Published online Sep 14, 2018. doi: 10.3748/wjg.v24.i34.3861
Peer-review started: May 25, 2018
First decision: June 21, 2018
Revised: July 5, 2018
Accepted: July 16, 2018
Article in press: July 16, 2018
Published online: September 14, 2018
Processing time: 112 Days and 2.1 Hours
To investigate the role of Delta-like ligand 4 (DLL4) on tumour growth in hepatitis B virus (HBV)-associated hepatocellular carcinoma (HCC) in vivo.
We suppressed DLL4 expression in an HBV expressing HCC cell line, HepG2.2.15 and analysed the growth ability of cells as subcutaneous tumours in nude mice. The expression of tumour angiogenesis regulators, VEGF-A and VEGF-R2 in tumour xenografts were examined by western blotting. The tumour proliferation and neovasculature were examined by immunohistochemistry. The viral replication and viral protein expression were measured by quantitative PCR and western blotting, respectively.
Eighteen days after implantation, tumour volume in mice implanted with shDLL4 HepG2.2.15 was significantly smaller than in mice implanted with control HepG2.2.15 (P < 0.0001). The levels of angiogenesis regulators, VEGF-A and VEGF-R2 were significantly decreased in implanted tumours with suppressed DLL4 compared with the control group (P < 0.001 and P < 0.05, respectively). Furthermore, the suppression of DLL4 expression in tumour cells reduced cell proliferation and the formation of new blood vessels in tumours. Unexpectedly, increased viral replication was observed after suppression of DLL4 in the tumours.
This study demonstrates that DLL4 is important in regulating the tumour growth of HBV-associated HCC as well as the neovascularization and suppression of HBV replication.
Core tip: We demonstrated that Delta-like ligand 4 (DLL4) is important for tumour growth of hepatitis B virus (HBV)-associated hepatocellular carcinoma (HCC) in a xenograft model. We found that the level of angiogenesis regulators, VEGF-A and VEGF-R2 were significantly decreased in HCC xenograft tumours with suppressed DLL4 compared with the control group. Consistent with these findings, the suppression of DLL4 expression in the tumour cells reduced cell proliferation and the formation of new blood vessels in the tumour. Furthermore, this is the first report that DLL4 in an HBV expressing HCC cell line plays a key role in regulating tumour growth, angiogenesis, and viral replication in a mouse model of xenograft transplantation.
- Citation: Kunanopparat A, Issara-Amphorn J, Leelahavanichkul A, Sanpavat A, Patumraj S, Tangkijvanich P, Palaga T, Hirankarn N. Delta-like ligand 4 in hepatocellular carcinoma intrinsically promotes tumour growth and suppresses hepatitis B virus replication. World J Gastroenterol 2018; 24(34): 3861-3870
- URL: https://www.wjgnet.com/1007-9327/full/v24/i34/3861.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i34.3861
Hepatocellular carcinoma (HCC) is the third most common cause of cancer-associated mortality. Approximately 80% of HCC is associated with chronic hepatitis viral infections[1]. Hepatitis B virus (HBV) infection is the most prevalent cause of HCC in developing countries. Although an HBV vaccine has successfully prevented HBV infection, there are still a large number of chronic hepatitis B patients who are at a high risk (maximum 100-fold increase over healthy individuals) of developing liver cancer[2,3]. The molecular mechanisms of HBV-associated HCC are poorly understood[4]. To date, sorafenib is the recommended drug for the treatment of HCC patients. However, the therapeutic outcome is still limited because liver cancer is often detected at advanced stages[5]. Therefore, a better understanding of the molecular mechanisms of tumour initiation and progression is needed for the further development of HCC therapy.
Notch signalling is an evolutionarily conserved pathway that regulates cell fate decision, embryonic development, tissue homeostasis, differentiation, proliferation, and apoptosis[6,7]. In mammals, the Notch pathway comprises of four Notch receptors (Notch1, 2, 3, 4) and five Notch ligands (Jagged1, 2, and DLL1, 3, 4). Activation of Notch signalling requires contact between a Notch ligand from the signal sending cells and a receptor on signal receiving cells to activate proteolytic cleavage and the subsequent translocation of the Notch intracellular domain to the nucleus where it translates target genes[8]. Dysregulation of Notch signalling has been reported in many types of cancer as either a tumour suppressor or tumour promoter depending on the type of cancer[9-11]. In HCC, the role of Notch signalling is still controversial. Many studies reported that Notch receptors were highly expressed in HCC compared with the adjacent human tumour tissue and that tumour growth was suppressed after the inhibition of Notch either by a gamma secretase inhibitor or by suppression of Notch target genes[12-16]. Several studies have also suggested that Notch is a tumour suppressor in HCC[15,17-19]. However, more evidence supports the pro-tumourigenic role of Notch in HCC carcinogenesis and progression, especially in HBV-associated HCC[20,21]. We previously reported that HBV regulatory protein HBx promoted HBV-associated HCC proliferation through Delta-like ligand 4 (DLL4) via the NF-κB pathway in HepG2, an HBV expressing HCC cell line[22].
Strong evidence indicates that DLL4 regulates angiogenesis and controls the balance of endothelial tip and stalk cell differentiation induced by VEGF[23]. DLL4 is highly expressed in tumour endothelial cells for tumour angiogenesis, which is the primary signal for tumour progression[24]. The inhibition of DLL4 in tumour endothelial cells suppressed tumour growth by inducing non-productive angiogenesis[25]. Currently, a DLL4 neutralizing antibody has been developed and is being tested in a clinical trial for anticancer therapy in various cancers[26,27]. However, the effect of DLL4 inhibition in HCC has not been explored. In this study, we investigated the role of DLL4 on tumour growth in HCC associated with HBV in a xenograft model and detailed the molecular mechanism of HCC.
The HBV-expressing HCC cell line (HepG2.2.15) and the HepG2 cell line were obtained from Professor Antonio Bertoletti [Singapore Institute for Clinical Sciences at Agency for Science, Technology and Research (A*Star)]. Cells were cultured in high glucose DMEM medium (Gibco, Carlsbad, CA, United States) supplemented with 10% foetal bovine serum (Gibco), 150 μg/mL of G418 (Gibco), and 1% of penicillin-streptomycin (Invitrogen, Carlsbad, CA, United States). Cultures were maintained at 37 °C in a 5% CO2 humidified incubator
HepG2.2.15 cells line was transfected with a set of DLL4 shRNA (Origene Technologies, MD, United States) targeting four shDLL4 cassettes in the pGFP-V-RS Vector (TG304977). The transient transfection used Lipofectamine 2000 (Invitrogen) at 2.5 μL for 1 μg of shRNA vector into 1 × 105 cells per well in a 12-well plate. After 48 h, DLL4 mRNA expression was determined in transfected cells. The highest efficacy of shRNA (5′-ACCAGAAGAAGGAGCTGGAAGTGGACTGT-3′) vector was used to generate stably transfected cells. For stably transfected cell lines, transiently transfected cells were plated into 96-well plates by limiting dilution and selected by the addition of 0.3 μg/mL puromycin to the culture medium for 4-5 wk. Puromycin-resistant clones with suppressed DLL4 were expanded, and DLL4 expression was analysed by western blot analysis and compared with the control. The clones with the highest degree of DLL4 suppression were used for tumour xenografts.
The study was approved by the Institutional Review Board of the Faculty of Medicine, Chulalongkorn University. All protocols were carried out in accordance with relevant guidelines and regulations. Male BALB/cMlac-nu mice aged four weeks were purchased from the National Laboratory Animal Center (Mahidol University, Thailand) and were acclimatized for two weeks before experimentation. Mice were maintained under 12 h light-dark cycle with 50% humidity and with free access to food and water. The shDLL4 HepG2.2.15 and control HepG2.2.15 cells were trypsinized at a concentration of 1 × 107 cells/mL. One millilitre of the cells was centrifuged and resuspended in 100 μL of Matrigel (Corning, NY, United States). The cell suspension was subcutaneously injected into the back left and right flanks of nude mice (n = 4-6). The tumour volume (cm3) was measured every three days until 18 d and 30 d using Vernier calipers and calculated using the formula: (length × width2)/6. The mice were weighed every three days and monitored for activity and mortality. All animals were euthanized by barbiturate overdose for tumour collection.
Total cell lysates were prepared in RIPA buffer (Cell Signaling Technology, MA, United States) containing protease inhibitor cocktail (Pierce, Thermo Fisher Scientific, MA, United States). After sonication, 20 μg of cell lysates were blotted and probed with primary antibodies to anti-DLL4, anti-cleaved Notch 1, anti-VEGFR2, anti-β actin (Cell Signaling Technology; 1:1000), anti-VEGF, anti-PreS1 HBV antigen, and anti-GAPDH (Santa Cruz Biotechnology, Dallas, TX, United States; 1:1000). Peroxidase-conjugated goat anti-rabbit immunoglobulin (Santa Cruz Biotechnology) and goat anti-mouse immunoglobulin (Cell Signaling Technology) were used as secondary antibodies. Immunoblot detection was performed using Super Signal West Femto Maximum Sensitivity Substrate (Pierce, Thermo Fisher Scientific). The protein intensity was estimated by the densitometry of scanned immunoblot bands using Image Studio Lite version 5.2 software (LI-COR Biosciences).
After the end of the experiment, the tumours were collected, fixed in 10% formalin solution, and embedded in a paraffin block. The tissue sections were cut with a microtome to obtain 4 μm thick paraffin sections, then deparaffinized and rehydrated in a series of xylenes and alcohols followed by retrieval of the antigenic epitopes. Antigen retrieval was performed in citrate buffer (pH 6, 100 °C for 20 min). The tissue sections were treated with 3% H2O2 for 15 min and blocked with normal serum for 30 min, then incubated with primary antibody in a humidity chamber at 4 °C overnight. The primary antibodies included anti-CD31 (Santa Cruz Biotechnologies; at a dilution of 1:500), and anti-Ki-67 (Ventana Medical Systems, Inc.; AZ, United States) (ready to use). ZytoChem Plus (HRP) Polymer anti-Rabbit (Zytomed Systems, Berlin, Germany) (ready to use) and rabbit anti-goat immunoglobulin-HRP (Dako; CA, United States), were used for the detection of primary antibodies. The immunoreaction was visualized with ultraView Universal DAB Detection Kit (Ventana Medical Systems, Inc.). The nuclei were counterstained with Mayer’s haematoxylin. Immunoreactions were measured in five microscopic fields per sample with 20 × objective magnification (Nikon Eclipse50i, Japan). The percentage of Ki67 was analysed by the ImmunoRatio web application[28].
Tumour vasculature imaging was performed as previously described[29]. Briefly, mice were anesthetised with an intraperitoneal injection of sodium pentobarbital (50 mg/kg BW). A catheter was inserted into the jugular vein for the application of fluorescence tracers. Then, the dorsal skin-fold chamber was removed, and the skin area around the chamber was fixed with modelling wax on a plate. To visualise the vascular lumen, a bolus of 0.1 mL of 5% fluorescein isothiocyanate-labelled dextran (FITC-dextran) was injected into the jugular vein. The tumour vasculature was visualised under a confocal microscope.
Total RNA was extracted from cell culture or xenograft tumour tissues using the RNeasy Mini kit (Qiagen, Hilden, Germany). One microgram of RNA was converted to cDNA using High Capacity cDNA Reverse Transcription Kits (Applied Biosystems, Carlsbad, CA, United States). Quantitative PCR amplification was performed with SYBR green (Applied Biosystems) on the Applied Biosystems 7500 Real-Time PCR System for 40 cycles. Derivation of the 2-ddCT method was applied for the relative quantification of mRNA expression. Beta-actin was used as an endogenous control. The primers used in this study are shown in Table 1.
| Genes | Primer sequence |
| β-actin | F-5′ACCAACTGGGACGACATGGAGAA-3′ |
| R-5′GTGGTGGTGAAGCTGTAGCC-3′ | |
| IFN-α | F-5′GCTTTACTGATGGTCCTGGTGGTG-3′ |
| R-5′GAGATTCTGCTCATTTGTGCCAG-3′ | |
| IFN-β | F-5′GAATGGGAGGCTTGAATACTGCCT-3′ |
| R-5′TAGCAAAGATGTTCTGGAGCATCTC-3′ | |
| TNF-α | F-5′CTTCTCCTTCCTGATCGTGG-3′ |
| R-5′GCTGGTTATCTCTCAGCTCCA-3′ | |
| HBx | F-5′CACCTCTCTTTACGCGGACT-3′ |
| R-5′GGTCGTTGACATTGCAGAGA-3′ | |
| HBV PreS1 | F-5′GGGTCACCATATTCTTGGGAAC-3′ |
| R-5′CCTGAGCCTGAGGGCTCCAC-3′ |
Genomic DNA was extracted from cell culture or xenograft tumour tissues using the QIAamp DNA Mini Kit (Qiagen). Fifty nanograms of DNA of all samples were amplified to determine preS1 HBV gene expression relative to a standard copy number of HBV. The quantitative PCR amplification was performed with SYBR green (Applied Biosystems) on the Applied Biosystems 7500 Real-Time PCR System.
One-way ANOVA and t-test were applied using Prism 5 software (GraphPad Software Inc., San Diego, CA, United States). The results are shown as the mean ± SD, and differences of P < 0.05 were accepted as the level of significance.
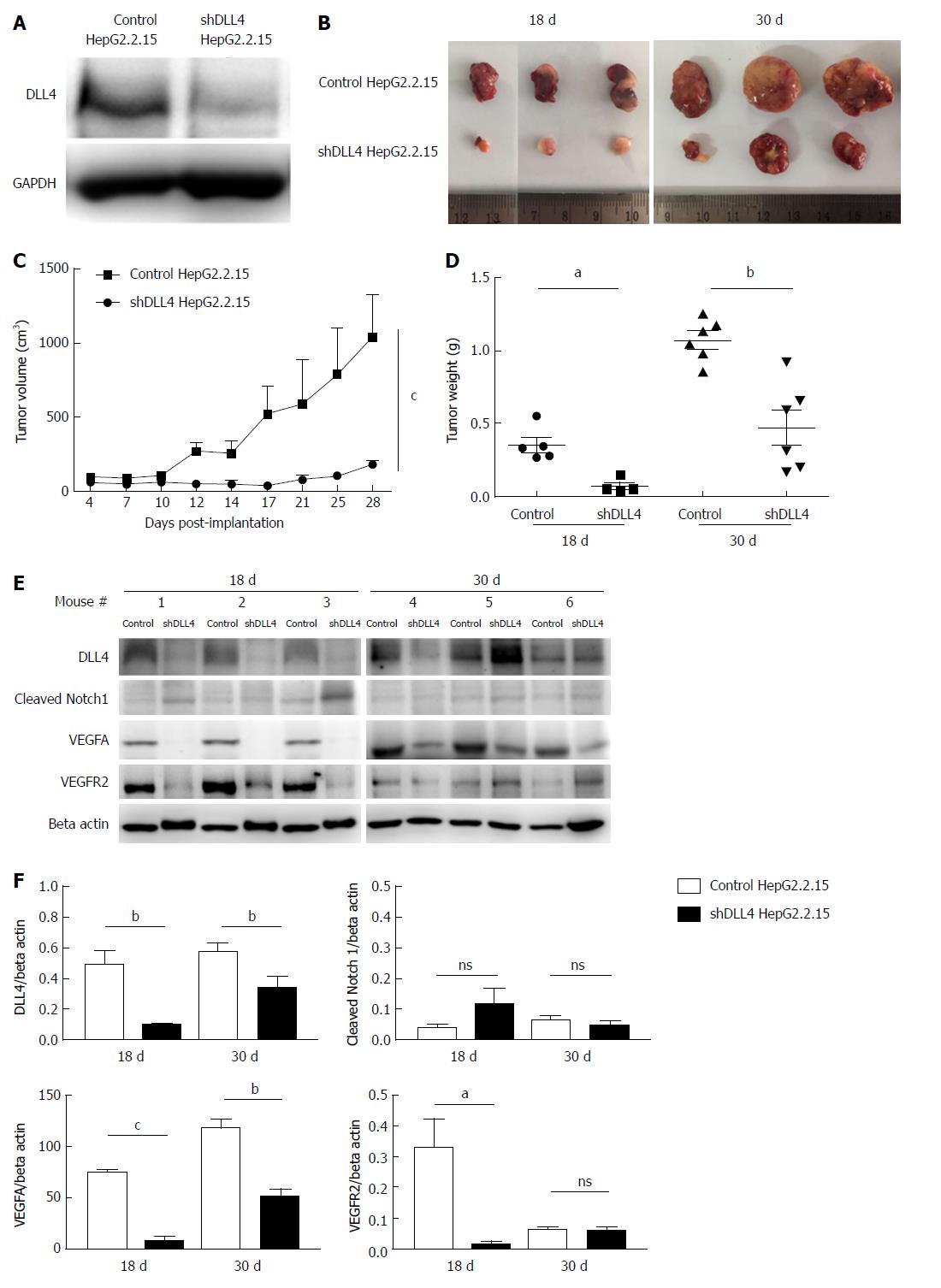
To investigate the role of DLL4 in HCC, we transfected a DLL4 suppression construct, shDLL4 to knockdown DLL4 expression or control pGFP-V-RS plasmids into the HepG2.2.15 cell line. Clones of transfected cells with stable knockdown of DLL4 were selected with puromycin by limiting dilution assay. Various clones were selected and DLL4 expression was determined by western blot and compared with the control (Figure 1A). Clones with the strongest DLL4 suppression were chosen for xenograft transplantation. At 18 d and 30 d after shDLL4 HepG2.2.15 transplantation, mice showed significantly reduced tumour growth (Figure 1B) for both tumour volume (n = 4-6, P < 0.0001) and tumour weight (n = 4-6, P < 0.05 at 18 d and P < 0.01 at 30 d) compared with mice transplanted with control HepG2.2.15 cells (Figure 1C and D).
We monitored the expression of DLL4 and cleaved Notch1 expression in tumours taken from xenograft mice. Interestingly, the expression of DLL4 recovered in one mouse (#5 Figure 1E) and cleaved Notch1 was not significantly different in the implanted HepG2.2.15 tumours with shDLL4 compared with controls (Figure 1E and F).
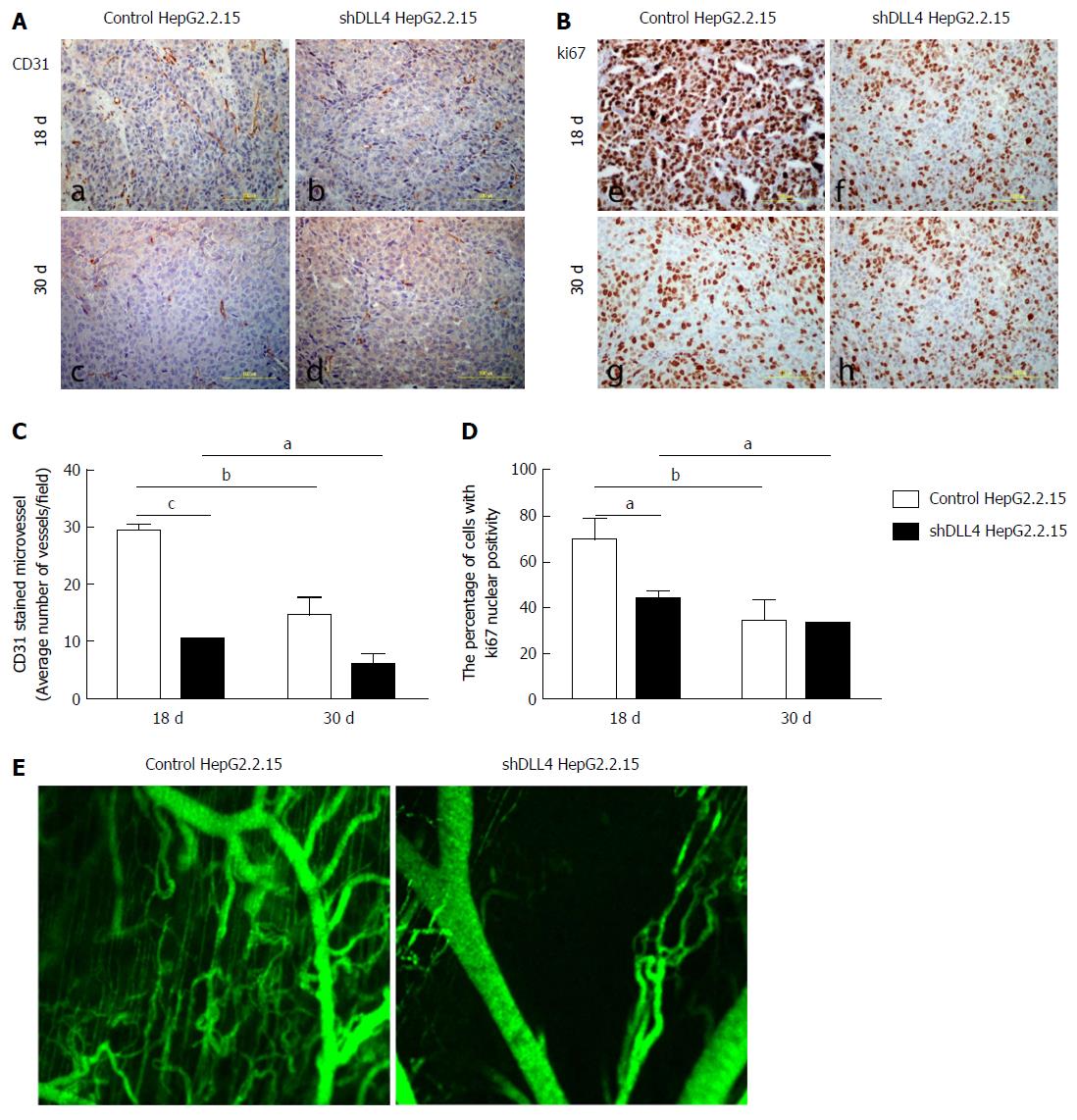
DLL4 is mainly expressed in vascular endothelium and is related to VEGF expression, which promotes tumour angiogenesis[30-32]. To examine whether DLL4-expressing HCC is associated with VEGF expression, we analysed VEGF expression from the tumour xenograft. Interestingly, at 18 d after transplantation, the expression of VEGF was significantly decreased in implanted HepG2.2.15 tumours with shDLL4 compared with the control tumour (P < 0.001) (Figure 1E and F). VEGFR2 and CD31, tumour vasculature markers, were also significantly reduced in shDLL4 HepG2.2.15 compared with control HepG2.2.15 (P < 0.05 and P < 0.001, respectively) (Figures 1E and F, 2A and B, respectively). CD31 expression, which indicates neovascularization, was also decreased consistent with the Ki67 expression.
Next, we analysed cell proliferation by Ki67 expression in tumour xenografts using immunohistochemistry. The suppression of DLL4 reduced the percentage of cells with Ki67 nuclear positivity as shown at day 18 post transplantation in Figure 2B and 2D (P < 0.05). The tumour vasculature was also decreased in shDLL4 HepG2.2.15 compared with control HepG2.2.15 (Figure 2E).
Interestingly, at 30 d after transplantation, the expression of VEGFR2, CD31, and Ki67 were not significantly different between shDLL4 HepG2.2.15 and control tumours. These data suggested that DLL4 may have an important role at the initiation stage of tumour proliferation.
We have previously shown that in vitro HBV activated Notch signalling by increasing DLL4 had no effect on HBV viral replication[22]. To confirm our observation in vivo, we monitored viral production in the tumour xenograft. Unexpectedly, we found that HBV viral DNA and HBx mRNA expression were significantly increased in shDLL4 HepG2.2.15 compared with control HepG2.2.15 (P < 0.05 and P < 0.01, respectively). HBV preS1 protein was also increased in shDLL4 HepG2.2.15 at 18 d and 30 d after implantation (Figure 3A-C). We therefore measured the amount of type I interferon and found no difference in the level of IFN-α, IFN-β, or TNF-α (Figure 3D-F). Taken together, we found that a decrease in DLL4 expression in the HBV expressing HCC cell line in vivo reduced tumour cell proliferation and increased viral replication.
In this study, we followed up on our previous observation that HBx induced DLL4 in the HCC cell line and regulated cell survival at least via the activation of Notch1[22]. The effects of DLL4 suppression in an HCC cell line was observed at two main levels: (1) was the effect on tumour growth and (2) was the effect on viral replication. As expected, HepG2.2.15 with reduced DLL4 expression grew poorly in immunocompromised nude mice, compared with the siRNA transfected control. The suppression effect of DLL4 remained intact 18 d after implantation but it was diminished in some, but not all mice, after 30 d. Interestingly, the level of cleaved Notch1 was not reduced in all tumours even when DLL4 was successfully suppressed. Similarly, the level of Hes1 and Hey1, two well-characterized Notch target genes, were also not affected by DLL4 suppression in tumours (data not shown). It is possible that other Notch receptors besides Notch1 and Notch ligands, such as JAG1 or DLL3, are activated in the tumours. Indeed, HBx expression induced JAG1 expression in the HCC cell line[33]. In one study, suppressing both DLL4 and Jagged1 increased the inhibitory effect on the proliferation and invasiveness of human gastric carcinoma[34]. Although we cannot rule out the possibility that Notch receptors/ligands are activated in the tumour, our observation clearly showed that DLL4 is important for the tumour growth of the implanted HepG2.2.15 cell line. However, there are studies that suggested that a high level of DLL4 is associated with inhibition of tumour growth and metastasis in HCC[35,36]. Notch receptors have been suggested to play a role in both oncogenes and tumour-suppressor genes in different cell types[37,38]. We hypothesized that DLL4 may act as an oncogene in the initiation stage of tumour development, and then act as a tumour suppressor in the late stage depending on the DLL4 isoform or other tumour microenvironments. However, the dual function of DLL4 as a tumour-suppressor and oncogene needs to be further clarified.
The most striking effect of DLL4 knockdown on HepG2.2.15 in vivo was the reduction of angiogenesis factors, VEGFA and VEGFR2 (Figure 1E). In our study, we detected VEGFA of tumour (human) origin and VEGFR2 of host (mouse) origin. When the tumour vasculature was visualized, a reduced vasculature was observed in DLL4 knockdown tumours, consistent with the reduced expression of angiogenesis factors. CD31, an endothelial cell marker, was also reduced. DLL4 was reported to be involved in tumour angiogenesis[39]. Suppressing DLL4 in tumours resulted in non-productive angiogenesis and the suppression of tumour growth[40]. Our observation is in line with these reports and confirmed the importance of DLL4 in angiogenesis during tumour growth. DLL4 on tumour cells interacts with Notch receptors on host stromal/endothelial cells and helps tumour angiogenesis, thus improving tumour vascular function[41]. This event leads to increased tumour growth in some, but not all, types of cancer cells such as glioblastoma and prostate cancer. Our result is consistent with this observation, and we have added HCC as another tumour cell that relies on DLL4 for vasculature formation. One intriguing observation from our study is that tumour (human) derived VEGF induces vasculature in the host (mouse).
Recent comprehensive and integrative genomic characterisation of hepatocellular carcinoma was performed on various HCC of different aetiologies. Although Notch receptors/ligands and the associated signalling molecules did not stand out as the prime mutated genes, the core Notch signalling was one of the pathways in HCC with enriched frequencies of functionally impactful mutations (ranked as No. 71)[42]. In HCV-related HCC, the Notch tumour signature genes (activation and deregulation) were found in 31.8% of patients (n = 91), suggesting a partial role of Notch signalling in promoting HCC in HCV infection[16]. This analysis highlighted the fact that Notch signalling may be involved in certain, but not all, subsets of HCC. In addition, it is not known whether HCC arising from other non-viral infections causes, but DLL4 has been linked to liver fibrosis and non-alcoholic steatohepatitis pathogenesis[43].
Tumour cell proliferation was significantly reduced when DLL4 was suppressed. This effect was determined by the reduction in Ki67, which was robust at an early stage (18 d post transplantation). There are two likely scenarios for the effect of DLL4 on tumour cell growth. One possibility is that reducing DLL4 expression decreased Notch signalling and as a result, cell growth in vivo was compromised. Many studies have reported the cell proliferation promoting effect of Notch in HCC[44-46]. Suppression of DLL4 in HepG2.2.15 reduced cell viability and interfered with cell cycle progression in vitro[22]. Another possibility is that the defect in angiogenesis within tumours played a key role in reducing cell proliferation. This effect together with reduced angiogenesis may contribute to severe growth retardation in vivo.
Unexpectedly, increased viral replication was observed in HepG2.2.15 upon DLL4 suppression in vivo. We previously reported that suppressing DLL4 in HepG2.2.15 in vitro did not alter viral replication[22]. This discrepancy highlights the more complex multi-cellular interactions in vivo. It is unclear how DLL4 suppression promoted HBV viral replication. However, there may be two possibilities: the extrinsic and/or the intrinsic effect. If suppressing DLL4 created an environment that was friendly to viral replication, such as by inducing less anti-viral cytokines IFNα/β or promoting skewed helper T cell polarization, then this is considered an extrinsic effect. In contrast, what we observed is that there was no significant difference in IFNα/β level between control and tumour with suppressed DLL4 and the mice lacked adaptive immune response. Thus, we concluded that the viral replication promoting effect must be intrinsic. Namely the intracellular environment with reduced DLL4 levels allowed the virus to replicate better. Currently, there is no evidence that this effect is dependent upon reduced Notch signalling.
There are several reports on the effect of viral infection and Notch ligand expression. In Dengue virus infection, DLL1 and DLL4 were upregulated in antigen presenting cells via the IFN-β signalling pathway, which in turn influenced helper T cell responses[47]. Respiratory syncytial virus also induced DLL4 expression in dendritic cells to direct helper T cell polarization[48]. In another report, Kaposi sarcoma herpesvirus induced the expression of DLL4 and JAG1 to alter cell cycle regulating genes in neighbouring cells[49]. However, there has been no report on the impact of DLL4 expression on HBV replication. Our observation that suppressing DLL4 decreased angiogenesis indicates it might induce hypoxic conditions within the tumour. Indeed, various studies have linked the expression of hypoxia-inducible factor and viral replication during carcinogenesis[50,51]. If hypoxia and cellular stress due to defective angiogenesis caused by DLL4 suppression is the cause for enhanced viral replication, then it is speculated that suppressing DLL4 expression may promote viral replication in other cell types as well. Whether increased HBV replication is the cause for reduced tumour growth is not determined.
Taken together, we report the novel findings that DLL4 in an HBV expressing HCC cell line regulated tumour growth, angiogenesis, and viral replication in a mouse model of xenograft transplantation. Therefore, DLL4 may be a good candidate for HCC therapy.
Hepatitis B virus (HBV)-associated hepatocellular carcinoma (HCC) has been studied for many decades. However, the molecular mechanism is still unclear. Notch signaling in HCC pathogenesis is controversial, but we found that HBx promoted HBV-associated HCC proliferation through Delta-like ligand 4 (DLL4) (Notch ligand) in an in vitro study. However, the effect of DLL4 inhibition in HCC has not been explored.
DLL4 has a potential function for angiogenesis that supports tumour growth. The understanding of DLL4 mechanism might lead to identifying a new target for HCC therapy.
We investigated the role of DLL4 on tumour growth in HCC associated with HBV in a xenograft model and detailed the molecular mechanism of HCC.
We inhibited the DLL4 expression in HBV-associated HCC, and then subcutaneously implanted in nude mice. We analysed the ability for tumour growth, angiogenesis regulators (VEGF-A, VEGF-R2) expression, neovasculature, and HBV expression in tumour xenografts.
The tumour volume, VEGF-A, and VEGF-R2 were significantly decreased in mice implanted with suppressed DLL4 HCC compared with the control group. The suppression of DLL4 expression in tumour cells reduced cell proliferation and the formation of new blood vessels in tumours. Unexpectedly, viral replication increased in DLL4 suppressed tumours.
This study demonstrates that DLL4 is important in regulating the tumour growth and neovascularization in HBV-associated HCC, as well as suppressing HBV replication in vivo.
This study showed that DLL4 is essential for the tumour growth of the implanted HBV-associated HCC cell line, especially in the initiation stage of tumour growth. However, the role of DLL4 as a tumour oncogene and tumour suppressor gene in HCC needs to further clarification.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Thailand
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C, C, C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Abd-Elsalam A, Abid S, Ahn KS, Li W S- Editor: Gong ZM L- Editor: Filipodia E- Editor: Yin SY
| 1. | Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893-2917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11128] [Cited by in RCA: 11835] [Article Influence: 845.4] [Reference Citation Analysis (4)] |
| 2. | El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264-1273.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2183] [Cited by in RCA: 2507] [Article Influence: 192.8] [Reference Citation Analysis (2)] |
| 3. | Yang HI, Lu SN, Liaw YF, You SL, Sun CA, Wang LY, Hsiao CK, Chen PJ, Chen DS, Chen CJ; Taiwan Community-Based Cancer Screening Project Group. Hepatitis B e antigen and the risk of hepatocellular carcinoma. N Engl J Med. 2002;347:168-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 924] [Cited by in RCA: 915] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 4. | Benhenda S, Cougot D, Buendia MA, Neuveut C. Hepatitis B virus X protein molecular functions and its role in virus life cycle and pathogenesis. Adv Cancer Res. 2009;103:75-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 132] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 5. | Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9016] [Cited by in RCA: 10266] [Article Influence: 603.9] [Reference Citation Analysis (2)] |
| 6. | Hori K, Sen A, Artavanis-Tsakonas S. Notch signaling at a glance. J Cell Sci. 2013;126:2135-2140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 327] [Cited by in RCA: 390] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 7. | Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965-973. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 784] [Cited by in RCA: 797] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 8. | Yamamoto S, Schulze KL, Bellen HJ. Introduction to Notch signaling. Methods Mol Biol. 2014;1187:1-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 77] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 9. | Koch U, Radtke F. Notch and cancer: a double-edged sword. Cell Mol Life Sci. 2007;64:2746-2762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 257] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 10. | South AP, Cho RJ, Aster JC. The double-edged sword of Notch signaling in cancer. Semin Cell Dev Biol. 2012;23:458-464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 120] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 11. | Timmerman LA, Grego-Bessa J, Raya A, Bertrán E, Pérez-Pomares JM, Díez J, Aranda S, Palomo S, McCormick F, Izpisúa-Belmonte JC. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004;18:99-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 686] [Cited by in RCA: 733] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 12. | Gao J, Dong Y, Zhang B, Xiong Y, Xu W, Cheng Y, Dai M, Yu Z, Xu H, Zheng G. Notch1 activation contributes to tumor cell growth and proliferation in human hepatocellular carcinoma HepG2 and SMMC7721 cells. Int J Oncol. 2012;41:1773-1781. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 13. | Gramantieri L, Giovannini C, Lanzi A, Chieco P, Ravaioli M, Venturi A, Grazi GL, Bolondi L. Aberrant Notch3 and Notch4 expression in human hepatocellular carcinoma. Liver Int. 2007;27:997-1007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 87] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 14. | Hu L, Xue F, Shao M, Deng A, Wei G. Aberrant expression of Notch3 predicts poor survival for hepatocellular carcinomas. Biosci Trends. 2013;7:152-156. [PubMed] |
| 15. | Ning L, Wentworth L, Chen H, Weber SM. Down-regulation of Notch1 signaling inhibits tumor growth in human hepatocellular carcinoma. Am J Transl Res. 2009;1:358-366. [PubMed] |
| 16. | Villanueva A, Alsinet C, Yanger K, Hoshida Y, Zong Y, Toffanin S, Rodriguez-Carunchio L, Solé M, Thung S, Stanger BZ. Notch signaling is activated in human hepatocellular carcinoma and induces tumor formation in mice. Gastroenterology. 2012;143:1660-1669.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 232] [Cited by in RCA: 255] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 17. | Qi R, An H, Yu Y, Zhang M, Liu S, Xu H, Guo Z, Cheng T, Cao X. Notch1 signaling inhibits growth of human hepatocellular carcinoma through induction of cell cycle arrest and apoptosis. Cancer Res. 2003;63:8323-8329. [PubMed] |
| 18. | Viatour P, Ehmer U, Saddic LA, Dorrell C, Andersen JB, Lin C, Zmoos AF, Mazur PK, Schaffer BE, Ostermeier A. Notch signaling inhibits hepatocellular carcinoma following inactivation of the RB pathway. J Exp Med. 2011;208:1963-1976. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 145] [Cited by in RCA: 173] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 19. | Wang C, Qi R, Li N, Wang Z, An H, Zhang Q, Yu Y, Cao X. Notch1 signaling sensitizes tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis in human hepatocellular carcinoma cells by inhibiting Akt/Hdm2-mediated p53 degradation and up-regulating p53-dependent DR5 expression. J Biol Chem. 2009;284:16183-16190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 81] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 20. | Sun Q, Wang R, Luo J, Wang P, Xiong S, Liu M, Cheng B. Notch1 promotes hepatitis B virus X protein-induced hepatocarcinogenesis via Wnt/β-catenin pathway. Int J Oncol. 2014;45:1638-1648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 21. | Wang F, Zhou H, Yang Y, Xia X, Sun Q, Luo J, Cheng B. Hepatitis B virus X protein promotes the growth of hepatocellular carcinoma by modulation of the Notch signaling pathway. Oncol Rep. 2012;27:1170-1176. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 22. | Kongkavitoon P, Tangkijvanich P, Hirankarn N, Palaga T. Hepatitis B Virus HBx Activates Notch Signaling via Delta-Like 4/Notch1 in Hepatocellular Carcinoma. PLoS One. 2016;11:e0146696. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 23. | Hellström M, Phng LK, Gerhardt H. VEGF and Notch signaling: the yin and yang of angiogenic sprouting. Cell Adh Migr. 2007;1:133-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 119] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 24. | Patel NS, Li JL, Generali D, Poulsom R, Cranston DW, Harris AL. Up-regulation of delta-like 4 ligand in human tumor vasculature and the role of basal expression in endothelial cell function. Cancer Res. 2005;65:8690-8697. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 273] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 25. | Ridgway J, Zhang G, Wu Y, Stawicki S, Liang WC, Chanthery Y, Kowalski J, Watts RJ, Callahan C, Kasman I. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature. 2006;444:1083-1087. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 743] [Cited by in RCA: 767] [Article Influence: 42.6] [Reference Citation Analysis (0)] |
| 26. | Chiorean EG, LoRusso P, Strother RM, Diamond JR, Younger A, Messersmith WA, Adriaens L, Liu L, Kao RJ, DiCioccio AT. A Phase I First-in-Human Study of Enoticumab (REGN421), a Fully Human Delta-like Ligand 4 (Dll4) Monoclonal Antibody in Patients with Advanced Solid Tumors. Clin Cancer Res. 2015;21:2695-2703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 110] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 27. | Smith DC, Eisenberg PD, Manikhas G, Chugh R, Gubens MA, Stagg RJ, Kapoun AM, Xu L, Dupont J, Sikic B. A phase I dose escalation and expansion study of the anticancer stem cell agent demcizumab (anti-DLL4) in patients with previously treated solid tumors. Clin Cancer Res. 2014;20:6295-6303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 129] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 28. | Tuominen VJ, Ruotoistenmäki S, Viitanen A, Jumppanen M, Isola J. ImmunoRatio: a publicly available web application for quantitative image analysis of estrogen receptor (ER), progesterone receptor (PR), and Ki-67. Breast Cancer Res. 2010;12:R56. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 384] [Cited by in RCA: 371] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 29. | Patumraj S, Yoysungnoen P, Kachonrattanadet P, Wirachwong P. Tumor neocapillary density in hepatocellular carcinoma cells implanted nude mice model. Clin Hemorheol Microcirc. 2005;33:137-144. [PubMed] |
| 30. | Miles KM, Seshadri M, Ciamporcero E, Adelaiye R, Gillard B, Sotomayor P, Attwood K, Shen L, Conroy D, Kuhnert F. Dll4 blockade potentiates the anti-tumor effects of VEGF inhibition in renal cell carcinoma patient-derived xenografts. PLoS One. 2014;9:e112371. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 45] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 31. | Vincent F, Bonnin P, Clemessy M, Contrerès JO, Lamandé N, Gasc JM, Vilar J, Hainaud P, Tobelem G, Corvol P. Angiotensinogen delays angiogenesis and tumor growth of hepatocarcinoma in transgenic mice. Cancer Res. 2009;69:2853-2860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 51] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 32. | Xiong YQ, Sun HC, Zhang W, Zhu XD, Zhuang PY, Zhang JB, Wang L, Wu WZ, Qin LX, Tang ZY. Human hepatocellular carcinoma tumor-derived endothelial cells manifest increased angiogenesis capability and drug resistance compared with normal endothelial cells. Clin Cancer Res. 2009;15:4838-4846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 164] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 33. | Gao J, Chen C, Hong L, Wang J, Du Y, Song J, Shao X, Zhang J, Han H, Liu J. Expression of Jagged1 and its association with hepatitis B virus X protein in hepatocellular carcinoma. Biochem Biophys Res Commun. 2007;356:341-347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 44] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 34. | Sun HW, Wu C, Tan HY, Wang QS. Combination DLL4 with Jagged1-siRNA can enhance inhibition of the proliferation and invasiveness activity of human gastric carcinoma by Notch1/VEGF pathway. Hepatogastroenterology. 2012;59:924-929. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 35. | Chen H, Yang L, Zang S, Zhuo L, Fang X, Zhang Y, Li K, Song K, A H. High level of Delta-like ligand 4 suppresses the metastasis of hepatocellular carcinoma. Int J Clin Exp Pathol. 2016;9:2989-2997. |
| 36. | Liu X, Zhou J, Zhou N, Zhu J, Feng Y, Miao X. SYNJ2BP inhibits tumor growth and metastasis by activating DLL4 pathway in hepatocellular carcinoma. J Exp Clin Cancer Res. 2016;35:115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 37. | Lobry C, Oh P, Mansour MR, Look AT, Aifantis I. Notch signaling: switching an oncogene to a tumor suppressor. Blood. 2014;123:2451-2459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 199] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 38. | Aster JC, Pear WS, Blacklow SC. The Varied Roles of Notch in Cancer. Annu Rev Pathol. 2017;12:245-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 342] [Cited by in RCA: 521] [Article Influence: 57.9] [Reference Citation Analysis (0)] |
| 39. | Li JL, Harris AL. Crosstalk of VEGF and Notch pathways in tumour angiogenesis: therapeutic implications. Front Biosci (Landmark Ed). 2009;14:3094-3110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 103] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 40. | Noguera-Troise I, Daly C, Papadopoulos NJ, Coetzee S, Boland P, Gale NW, Lin HC, Yancopoulos GD, Thurston G. Blockade of Dll4 inhibits tumour growth by promoting non-productive angiogenesis. Nature. 2006;444:1032-1037. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 768] [Cited by in RCA: 811] [Article Influence: 45.1] [Reference Citation Analysis (0)] |
| 41. | Li JL, Sainson RC, Shi W, Leek R, Harrington LS, Preusser M, Biswas S, Turley H, Heikamp E, Hainfellner JA. Delta-like 4 Notch ligand regulates tumor angiogenesis, improves tumor vascular function, and promotes tumor growth in vivo. Cancer Res. 2007;67:11244-11253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 283] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 42. | Cancer Genome Atlas Research Network. Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell. 2017;169:1327-1341.e23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1578] [Cited by in RCA: 1729] [Article Influence: 216.1] [Reference Citation Analysis (1)] |
| 43. | Kawaguchi K, Honda M, Kaneko S. The Role of Notch Signaling in Liver Diseases: Contribution to Development and Cancer. Int J Cancer Clin Res. 2017;4:079. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 44. | Lu J, Xia Y, Chen K, Zheng Y, Wang J, Lu W, Yin Q, Wang F, Zhou Y, Guo C. Oncogenic role of the Notch pathway in primary liver cancer. Oncol Lett. 2016;12:3-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 45. | Strazzabosco M, Fabris L. Notch signaling in hepatocellular carcinoma: guilty in association! Gastroenterology. 2012;143:1430-1434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 46. | Zhu B, Sun L, Luo W, Li M, Coy DH, Yu L, Yu W. Activated Notch signaling augments cell growth in hepatocellular carcinoma via up-regulating the nuclear receptor NR4A2. Oncotarget. 2017;8:23289-23302. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 47. | Li Y, Wu S, Pu J, Huang X, Zhang P. Dengue virus up-regulates expression of notch ligands Dll1 and Dll4 through interferon-β signalling pathway. Immunology. 2015;144:127-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 48. | Schaller MA, Neupane R, Rudd BD, Kunkel SL, Kallal LE, Lincoln P, Lowe JB, Man Y, Lukacs NW. Notch ligand Delta-like 4 regulates disease pathogenesis during respiratory viral infections by modulating Th2 cytokines. J Exp Med. 2007;204:2925-2934. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 88] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 49. | Emuss V, Lagos D, Pizzey A, Gratrix F, Henderson SR, Boshoff C. KSHV manipulates Notch signaling by DLL4 and JAG1 to alter cell cycle genes in lymphatic endothelia. PLoS Pathog. 2009;5:e1000616. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 53] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 50. | Cuninghame S, Jackson R, Zehbe I. Hypoxia-inducible factor 1 and its role in viral carcinogenesis. Virology. 2014;456-457:370-383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 51. | Wilson GK, Tennant DA, McKeating JA. Hypoxia inducible factors in liver disease and hepatocellular carcinoma: current understanding and future directions. J Hepatol. 2014;61:1397-1406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 148] [Article Influence: 13.5] [Reference Citation Analysis (0)] |