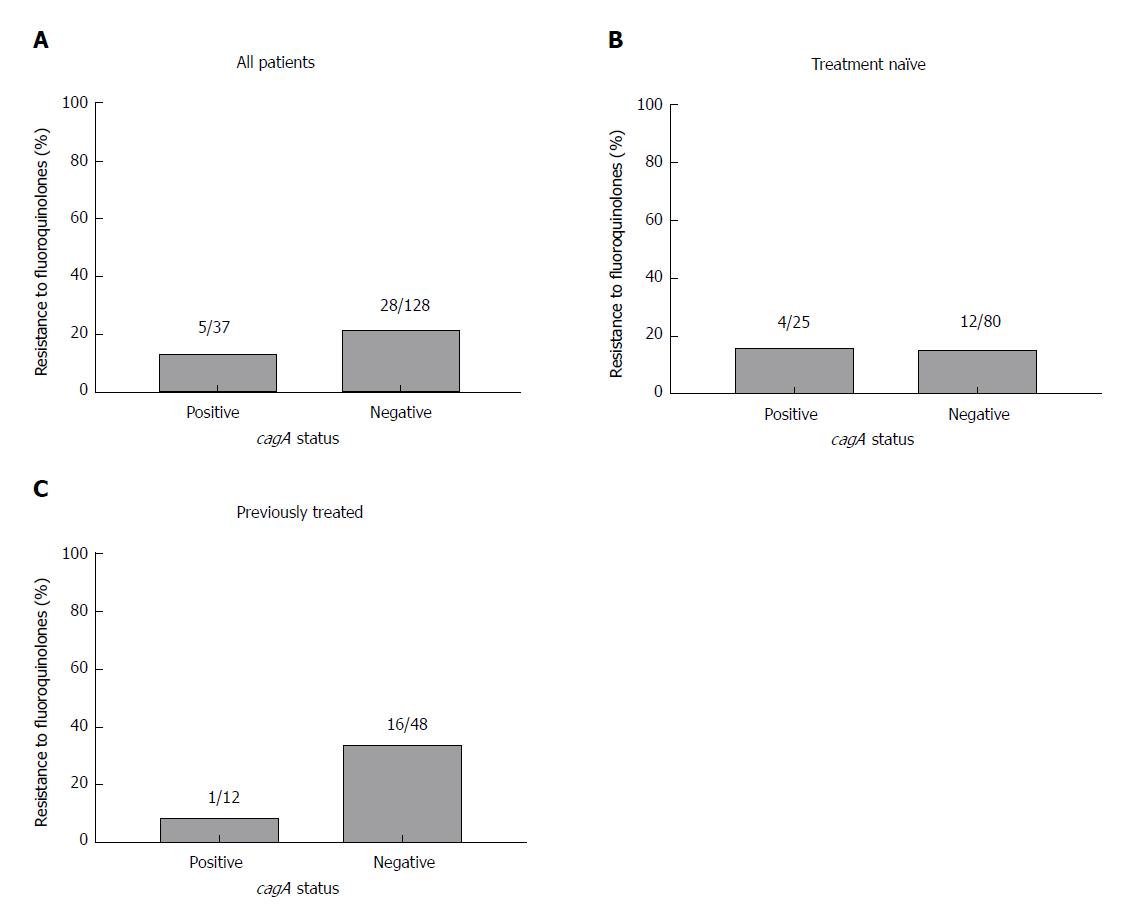
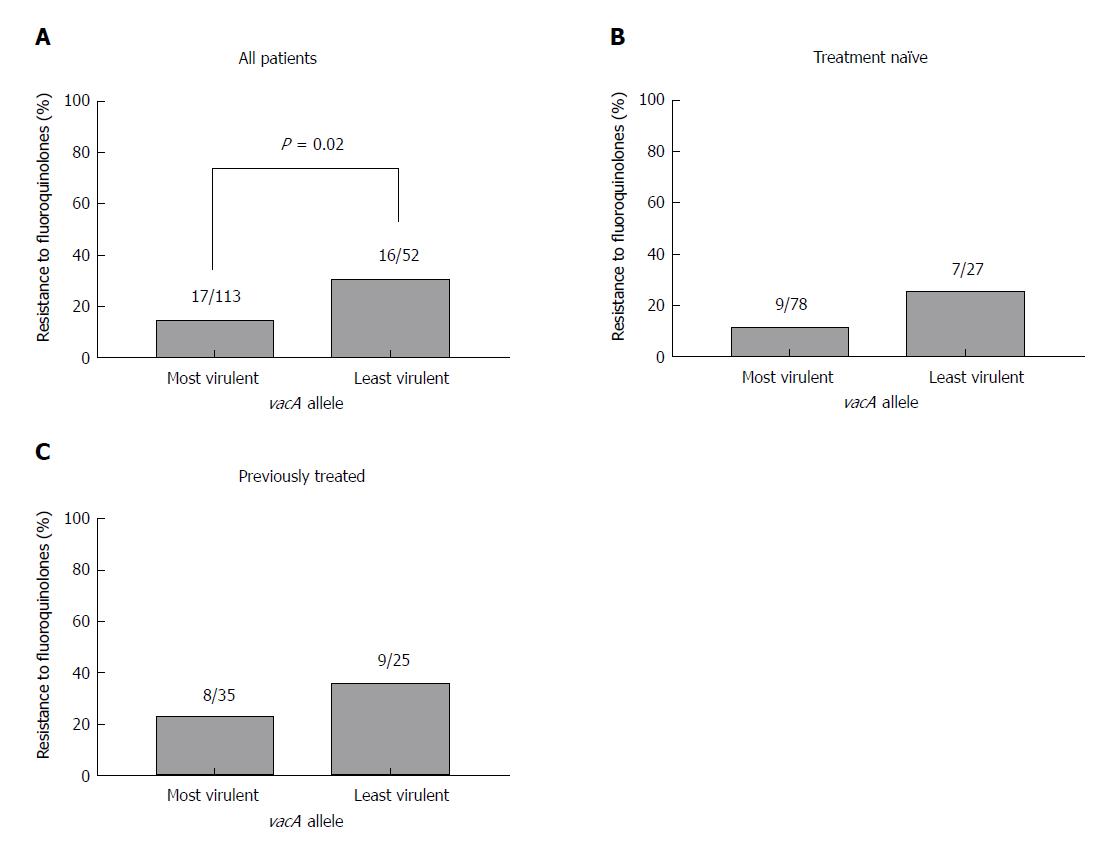
The frequency of resistance to fluoroquinolones in each virulence factor genotype was also examined. CagA status was not significantly associated with fluoroquinolone resistance when all patients were analysed (Figure 3A) or when the patients were sub-divided into those with primary infections (Figure 3B) and those previously treated (Figure 3C). While there was a significant association between the less virulent vacA s2 genotype and fluoroquinolone resistance when all patients were included [15% (n = 17/113) vs 30.8% (n = 16/52); χ2 = 5.5; P = 0.02; Pearson χ2 test; Figure 4A], this did not reach statistical significance in treatment naïve patients (Figure 4B) or those previously treated (Figure 4C).
Taken together, these findings indicate that the absence of cagA and the less virulent vacA genotypes (S2/M1 and S2/M2) may be predictors of primary clarithromycin resistance in treatment-naïve patients.
This study aimed to provide an update on the prevalence of antibiotic resistance and distribution of virulence factor genotypes in H. pylori strains in Ireland. In addition we investigated whether virulence factor genotypes are associated with antibiotic susceptibility. Primary clarithromycin resistance among our patients was high at 50.5% and even higher in those previously treated at 78%. Among patients infected with a resistant strain, the most common point mutation conferring clarithromycin resistance was A2147G, in keeping with other studies[14-19]. Our primary clarithromycin resistance rate is high compared to rates reported in Europe, Asia Pacific and other countries[5,19-21]. Variations in H. pylori antibiotic resistance rates among different populations are influenced by previous antibiotic use, with studies demonstrating that previous exposure to macrolides increases the risk of clarithromycin resistant H. pylori infection[5,22]. The sharp increase in primary clarithromycin resistance from 3.9% in 1997 to 9.3% in 2008[23], to the current rate of 50.5% in 2017 is a cause for concern and is reflected in the poor eradication rate (56.8% ITT) for 7 days clarithromycin-based triple therapy recently reported from our centre[3]. In an effort to address increasing antibiotic resistance and falling eradication rates, the Irish H. pylori Working Group have recently highlighted the need for more widespread antibiotic resistance surveillance and extended H. pylori treatment durations[24]. It should be noted that antibiotic resistance was determined at the genetic level in the current study compared to culture and Etests in the earlier Irish surveys.
The primary and secondary rates of fluoroquinolone resistance were 15.2% and 28.3%, respectively. The primary rate of levofloxacin resistance has only risen slightly since the last Irish survey in 2008-2009, which reported a rate of 12%[25], and is in keeping with the 14.1% rate reported in Europe[5]. The most common point mutation conferring resistance to fluoroquinolones in our patients was gyr91 D91Y. This contrasts with other studies in which gyr91 D91N and gyr87 N87K mutations were reported with highest frequency[14,16-18].
In our cohort, the overall frequency of H. pylori infections with strains containing the cagA gene was 22.4%. This has decreased since the distribution of the cagA genotype was last investigated in Ireland in 2009, with a frequency of 68% reported[12]. It is also lower than distributions reported in Cuba and Iran[26,27]. There is a well-known association between cagA-positive strains of H. pylori and peptic ulcer disease[28,29]. This relatively low frequency of cagA-positive genotype is not surprising given that the prevalence of peptic ulcer disease was also low in our cohort at 12.7% (Table 2), which is a decrease on the prevalence of peptic ulcer disease reported in the previous Irish study (17%[12]). The most prevalent vacA genotype in our cohort was S1/M2, followed by S2/M2, S1/M1 and S2/M1. This pattern is similar to the pattern reported in Ireland in 2009 as well as the studies mentioned above[12,26,27].
Interestingly, the frequency of the more virulent S1 genotype was significantly lower in those previously treated than the treatment-naïve group (58.3% vs 74.3%). Additionally, the frequency of the least virulent S2/M2 genotype was significantly higher in those previously treated previously (36.7% vs 21%). This is in accordance with a hypothesis described previously which suggests that more virulent strains elicit a stronger inflammatory response, enabling increased blood flow to the site of infection, therefore enhancing delivery of antibiotics and the potential for successful eradication[30]. Another potential explanation is that a more virulent strain of H. pylori may replicate faster and is therefore more susceptible to antibiotics, whose mechanisms of action are to inhibit bacterial replication[31].
We found an inverse relationship between the virulence of the infecting strain and the presence of clarithromycin resistance: the absence of cagA, and the less virulent vacA genotypes (S2/M1 and S2/M2), may be indicators of clarithromycin resistance, in particular in treatment-naïve patients. The association between virulence factors and antibiotic resistance in H. pylori has been evaluated in other studies, with controversial results. Absence of cagA was found to be a risk factor for metronidazole resistance[12] and other studies have found an association between clarithromycin resistance mutations and the less virulent vacA genotypes[32,33]. Another report revealed that cagE and vacA S1 correlated with clarithromycin and metronidazole resistance[34], while others found that neither cagA nor vacA was associated with resistance[29,35-37]. There may be no direct causation involving the presence of less virulent strains of H. pylori and antibiotic resistance. Rather, the presence or absence of virulence factors may cause physiological effects which create an environment in which antibiotic resistant strains of H. pylori can flourish as outlined above[31]. As less virulent strains are less immunogenic, an inadequate delivery of antibiotics may reach infected areas in the stomach and as a result, antimicrobial resistant strains may be selected for in the population of less virulent strains. It has been shown that a cagA- strain may tend to acquire drug resistance in vitro[12]. Indeed, studies have shown that virulence factor genotype may also influence treatment outcome. A number of studies have reported the presence or absence of cagA and vacA as predictors of eradication of H. pylori[36,38-40]. Wang et al[39] conducted a meta-analysis of 25 studies and found that infection with cagA positive, vacA S1 strains were associated with H. pylori eradication.
In conclusion, this study found that the cagA negative and vacA S1/M2 genotypes were the most dominant in H. pylori strains in Ireland. A surprisingly high rate of primary genotypic clarithromycin resistance was observed (50.5%), with a primary genotypic fluoroquinolone resistance rate of 15.2%. It was also found that there is a relationship between the less virulent strains of H. pylori (cagA-negative and vacA S2) and primary clarithromycin resistance. It is well known that the prevalence of antibiotic resistance is increasing worldwide while eradication rates of H. pylori are decreasing. The relationship between less virulent strains of H. pylori and presence of antibiotic resistance found herein could potentially be an avenue to explore in the effort to improve eradication rates.
Helicobacter pylori (H. pylori) causes chronic gastritis, gastric and duodenal ulcers, gastric adenocarcinoma and mucosa-associated lymphoid tissue lymphoma. Disease outcome is related to both host and bacterial factors. Eradication is recommended in all symptomatic patients and those at risk of gastric cancer. However, eradication rates for current therapies are falling due to the emergence of antibiotic resistant H. pylori strains. H. pylori is a highly heterogeneous bacterium and its virulence varies geographically. Virulence factors contribute to the pathogenicity of the bacteria and have been suggested to influence treatment outcome.
In response to the increasing problem of H. pylori antibiotic resistance, local antibiotic resistance surveillance is recommended to guide clinicians in their choice of H. pylori therapy. Knowledge of local antimicrobial resistance rates and the prevalence of virulent infections will influence strategies for optimising the management of H. pylori infection.
This study aimed to provide an update on the prevalence of antibiotic resistance in Ireland, in particular for the antibiotics clarithromycin and fluoroquinolones. The virulence of the infecting strains was assessed by investigating cagA and vacA status. In addition the relationship between virulence factor status and antibiotic resistance was evaluated.
DNA was extracted from antral and corpus biopsies obtained from H. pylori-infected patients. Genotyping for clarithromycin and fluoroquinolone-mediating mutations was performed using the Genotype HelicoDR assay. CagA and vacA genotypes were investigated using PCR and agarose gel electrophoresis.
Primary resistance to clarithromycin was high at 50.5%. Primary resistance to fluoroquinolones was 15.2%. Primary resistance to both antibiotics was 12.4%. A cagA-positive genotype was detected in 22.4% of patient samples. The dominant vacA genotype was S1/M2 at 44.8%, followed by S2/M2 at 26.7%, S1/M1 at 23.6% and S2/M1 at 4.8%. Primary clarithromycin resistance was significantly lower in cagA-positive strains than in cagA-negative strains (32% vs 56.3%). Similarly, in patients infected with more virulent H. pylori strains bearing the vacA s1 genotype, primary clarithromycin resistance was significantly lower than in those infected with less virulent strains bearing the vacA s2 genotype, (41% vs 77.8%). In summary, genotypic H. pylori clarithromycin resistance is high and cagA-negative strains are dominant in our population. Less virulent (cagA-negative and vacA S2-containing) strains of H. pylori are associated with primary clarithromycin resistance.
Given the high rate of primary clarithromycin resistance detected in our study, the use of alternatives to clarithromycin-based triple therapy should be considered for first line H. pylori treatment in our cohort. In order to validate the association between less virulent strains and clarithromycin resistance, the influence of virulence factor genotype on treatment outcome should be assessed.
The authors would like to acknowledge Mark Feighery, Ciara Treacy and Edwin Fahy for technical assistance.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Ireland
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Chiba T, Rodrigo L, Sugimoto M S- Editor: Wang XJ L- Editor: A E- Editor: Huang Y
| 1. | Malfertheiner P, Megraud F, O'Morain CA, Gisbert JP, Kuipers EJ, Axon AT, Bazzoli F, Gasbarrini A, Atherton J, Graham DY. Management of Helicobacter pylori infection-the Maastricht V/Florence Consensus Report. Gut. 2017;66:6-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2220] [Cited by in RCA: 1983] [Article Influence: 247.9] [Reference Citation Analysis (1)] |
| 2. | Leja M, Axon A, Brenner H. Epidemiology of Helicobacter pylori infection. Helicobacter. 2016;21 Suppl 1:3-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 76] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 3. | Haider RB, Brennan DE, Omorogbe J, Holleran G, Hall B, O'Morain C, Breslin N, O'Connor HJ, Smith SM, McNamara D. A randomized-controlled study to compare the efficacy of sequential therapy with standard triple therapy for Helicobacter pylori eradication in an Irish population. Eur J Gastroenterol Hepatol. 2015;27:1265-1269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 4. | Graham DY, Fischbach L. Helicobacter pylori treatment in the era of increasing antibiotic resistance. Gut. 2010;59:1143-1153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 652] [Cited by in RCA: 736] [Article Influence: 49.1] [Reference Citation Analysis (1)] |
| 5. | Megraud F, Coenen S, Versporten A, Kist M, Lopez-Brea M, Hirschl AM, Andersen LP, Goossens H, Glupczynski Y; Study Group participants. Helicobacter pylori resistance to antibiotics in Europe and its relationship to antibiotic consumption. Gut. 2013;62:34-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 653] [Cited by in RCA: 632] [Article Influence: 52.7] [Reference Citation Analysis (3)] |
| 6. | Smith SM, O'Morain C, McNamara D. Antimicrobial susceptibility testing for Helicobacter pylori in times of increasing antibiotic resistance. World J Gastroenterol. 2014;20:9912-9921. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 114] [Cited by in RCA: 112] [Article Influence: 10.2] [Reference Citation Analysis (1)] |
| 7. | Fischbach L, Evans EL. Meta-analysis: the effect of antibiotic resistance status on the efficacy of triple and quadruple first-line therapies for Helicobacter pylori. Aliment Pharmacol Ther. 2007;26:343-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 246] [Cited by in RCA: 282] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 8. | Broutet N, Tchamgoué S, Pereira E, Lamouliatte H, Salamon R, Mégraud F. Risk factors for failure of Helicobacter pylori therapy--results of an individual data analysis of 2751 patients. Aliment Pharmacol Ther. 2003;17:99-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 163] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 9. | Uotani T, Miftahussurur M, Yamaoka Y. Effect of bacterial and host factors on Helicobacter pylori eradication therapy. Expert Opin Ther Targets. 2015;19:1637-1650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 10. | Atherton JC, Cao P, Peek RM Jr, Tummuru MK, Blaser MJ, Cover TL. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771-17777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1078] [Cited by in RCA: 1108] [Article Influence: 36.9] [Reference Citation Analysis (0)] |
| 11. | Rhead JL, Letley DP, Mohammadi M, Hussein N, Mohagheghi MA, Eshagh Hosseini M, Atherton JC. A new Helicobacter pylori vacuolating cytotoxin determinant, the intermediate region, is associated with gastric cancer. Gastroenterology. 2007;133:926-936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 283] [Cited by in RCA: 312] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 12. | Taneike I, Nami A, O'Connor A, Fitzgerald N, Murphy P, Qasim A, O'Connor H, O'Morain C. Analysis of drug resistance and virulence-factor genotype of Irish Helicobacter pylori strains: is there any relationship between resistance to metronidazole and cagA status? Aliment Pharmacol Ther. 2009;30:784-790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 21] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 13. | Brennan DE, Omorogbe J, Hussey M, Tighe D, Holleran G, O'Morain C, Smith SM, McNamara D. Molecular detection of Helicobacter pylori antibiotic resistance in stool vs biopsy samples. World J Gastroenterol. 2016;22:9214-9221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 38] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (2)] |
| 14. | Pastukh N, Binyamin D, On A, Paritsky M, Peretz A. GenoType® HelicoDR test in comparison with histology and culture for Helicobacter pylori detection and identification of resistance mutations to clarithromycin and fluoroquinolones. Helicobacter. 2017;22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 15. | Cambau E, Allerheiligen V, Coulon C, Corbel C, Lascols C, Deforges L, Soussy CJ, Delchier JC, Megraud F. Evaluation of a new test, genotype HelicoDR, for molecular detection of antibiotic resistance in Helicobacter pylori. J Clin Microbiol. 2009;47:3600-3607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 128] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 16. | Lee JW, Kim N, Nam RH, Park JH, Choi YJ, Kim JM, Kim JS, Jung HC. GenoType HelicoDR test in the determination of antimicrobial resistance of Helicobacter pylori in Korea. Scand J Gastroenterol. 2014;49:1058-1067. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 17. | Miendje Deyi VY, Burette A, Bentatou Z, Maaroufi Y, Bontems P, Lepage P, Reynders M. Practical use of GenoType® HelicoDR, a molecular test for Helicobacter pylori detection and susceptibility testing. Diagn Microbiol Infect Dis. 2011;70:557-560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 18. | Sanches BS, Martins GM, Lima K, Cota B, Moretzsohn LD, Ribeiro LT, Breyer HP, Maguilnik I, Maia AB, Rezende-Filho J. Detection of Helicobacter pylori resistance to clarithromycin and fluoroquinolones in Brazil: A national survey. World J Gastroenterol. 2016;22:7587-7594. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 31] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 19. | Tanih NF, Ndip RN. Molecular Detection of Antibiotic Resistance in South African Isolates of Helicobacter pylori. Gastroenterol Res Pract. 2013;2013:259457. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 20. | Macías-García F, Llovo-Taboada J, Díaz-López M, Bastón-Rey I, Domínguez-Muñoz JE. High primary antibiotic resistance of Helicobacter Pylori strains isolated from dyspeptic patients: A prevalence cross-sectional study in Spain. Helicobacter. 2017;22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 21. | Kuo YT, Liou JM, El-Omar EM, Wu JY, Leow AHR, Goh KL, Das R, Lu H, Lin JT, Tu YK. Primary antibiotic resistance in Helicobacter pylori in the Asia-Pacific region: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2017;2:707-715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 233] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 22. | McNulty CA, Lasseter G, Shaw I, Nichols T, D'Arcy S, Lawson AJ, Glocker E. Is Helicobacter pylori antibiotic resistance surveillance needed and how can it be delivered? Aliment Pharmacol Ther. 2012;35:1221-1230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 55] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 23. | O'Connor A, Taneike I, Nami A, Fitzgerald N, Murphy P, Ryan B, O'Connor H, Qasim A, Breslin N, O'Moráin C. Helicobacter pylori resistance to metronidazole and clarithromycin in Ireland. Eur J Gastroenterol Hepatol. 2010;22:1123-1127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 24. | Smith S, Boyle B, Brennan D, Buckley M, Crotty P, Doyle M, Farrell R, Hussey M, Kevans D, Malfertheiner P. The Irish Helicobacter pylori Working Group consensus for the diagnosis and treatment of H. pylori infection in adult patients in Ireland. Eur J Gastroenterol Hepatol. 2017;29:552-559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 25. | O'Connor A, Taneike I, Nami A, Fitzgerald N, Ryan B, Breslin N, O'Connor H, McNamara D, Murphy P, O'Morain C. Helicobacter pylori resistance rates for levofloxacin, tetracycline and rifabutin among Irish isolates at a reference centre. Ir J Med Sci. 2013;182:693-695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 26. | Feliciano O, Gutierrez O, Valdés L, Fragoso T, Calderin AM, Valdes AE, Llanes R. Prevalence of Helicobacter pylori vacA, cagA, and iceA Genotypes in Cuban Patients with Upper Gastrointestinal Diseases. Biomed Res Int. 2015;2015:753710. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 27. | Pajavand H, Alvandi A, Mohajeri P, Bakhtyari S, Bashiri H, Kalali B, Gerhard M, Najafi F, Abiri R. High Frequency of vacA s1m2 Genotypes Among Helicobacter pylori Isolates From Patients With Gastroduodenal Disorders in Kermanshah, Iran. Jundishapur J Microbiol. 2015;8:e25425. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 28. | van Doorn LJ, Schneeberger PM, Nouhan N, Plaisier AP, Quint WG, de Boer WA. Importance of Helicobacter pylori cagA and vacA status for the efficacy of antibiotic treatment. Gut. 2000;46:321-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 107] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 29. | Godoy AP, Ribeiro ML, Benvengo YH, Vitiello L, Miranda Mde C, Mendonça S, Pedrazzoli J Jr. Analysis of antimicrobial susceptibility and virulence factors in Helicobacter pylori clinical isolates. BMC Gastroenterol. 2003;3:20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 75] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 30. | Khan A, Farooqui A, Manzoor H, Akhtar SS, Quraishy MS, Kazmi SU. Antibiotic resistance and cagA gene correlation: a looming crisis of Helicobacter pylori. World J Gastroenterol. 2012;18:2245-2252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 25] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 31. | Sugimoto M, Yamaoka Y. Virulence factor genotypes of Helicobacter pylori affect cure rates of eradication therapy. Arch Immunol Ther Exp (Warsz). 2009;57:45-56. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 58] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 32. | Boyanova L, Markovska R, Yordanov D, Gergova G, Mitov I. Clarithromycin Resistance Mutations in Helicobacter pylori in Association with Virulence Factors and Antibiotic Susceptibility of the Strains. Microb Drug Resist. 2016;22:227-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 33. | Elviss NC, Owen RJ, Xerry J, Walker AM, Davies K. Helicobacter pylori antibiotic resistance patterns and genotypes in adult dyspeptic patients from a regional population in North Wales. J Antimicrob Chemother. 2004;54:435-440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 34. | Karabiber H, Selimoglu MA, Otlu B, Yildirim O, Ozer A. Virulence factors and antibiotic resistance in children with Helicobacter pylori gastritis. J Pediatr Gastroenterol Nutr. 2014;58:608-612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 34] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 35. | López-Brea M, Martínez MJ, Domingo D, Sánchez I, Alarcón T. Metronidazole resistance and virulence factors in Helicobacter pylori as markers for treatment failure in a paediatric population. FEMS Immunol Med Microbiol. 1999;24:183-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 36. | Liou JM, Chang CY, Chen MJ, Chen CC, Fang YJ, Lee JY, Wu JY, Luo JC, Liou TC, Chang WH. The Primary Resistance of Helicobacter pylori in Taiwan after the National Policy to Restrict Antibiotic Consumption and Its Relation to Virulence Factors-A Nationwide Study. PLoS One. 2015;10:e0124199. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 70] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 37. | van Doorn LJ, Glupczynski Y, Kusters JG, Mégraud F, Midolo P, Maggi-Solcà N, Queiroz DM, Nouhan N, Stet E, Quint WG. Accurate prediction of macrolide resistance in Helicobacter pylori by a PCR line probe assay for detection of mutations in the 23S rRNA gene: multicenter validation study. Antimicrob Agents Chemother. 2001;45:1500-1504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 120] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 38. | Suzuki T, Matsuo K, Sawaki A, Ito H, Hirose K, Wakai K, Sato S, Nakamura T, Yamao K, Ueda R. Systematic review and meta-analysis: importance of CagA status for successful eradication of Helicobacter pylori infection. Aliment Pharmacol Ther. 2006;24:273-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 45] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 39. | Wang D, Li Q, Gong Y, Yuan Y. The association between vacA or cagA status and eradication outcome of Helicobacter pylori infection: A meta-analysis. PLoS One. 2017;12:e0177455. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 40. | Figura N, Moretti E, Vaglio L, Langone F, Vernillo R, Vindigni C, Giordano N. Factors modulating the outcome of treatment for the eradication of Helicobacter pylori infection. New Microbiol. 2012;35:335-340. [PubMed] |