Published online Jan 21, 2018. doi: 10.3748/wjg.v24.i3.371
Peer-review started: August 9, 2017
First decision: August 29, 2017
Revised: October 16, 2017
Accepted: November 21, 2017
Article in press: November 21, 2017
Published online: January 21, 2018
Processing time: 163 Days and 7.7 Hours
In our previous study, we have built a nine-gene (GPC3, HGF, ANXA1, FOS, SPAG9, HSPA1B, CXCR4, PFN1, and CALR) expression detection system based on the GeXP system. Based on peripheral blood and GeXP, we aimed to analyze the results of genes expression by different multi-parameter analysis methods and build a diagnostic model to classify hepatocellular carcinoma (HCC) patients and healthy people.
Logistic regression analysis, discriminant analysis, classification tree analysis, and artificial neural network were used for the multi-parameter gene expression analysis method. One hundred and three patients with early HCC and 54 age-matched healthy normal controls were used to build a diagnostic model. Fifty-two patients with early HCC and 34 healthy people were used for validation. The area under the curve, sensitivity, and specificity were used as diagnostic indicators.
Artificial neural network of the total nine genes had the best diagnostic value, and the AUC, sensitivity, and specificity were 0.943, 98%, and 85%, respectively. At last, 52 HCC patients and 34 healthy normal controls were used for validation. The sensitivity and specificity were 96% and 86%, respectively.
Multi-parameter analysis methods may increase the diagnostic value compared to single factor analysis and they may be a trend of the clinical diagnosis in the future.
Core tip: We aimed to analyze the results of expression of nine genes, which we identified previously, by different multi-parameter analysis methods and build a diagnostic model to classify hepatocellular carcinoma patients and healthy people. Logistic regression analysis, discriminant analysis, classification tree analysis, and artificial neural network were used for the multi-parameter gene expression analysis.
- Citation: Xie H, Xue YQ, Liu P, Zhang PJ, Tian ST, Yang Z, Guo Z, Wang HM. Multi-parameter gene expression profiling of peripheral blood for early detection of hepatocellular carcinoma. World J Gastroenterol 2018; 24(3): 371-378
- URL: https://www.wjgnet.com/1007-9327/full/v24/i3/371.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i3.371
Hepatocellular carcinoma (HCC) is one of the most common cancers in the world[1]. Chronic infection with hepatitis B or C virus, dietary aflatoxin B1 intake, and alcohol abuse clearly show a significant correlation with the incidence of HCC. In China, more than 90% of HCC patients are reported to experience chronic HBV infection[2]. Currently, HCC is often diagnosed at an advanced stage and has a poor prognosis. Clinical practice has demonstrated that early diagnosis of HCC can significantly increase the survival time. Many biomarkers have been proposed, and some are currently used in clinical diagnosis[3,4]; however, even alpha-fetoprotein, the most widely used biomarker for HCC diagnosis, has a poor diagnostic value[5]. Although pathology is used as a gold standard for diagnosis of HCC, it is invasive, and tissue samples are not easily obtained. Therefore, a non-invasive, accurate, and fast method for early detection of HCC is urgently needed.
Peripheral blood samples, which are easily and repeatedly obtained in the clinical setting, have been demonstrated to be valuable for disease prediction and classification, drug response evaluation, and toxicity classification[6-8]. These features make peripheral blood samples attractive to aid in the early detection of HCC[9]. As we know, HCC is a complex multi-gene and multi-factorial disease, and a single biomarker is not adequate to reflect the HCC status. A panel of biomarkers is a promising method for early detection of HCC, and now some panels have been used for cancer prediction[10,11]. A single gene analysis method is not sufficient when gene expression is used for diagnosis. Multi-parameter analysis methods, such as logistic regression analysis (LRA), discriminant analysis (DA), classification tree analysis (CTA), and artificial neural network (ANN), which can analyze multiple factors, have been shown to increase the diagnostic sensitivity and specificity and may be promising analysis methods for multi-parameter analysis[12-14].
In our previous study, we used Affymetrix to screen differential gene expression and built a 9-gene (GPC3, HGF, ANXA1, FOS, SPAG9, HSPA1B, CXCR4, PFN1, and CALR) expression detection system based on the GenomeLab GeXP Genetic Analysis system[15], known as GeXP, which can detect up to 35 genes in one reaction[16]. Based on peripheral blood and GeXP, we compared multi-parameter gene expression using various multi-parameter analysis methods and built a diagnostic model to classify early-stage HCC patients and healthy people.
The study was reviewed and approved by the 302 Hospital of People’s Liberation Army Institutional Review Board. After obtaining patient consent, blood samples from 103 early-stage HCC patients with chronic HBV infection were collected at our hospital. Fifty-four age-matched healthy normal control samples were collected from the people who underwent a health examination. Both samples were used to build the diagnostic model. Fifty-two early-stage HCC patients and 34 healthy people were used for validation. The disease status of early-stage HCC patients was confirmed by histopathological analysis, and tumors were staged according to the Barcelona Clinic Liver Cancer (BCLC) staging classification as either T1 (single lesion < 2 cm in diameter) or T2 (single lesion between 2 and 5 cm in diameter or < 3 lesions, each of which was < 3 cm in diameter)[17]. In addition, peripheral blood from HCC patients was collected before any therapy. The clinical characteristics of all the samples used for this study are shown in Table 1.
| Variable | Building model | Model validation | ||
| HCC | Control | HCC | Control | |
| Number of patients | 103 | 54 | 52 | 34 |
| Male | 54 | 29 | 29 | 19 |
| Female | 49 | 25 | 23 | 15 |
| Age (mean ± SD) | 54 ± 12 | 49 ± 11 | 51 ± 14 | 52 ± 9 |
| Tumor size (> 3 cm) | 29 | 0 | 13 | 0 |
| Tumor size (< 3 cm) | 74 | 0 | 39 | 0 |
| Number of nodules (Unilocular) | 36 | 0 | 21 | 0 |
| Number of nodules (Multilocular) | 67 | 0 | 31 | 0 |
| Cirrhosis | 89 | 0 | 36 | 0 |
| BCLC stage (T1) | 27 | 0 | 19 | 0 |
| BCLC stage (T2) | 76 | 0 | 32 | 0 |
Peripheral blood (2.5 mL) was collected and added into the PAXGene blood RNA tubes (Qiagen, Valencia, CA, United States). After inverting, the tubes were stored at -80 °C until total RNA extraction.
Total RNA was isolated using the PAXGene Blood RNA Kit (Qiagen, Valencia, CA, United States) according to the manufacturer’s instructions. The quality and quantity of RNA were measured by agarose gel electrophoresis and DU 800 spectrophotometry (Beckman Coulter, Fullerton, CA, United States), respectively. The primers for the nine genes were shown in our previous study. Reverse and forward primers were diluted 1:200 and 1:500 in nuclease-free water to a final concentration of 500 nmol/L and 200 nmol/L, respectively. Total RNA (50 ng) was used for reverse transcription (RT) with chimeric reverse primers in a single reaction. The concentrations of the primers were diluted at a ratio of 1:8. The RT reactions were performed using the following parameters: 48 °C for 1 min, 42 °C for 60 min, 95 °C for 5 min, and then held at 4 °C.
Polymerase chain reaction (PCR) reaction (10 μL) was performed in the 96-well PCR Detection Plate using the following parameters: 95 °C for 10 min, followed by 35 cycles of 94 °C for 30 s, 55 °C for 30 s, and 68 °C for 1 minute. The PCR products were diluted using nuclease-free water and added to the detection plate, which contained sample loading solution and DNA Size Standard 400. The GeXP system was used to match each gene fragment size and to measure the fluorescent dye signal strength in arbitrary units, and then the dataset was normalized to the housekeeping gene B2M. Finally, the dataset was log transformed.
LRA[18,19], DA[20,21], CTA[22,23], and ANN analysis[24,25] were used as the multi-parameter gene expression analysis methods. In LRA, the “Forward: Conditional” method was used to select variables. The stepwise probability was “Entry 0.05” and “Removal 0.10”, the “Probabilities” was saved, and then “CI for exp (B) 95%” was shown. The “Probabilities” of the HCC group were used for receiver operating characteristic (ROC) analysis, and the cutoff value was based on the Youden Index. In DA, “Use stepwise method” was used to select variables. The stepwise criterion was the “F value”. The “Entry” was 3.84, and the “Removal” was 2.71. The “Probabilities of group membership” was saved as a new variable, “Dis_n,” which represented the probability of HCC, and then the “Probabilities of group membership” was used for ROC analysis. In the CTA, “Exhaustive CHAID” was used as a growth method, and 20% of the samples were selected as the validation samples. The “Predicted value” was saved as a new variable for the ROC analysis. In the ANN analysis, the ROC curve was saved, and 80% of the samples were used as the training group and 20% as the test group. After comparison, the best diagnostic model was chosen, and 52 HCC samples and 34 healthy normal controls were used to validate the model. The ROC curve, area under the curve (AUC), sensitivity, and specificity were used as diagnostic indicators.
A total of nine genes (GPC3, HGF, ANXA1, FOS, SPAG9, HSPA1B, CXCR4, PFN1, and CALR) were used for HCC detection. We used the ROC curve of the nine genes to evaluate the diagnostic value and then analyzed the P-value and 95% confidence interval (CI). The diagnostic value of the AUC, the P-value, 95% confidence interval, cutoff value, sensitivity, and specificity are shown in Table 2. The ROC curves from four genes (HGF, ANXA1, SPAG9, and PFN1) showed P-values less than 0.05. According to the P-values, four genes (HGF, ANXA1, SPAG9, and PFN1) were chosen to have diagnostic value. In the multi-parameter analysis, both the 9-gene and the 4-gene sets were used.
| Gene | AUC | P value | 95%CI for AUC | Cutoff | Sen | Spe | |
| Lower | Upper | ||||||
| HGF | 0.620 | 0.014 | 0.532 | 0.708 | 0.102 | 0.408 | 0.926 |
| ANXA1 | 0.697 | < 0.001 | 0.614 | 0.780 | 0.919 | 0.728 | 0.593 |
| SPAG9 | 0.761 | < 0.001 | 0.681 | 0.842 | 0.477 | 0.942 | 0.481 |
| PFN1 | 0.700 | < 0.001 | 0.620 | 0.781 | 0.383 | 0.437 | 0.907 |
In the LRA for HCC detection, we compared the diagnostic value of the full 9-gene set (GPC3, HGF, ANXA1, FOS, SPAG9, HSPA1B, CXCR4, PFN1, and CALR) and the 4-gene set (HGF, ANXA1, SPAG9, and PFN1). From the full 9-gene set, five genes were selected using a “Forward: Conditional” method. The diagnostic formula was as follows:
Y = -4.089 + 21.269 XANXA1 - 5.339 XFOS + 32.543 XSPAG9 - 2.743 XCXCR4 + 9.524 XPFN1 (Y = logit P)
In the 4-gene set, three genes were selected, and the diagnostic formula was as follows:
Y = -4.826 + 13.172 XANXA1 + 15.353 XSPAG9 + 8.755 XPFN1 (Y = logit P)
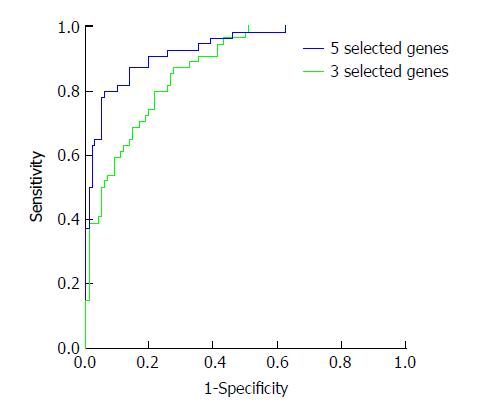
Then, the probability was used for evaluating the diagnostic value. The ROC curves are shown in Figure 1. The AUC of the five selected genes was 0.933, and that of the three selected genes was 0.878. This indicated that the five selected genes had better diagnostic value, and when the cutoff of logit P was 0.548, the sensitivity and specificity were 94% and 80%, respectively. According to the diagnostic formula, we can know the risk of the detected sample. In addition, the odds ratio (OR) of the five selected genes was also analyzed and is shown in Table 3. An OR greater than 1 indicates that the factor is a risk factor for the disease, and an OR less than 1 means it is a protective factor. Thus, the ANXA1, SPAG9, and PFN1 genes were risk factors. When their gene expression is higher, it may increase the incidence of HCC. The FOS and CXCR4 genes were found to be protective factors. When their gene expression is higher, it may decrease the incidence of HCC.
| Gene | P | OR | 95% CI for OR | |
| Lower | Upper | |||
| ANXA1 | < 0.001 | 1.73E+09 | 1.67E+05 | 1.79E+13 |
| FOS | 0.012 | 4.80E-03 | 7.30E-05 | 3.16E-01 |
| SPAG9 | < 0.001 | 1.36E+14 | 3.19E+07 | 5.80E+20 |
| CXCR4 | < 0.001 | 6.44E-02 | 1.47E-02 | 2.81E-01 |
| PFN1 | < 0.001 | 1.37E+04 | 9.82E+01 | 1.91E+06 |
In the DA, the full 9-gene set was analyzed by Bayes DA. Six genes were selected by “Use stepwise method”. The following formulas were used separately:
Y1 = -7.306 + 8.078 XGPC3 + 20.770 XANXA1 - 3.414 XFOS + 12.652 XSPAG9 + 1.842 XCXCR4 + 18.248 XPFN1
Y2 = -5.612 + 15.760 XGPC3 + 4.432 XANXA1 + 1.585 XFOS - 10.298 XSPAG9 + 4.320 XCXCR4 + 12.370 XPFN1
In the 4-gene set, three genes entered the diagnostic formulas. The formulas were as follows:
Y1 = -6.665 + 21.814 XANXA1 + 18.524 XSPAG9 + 16.663 XPFN1
Y2 = -2.806 + 13.784 XANXA1 + 8.081 XSPAG9 + 10.393 XPFN1
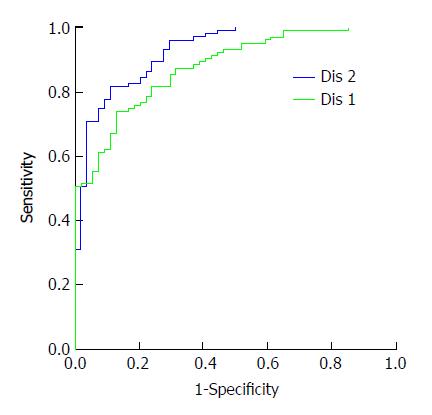
The probability of the healthy normal group and the HCC group analyzed by the three selected genes was saved as a new variable named Dis 1. The probability of the six selected genes was saved as Dis 2. The new variables Dis 1 and Dis 2 were used for the ROC analysis, as shown in Figure 2. The AUC of Dis 1 was 0.877 and of Dis 2 was 0.926. This result meant that the six selected genes had better diagnostic value for HCC, and when the cutoff of Y was 0.628, the sensitivity and specificity were 82% and 89%, respectively.
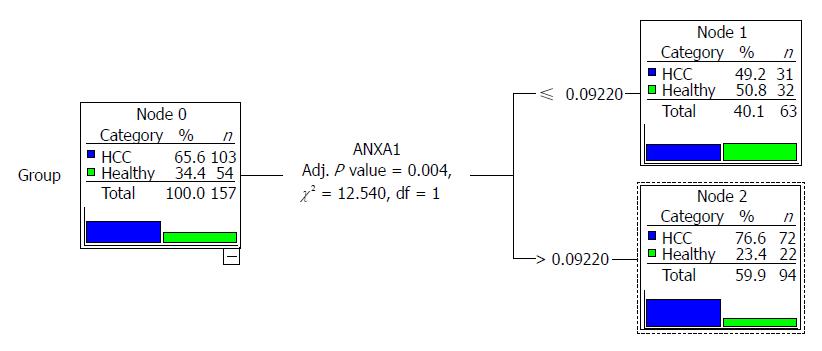
We compared the diagnostic value of the full 9-gene set and the 4-gene set by CTA; however, only ANXA1 entered the classification tree. The classification tree of the full 9-gene set was the same as the 4-gene set, as shown in Figure 3. When the cutoff was 0.629, the sensitivity and specificity were 70% and 60%, respectively. The total accuracy was only 66.20%.
The full 9-gene set and the 4-gene set were then analyzed by the ANN method; their network had one hidden layer which contained five units. In the training group of the full 9-gene set, the percent incorrect prediction was 12.3%, and for the testing group, it was 9.1%. In the 4-gene set, the percentages were 20.9% and 8.7%, respectively. In addition, the full 9-gene set had better predictive probabilities than the 4-gene set. The AUC of the full 9-gene set was 0.943, greater than that of the 4-gene set, which was 0.877.
Multiple multi-parameter analysis methods were used to build models, and the AUC was used to evaluate the diagnostic value. When a single gene was used, the AUC of SPAG9 was greater than those of the other genes. The AUCs of LRA, DA, and ANN were greater than that of SPAG9, and the AUC of CTA was less than that of SPAG9. Among the methods, the ANN of the full 9-gene set had the best diagnostic value; the AUC, sensitivity, and specificity were 0.943, 98%, and 85%, respectively. Finally, 52 HCC patients and 34 healthy normal controls were used for validation. The sensitivity and specificity were 96% and 86%, respectively. Above all, multi-parameter analysis methods may increase the diagnostic value compared to single-factor analysis and this approach may be a trend for future clinical diagnostic methods.
Clinical peripheral blood samples can be obtained easily and in a minimally invasive way. Studies have shown that mRNA in peripheral blood has the potential to be used for the early detection of cancers. There are many mRNA detection methods; however, the most commonly used method, real-time PCR, is limited by the number of genes and the amplification efficiency. The Beckman Coulter (Fullerton, CA, United States) GenomeLab GeXP Genetic Analysis system was designed ideally for up to 35 genes per reaction and can be used to detect 192 samples simultaneously in one single detection[26]. In addition, the GeXP system uses a universal priming strategy to decrease the variations in amplification efficiency across multiple genes[16,27]. Both strengths make GeXP an ideal multiple-gene expression detection method as well as a useful validation tool that is more similar to large-scale gene analysis methods, such as microarrays, than real-time PCR. We combined gene-chip analysis, peripheral blood, the GeXP detection system, and bioinformatics by using gene screening, model building, and bioinformatics analysis to build a gene expression profiling standard operating procedure for the early detection of cancer.
Studies have demonstrated that multi-parameter analysis can increase the sensitivity and specificity and is considered promising for future diagnostic methods. Many multi-parameter methods have been used for cancer early detection. In our study, logistic analysis increased the AUC to 0.933. Out of the nine genes, five (ANXA1, FOS, SPAG9, CXCR4, and PFN1) entered the diagnostic formula; however, FOS and CXCR4, which had poor diagnostic value, also entered the formula. This demonstrated that the genes showed significant differences between groups, and even if they had AUC values of less than 0.5, they may contribute to the logistic analysis. In the 4-gene set, three genes (ANXA1, SPAG9, and PFN1) entered the formula, but the AUC was less than that of the full 9-gene set. This demonstrated that genes with more significant differences may result in better diagnostic values. The logistic had a formula which can get a continuous value: logit (P). With a gene expression panel, we can predict the P-value to differentiate healthy people and HCC patients, which may provide a clinical indication for both physicians and patients.
After OR analysis, the ANXA1, SPAG9, and PFN1 genes were detected as risk factors. The FOS and CXCR4 genes were protective factors. Annexin1 (ANXA1) is a member of the annexin family of phospholipid-binding and calcium-binding proteins with a well demonstrated role in early delayed inhibitory feedback of glucocorticoids in the hypothalamus and pituitary gland[28]. Studies have demonstrated that ANXA1 is involved in tumorigenesis and can increase the incidence of HCC[29]. SPAG9 (sperm associated antigen 9) is a gene encoding c-Jun-amino-terminal kinase-interacting protein 4. This enzyme is a scaffolding protein that connects the mitogen-activated protein kinases to related transcription factor targets for the activation of JNK signaling pathways[30]. SPAG9 was also demonstrated to be a biomarker for breast cancer and cervical carcinoma[31,32]. Profilin-1 (PFN1) has been regarded as a tumor-suppressor molecule for breast cancer, and it can enhance ADP-to-ATP exchange on G-actin. In addition, it can also act as a shuttle to deliver ATP-bound G-actin to facilitate actin polymerization[33]. Studies have demonstrated that PFN1 is overexpressed in cancer cells by up-regulating PTEN and down-regulating AKT, and it is also an inhibitor of mammary carcinoma aggressiveness[34]. If PFN1 is silenced, it can inhibit endothelial cell proliferation, migration, and morphogenesis[35]. All three genes may contribute to the development of HCC.
In the DA, the AUC was similar to the logistic analysis. This demonstrated that it may be a valuable analysis method; however, because it had strict demand on the data distribution, its application was greatly limited. In our study, although we got the discriminant formula, it may have high bias because the dataset was not normally distributed. In the CTA, only one gene had diagnostic value. This finding demonstrated that they were not suitable for our study. ANN analysis has been demonstrated to provide better diagnostic value in disease prediction and cancer early detection. In our study, ANN had the best diagnostic value compared to the other analysis methods, and the predicted probabilities of the groups were also shown. In addition, in the training group for the full 9-gene set, the percent incorrect prediction was 12.3%, and for the testing group, it was 9.1%. All of these demonstrate that the ANN model we built was successful.
In our previous study, we screened the mRNA in peripheral blood samples by Affymetrix GeneChip analysis, and nine genes (GPC3, HGF, ANXA1, FOS, SPAG9, HSPA1B, CXCR4, PFN1, and CALR) were used for differentiating the healthy normal control and HCC groups. We have now built an ANN detection system. The sensitivity and specificity were 96% and 86%, respectively, which were greater than those of single gene analysis.
We have built a 9-gene (GPC3, HGF, ANXA1, FOS, SPAG9, HSPA1B, CXCR4, PFN1 and CALR) expression detection system based on the GeXP system in our previous study. We aimed to analyze the results of gene expression by different multi-parameter analysis methods and build a diagnostic model to classify hepatocellular carcinoma (HCC) patients and healthy people.
Although pathology is used as a golden standard for diagnosis of HCC, it is invasive and tissue sample is not easily obtained. Therefore, a non-invasive, accurate, and fast method for early detection of HCC is pressing.
A non-invasive, accurate, and fast method for early detection of HCC may be provided by our research based on the mRNA in peripheral blood.
We have successfully built an artificial neural network detection system combining detection system and bioinformatics together for differentiating the healthy normal group and HCC group. The sensitivity and specificity were separately 96% and 86%, respectively, which were greater than those of single-gene analysis.
Artificial neural network of the total nine genes had the best diagnostic value, and the AUC, sensitivity, and specificity were 0.943, 98%, and 85%, respectively. At last, 52 HCC patients and 34 healthy normal controls were used for validation. The sensitivity and specificity were 96% and 86%, respectively.
Based on the mRNA in peripheral blood, a multi-parameter analysis method was used to analyze multiple genes, which may increase the diagnostic value compared to the single factor analysis for the early detection of HCC, and it may be a trend of the clinical diagnosis in the future. It may provide a non-invasive, accurate, and fast method for early detection of HCC.
The GeXP system uses a universal priming strategy to decrease the variations in amplification efficiency across multiple genes, and it is an ideal multiple-gene expression detection method as well as a useful validation tool that is more similar to large-scale gene analysis methods. Combination of the peripheral blood, GeXP detection system, and bioinformatics together may be the future strategy to build an assistant detection method for cancer.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Boeckxstaens GE, Lee MW S- Editor: Chen K L- Editor: Wang TQ E- Editor: Huang Y
| 1. | Parkin DM, Bray FI, Devesa SS. Cancer burden in the year 2000. The global picture. Eur J Cancer. 2001;37 Suppl 8:S4-S66. [PubMed] |
| 2. | Gao JD, Shao YF, Xu Y, Ming LH, Wu ZY, Liu GT, Wang XH, Gao WH, Sun YT, Feng XL. Tight association of hepatocellular carcinoma with HBV infection in North China. Hepatobiliary Pancreat Dis Int. 2005;4:46-49. [PubMed] |
| 3. | Berretta M, Cavaliere C, Alessandrini L, Stanzione B, Facchini G, Balestreri L, Perin T, Canzonieri V. Serum and tissue markers in hepatocellular carcinoma and cholangiocarcinoma: clinical and prognostic implications. Oncotarget. 2017;8:14192-14220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 4. | Xu J, Li J, Zheng TH, Bai L, Liu ZJ. MicroRNAs in the Occurrence and Development of Primary Hepatocellular Carcinoma. Adv Clin Exp Med. 2016;25:971-975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 42] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 5. | El-Serag HB, Marrero JA, Rudolph L, Reddy KR. Diagnosis and treatment of hepatocellular carcinoma. Gastroenterology. 2008;134:1752-1763. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 768] [Cited by in RCA: 824] [Article Influence: 48.5] [Reference Citation Analysis (0)] |
| 6. | Wang Y, Barbacioru CC, Shiffman D, Balasubramanian S, Iakoubova O, Tranquilli M, Albornoz G, Blake J, Mehmet NN, Ngadimo D. Gene expression signature in peripheral blood detects thoracic aortic aneurysm. PLoS One. 2007;2:e1050. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 76] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 7. | Kuzman MR, Medved V, Terzic J, Krainc D. Genome-wide expression analysis of peripheral blood identifies candidate biomarkers for schizophrenia. J Psychiatr Res. 2009;43:1073-1077. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 52] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 8. | Alonso V, Neves AF, Marangoni K, Faria PC, Freschi AP, Capaneli AC, Meola J, Goulart LR. Gene expression profile in the peripheral blood of patients with prostate cancer and benign prostatic hyperplasia. Cancer Detect Prev. 2009;32:336-337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 9. | Yin CQ, Yuan CH, Qu Z, Guan Q, Chen H, Wang FB. Liquid Biopsy of Hepatocellular Carcinoma: Circulating Tumor-Derived Biomarkers. Dis Markers. 2016;2016:1427849. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 48] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 10. | Paczesny S, Krijanovski OI, Braun TM, Choi SW, Clouthier SG, Kuick R, Misek DE, Cooke KR, Kitko CL, Weyand A. A biomarker panel for acute graft-versus-host disease. Blood. 2009;113:273-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 297] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 11. | Aarøe J, Lindahl T, Dumeaux V, Saebø S, Tobin D, Hagen N, Skaane P, Lönneborg A, Sharma P, Børresen-Dale AL. Gene expression profiling of peripheral blood cells for early detection of breast cancer. Breast Cancer Res. 2010;12:R7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 79] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 12. | Han M, Liew CT, Zhang HW, Chao S, Zheng R, Yip KT, Song ZY, Li HM, Geng XP, Zhu LX. Novel blood-based, five-gene biomarker set for the detection of colorectal cancer. Clin Cancer Res. 2008;14:455-460. [PubMed] |
| 13. | Liew CC, Ma J, Tang HC, Zheng R, Dempsey AA. The peripheral blood transcriptome dynamically reflects system wide biology: a potential diagnostic tool. J Lab Clin Med. 2006;147:126-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 457] [Cited by in RCA: 525] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 14. | Osman I, Bajorin DF, Sun TT, Zhong H, Douglas D, Scattergood J, Zheng R, Han M, Marshall KW, Liew CC. Novel blood biomarkers of human urinary bladder cancer. Clin Cancer Res. 2006;12:3374-3380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 92] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 15. | Zhang PJ, Run WW, P L, Wang CB, Deng XX, Wang BB, Chen BB, J J, Liu HY, Dong ZN. Peripheral blood mRNA expression patterns to differentiate hepatocellular carcinoma from other hepatic diseases. Front Biosci (Elite Ed). 2012;4:620-630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 16. | Rai AJ, Kamath RM, Gerald W, Fleisher M. Analytical validation of the GeXP analyzer and design of a workflow for cancer-biomarker discovery using multiplexed gene-expression profiling. Anal Bioanal Chem. 2009;393:1505-1511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 55] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 17. | Llovet JM, Fuster J, Bruix J; Barcelona-Clínic Liver Cancer Group. The Barcelona approach: diagnosis, staging, and treatment of hepatocellular carcinoma. Liver Transpl. 2004;10:S115-S120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 500] [Cited by in RCA: 514] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 18. | Zhou X, Liu KY, Wong ST. Cancer classification and prediction using logistic regression with Bayesian gene selection. J Biomed Inform. 2004;37:249-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 88] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 19. | Elemeery MN, Badr AN, Mohamed MA, Ghareeb DA. Validation of a serum microRNA panel as biomarkers for early diagnosis of hepatocellular carcinoma post-hepatitis C infection in Egyptian patients. World J Gastroenterol. 2017;23:3864-3875. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 34] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 20. | Sergeev AS, Agapova RK, Bogadel’nikova IV, Perel’man MI. [The use of discrete characters in discriminant analysis for diagnosis of pulmonary tuberculosis and for classification of patients differing in treatment efficiency based on polymorphisms at nine codominant loci-HP, GC, TF, PI, PGM1, GLO1, C3, ACP1 and ESD]. Genetika. 2003;39:996-1002. [PubMed] |
| 21. | Cox IJ, Aliev AE, Crossey MM, Dawood M, Al-Mahtab M, Akbar SM, Rahman S, Riva A, Williams R, Taylor-Robinson SD. Urinary nuclear magnetic resonance spectroscopy of a Bangladeshi cohort with hepatitis-B hepatocellular carcinoma: A biomarker corroboration study. World J Gastroenterol. 2016;22:4191-4200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 23] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 22. | Robin X, Turck N, Hainard A, Lisacek F, Sanchez JC, Müller M. Bioinformatics for protein biomarker panel classification: what is needed to bring biomarker panels into in vitro diagnostics? Expert Rev Proteomics. 2009;6:675-689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 46] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 23. | Min JH, Kim YK, Choi SY, Jeong WK, Lee WJ, Ha SY, Ahn S, Ahn HS. Differentiation between cholangiocarcinoma and hepatocellular carcinoma with target sign on diffusion-weighted imaging and hepatobiliary phase gadoxetic acid-enhanced MR imaging: Classification tree analysis applying capsule and septum. Eur J Radiol. 2017;92:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 24. | Vohradský J. Neural network model of gene expression. FASEB J. 2001;15:846-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 92] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 25. | Wu CF, Wu YJ, Liang PC, Wu CH, Peng SF, Chiu HW. Disease-free survival assessment by artificial neural networks for hepatocellular carcinoma patients after radiofrequency ablation. J Formos Med Assoc. 2017;116:765-773. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 26. | Chen QR, Vansant G, Oades K, Pickering M, Wei JS, Song YK, Monforte J, Khan J. Diagnosis of the small round blue cell tumors using multiplex polymerase chain reaction. J Mol Diagn. 2007;9:80-88. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 40] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 27. | Nagel MA, Gilden D, Shade T, Gao B, Cohrs RJ. Rapid and sensitive detection of 68 unique varicella zoster virus gene transcripts in five multiplex reverse transcription-polymerase chain reactions. J Virol Methods. 2009;157:62-68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 30] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 28. | Gerke V, Creutz CE, Moss SE. Annexins: linking Ca2+ signalling to membrane dynamics. Nat Rev Mol Cell Biol. 2005;6:449-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1036] [Cited by in RCA: 1124] [Article Influence: 56.2] [Reference Citation Analysis (0)] |
| 29. | Ahn SH, Sawada H, Ro JY, Nicolson GL. Differential expression of annexin I in human mammary ductal epithelial cells in normal and benign and malignant breast tissues. Clin Exp Metastasis. 1997;15:151-156. [PubMed] |
| 30. | Engström W, Ward A, Moorwood K. The role of scaffold proteins in JNK signalling. Cell Prolif. 2010;43:56-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 48] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 31. | Kanojia D, Garg M, Gupta S, Gupta A, Suri A. Sperm-associated antigen 9, a novel biomarker for early detection of breast cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:630-639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 68] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 32. | Garg M, Kanojia D, Salhan S, Suri S, Gupta A, Lohiya NK, Suri A. Sperm-associated antigen 9 is a biomarker for early cervical carcinoma. Cancer. 2009;115:2671-2683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 52] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 33. | Shen K, Xi Z, Xie J, Wang H, Xie C, Lee CS, Fahey P, Dong Q, Xu H. Guttiferone K suppresses cell motility and metastasis of hepatocellular carcinoma by restoring aberrantly reduced profilin 1. Oncotarget. 2016;7:56650-56663. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 34. | Zou L, Jaramillo M, Whaley D, Wells A, Panchapakesa V, Das T, Roy P. Profilin-1 is a negative regulator of mammary carcinoma aggressiveness. Br J Cancer. 2007;97:1361-1371. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 90] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 35. | Das T, Bae YH, Wells A, Roy P. Profilin-1 overexpression upregulates PTEN and suppresses AKT activation in breast cancer cells. J Cell Physiol. 2009;218:436-443. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 46] [Article Influence: 2.9] [Reference Citation Analysis (0)] |