Published online Jun 7, 2018. doi: 10.3748/wjg.v24.i21.2261
Peer-review started: March 28, 2018
First decision: April 19, 2018
Revised: April 24, 2018
Accepted: April 26, 2018
Article in press: April 26, 2018
Published online: June 7, 2018
Processing time: 68 Days and 17.5 Hours
Hepatitis B virus (HBV) is still a worldwide health concern. While divergent factors are involved in its pathogenesis, it is now clear that HBV RNAs, principally templates for viral proteins and viral DNAs, have diverse biological functions involved in HBV pathogenesis. These functions include viral replication, hepatic fibrosis and hepatocarcinogenesis. Depending on the sequence similarities, HBV RNAs may act as sponges for host miRNAs and may deregulate miRNA functions, possibly leading to pathological consequences. Some parts of the HBV RNA molecule may function as viral-derived miRNA, which regulates viral replication. HBV DNA can integrate into the host genomic DNA and produce novel viral-host fusion RNA, which may have pathological functions. To date, elimination of HBV-derived covalently closed circular DNA has not been achieved. However, RNA transcription silencing may be an alternative practical approach to treat HBV-induced pathogenesis. A full understanding of HBV RNA transcription and the biological functions of HBV RNA may open a new avenue for the development of novel HBV therapeutics.
Core tip: Recently, it has been shown that hepatitis B virus (HBV) RNAs have diverse biological functions in the pathogenesis of HBV. HBV RNAs may work as sponges for host miRNAs and deregulate miRNA functions. Novel viral-host fusion RNA may be produced from HBV-DNA integration sites, which may also have pathological functions. Understanding HBV RNA transcription and the biological functions of HBV-related RNAs may open a new avenue for the development of novel HBV therapeutics that target HBV RNAs.
- Citation: Sekiba K, Otsuka M, Ohno M, Yamagami M, Kishikawa T, Suzuki T, Ishibashi R, Seimiya T, Tanaka E, Koike K. Hepatitis B virus pathogenesis: Fresh insights into hepatitis B virus RNA. World J Gastroenterol 2018; 24(21): 2261-2268
- URL: https://www.wjgnet.com/1007-9327/full/v24/i21/2261.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i21.2261
Hepatitis B virus (HBV) is a small enveloped DNA virus that belongs to the Hepadnaviridae virus family. HBV may establish a chronic infection in the liver, which can, in turn, lead to cirrhosis and hepatocellular carcinoma (HCC). Although HBV has infected humans for at least 500 years[1], the virus was not discovered until 1966[2], and in 1970 Dane et al[3] identified the virus particle by electron microscopy. Since then, an antiviral therapy has been developed; these anti-HBV drugs are nucleos(t)ide analogs that can sufficiently suppress viral DNA load in most cases[4-9]. Moreover, vaccination programs have already been established to prevent HBV infection[10]. However, these are not sufficient to eradicate HBV. In fact, an estimated 257 million people are still chronically infected, and 887 thousand people die annually, primarily from the complications of HBV, which include cirrhosis and HCC[11-13].
Recently, RNAs, especially non-coding RNAs, have been revealed to have diverse functions[14]. We and others previously reported that viral RNAs not only work as templates for protein synthesis and viral DNA replication in the case of HBV but also exhibit biological functions involved in its pathogenesis[15,16]. In this context, even when HBV DNA is maintained at a relatively low level by nucleos(t)ide analogs, viral RNAs alone may harm the host, leading to cirrhosis or HCC. Thus, understanding the functions of HBV RNAs may act as a platform for the future development of HBV therapeutics. In this paper, we review current knowledge on the biological impact of HBV RNAs on host cells.
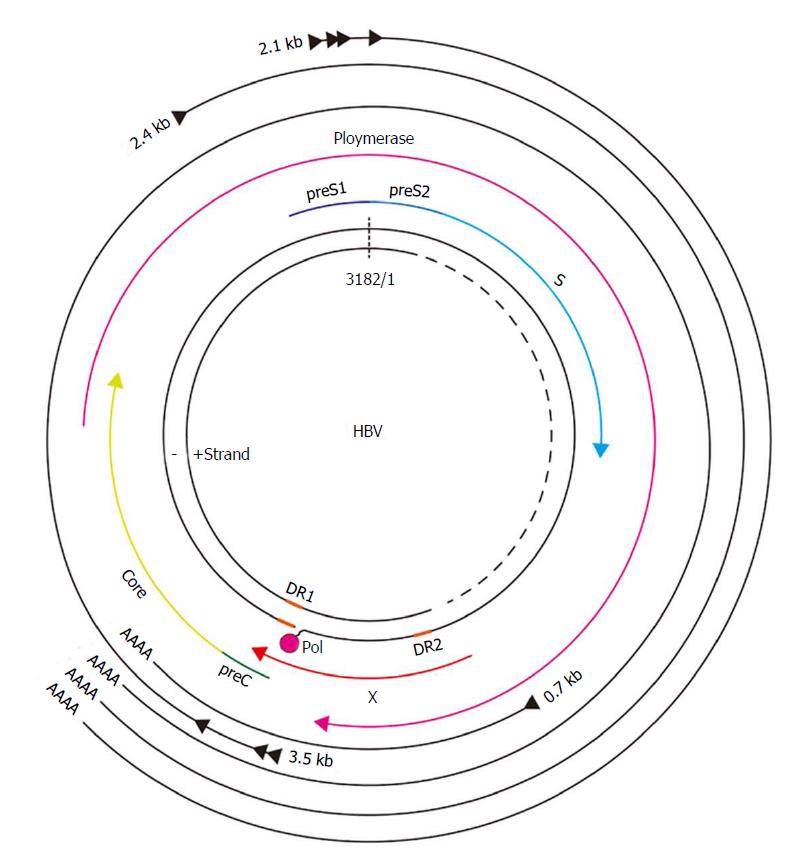
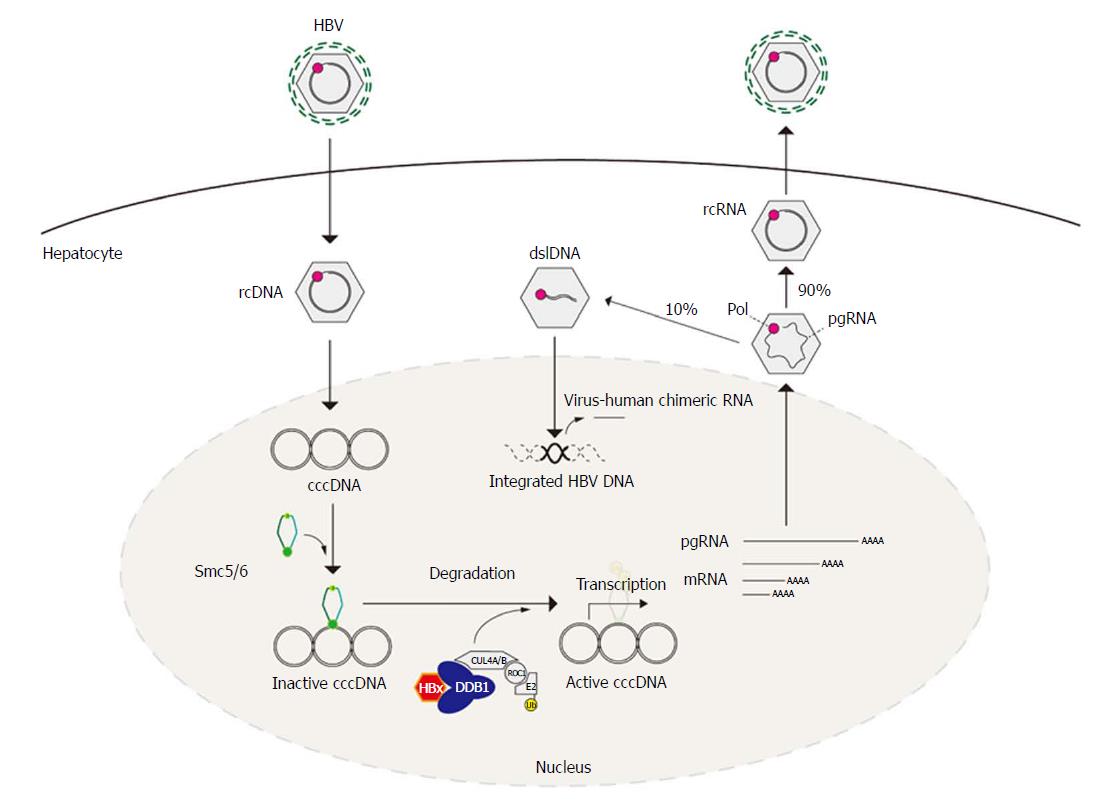
The HBV genome has four overlapping open reading frames: 3.5 kb pre-C/C or pre-genomic RNA (pgRNA), 2.4 kb pre-S, and 0.7 kb X mRNA (Figure 1). Viral particles with a 3.2-kb-long partially double-stranded relaxed circular DNA (rcDNA) genome invade the cell through the sodium taurocholate co-transporting polypeptide (NTCP) receptor. After un-coating of surface antigen, the core particles transport the genome to the hepatocyte nucleus. Then, covalently closed circular DNA (cccDNA) is molded from rcDNA. The cccDNA plays a role as a template in the transcription of HBV RNA (Figure 2)[17].
The viral genes are transcribed by the cellular RNA polymerase II from cccDNA. Two enhancers designated enhancer I (EnhI) and enhancer II (EnhII) have been identified in the HBV genome, which drive and regulate the expression of the complete viral transcripts[18]. Moreover, recently, various host proteins were revealed to be involved in the process of HBV RNA transcription from cccDNA, and the most representative host proteins are structural maintenance of chromosomes (Smc) proteins Smc5 and Smc6. Because Smc5/6 inhibit HBV RNA transcription from cccDNA, the efficient transcription of HBV RNA from cccDNA requires the degradation of Smc5/6. HBV regulatory protein X (HBx) hijacks the host Cullin 4-ROC1 RING E3 ubiquitin ligase (CRL4) complex to target Smc5/6 co-localized with nuclear domain 10 (ND10) for ubiquitination, which, in turn, promotes HBV transcription[19-21]. Thus, the existence of HBV RNAs means the degradation of Smc5/6. Because Smc5/6 is related to DNA repair[22], this degradation may eventually lead to carcinogenesis. Therefore, this ubiquitination pathway has strong potential as a novel therapeutic target in interventions for HBV pathogenesis.
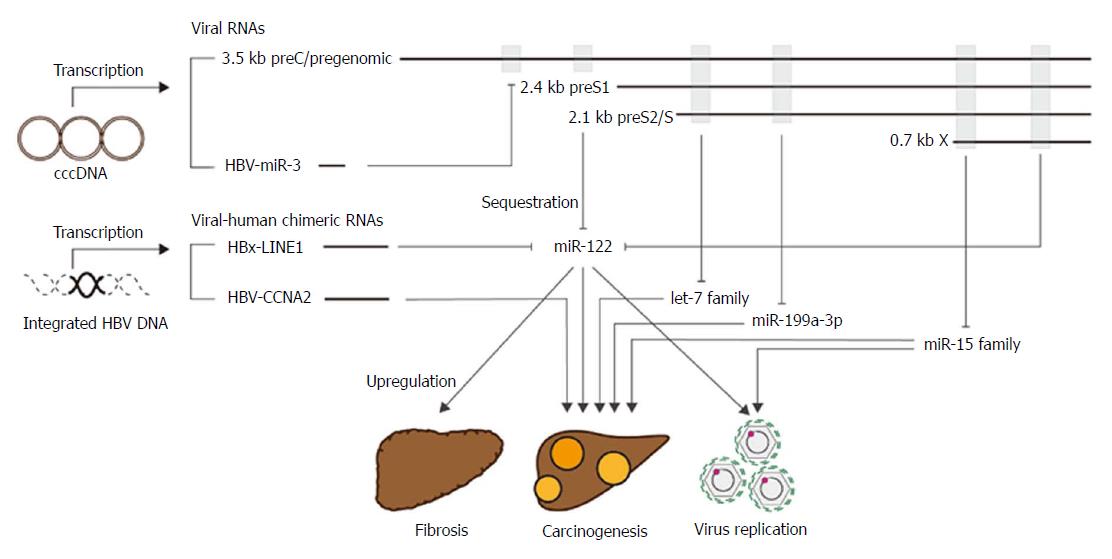
MicroRNAs (miRNAs) are short, single-stranded, non-coding RNAs. Mature miRNAs are recruited into the Ago2-related RNA-induced silencing complex (RISC) and act to suppress the gene expression of target mRNAs. Depending on the target mRNA, miRNAs are responsible for various biological functions[23]. Recent studies have shown that HBV RNAs have several regions complementary to miRNAs, and act as miRNA sponges to upregulate the expression of miRNA targets; this results in the induction of HBV pathogenesis[15,24]. A list of miRNAs that could be trapped by HBV RNAs and may be involved in HBV pathogenesis is shown in Figure 3. In the following paragraphs, we discuss the potential biological roles of miRNAs in HBV pathogenesis.
Although our knowledge of the direct relationship between HBV RNAs and viral replications are limited, HBV RNAs may promote viral replication via sequestering cellular miRNAs, such as miR-122 and miR-15 family[25,26].
miR-122 is highly and specifically expressed in hepatocytes. It plays multiple roles in the control of lipid metabolism, iron homeostasis, and the circadian rhythm and has anti-inflammatory and anti-tumorigenic functions[27-31]. The expression level of miR-122 is decreased in HBV-producing cells and in liver tissue from chronic hepatitis B patients[25,32,33]. Furthermore, there is an inverse correlation between miR-122 expression level and HBV replication[32]. Previously, it was found that the expression levels of pri-miR-122 and pre-miR122, the precursors of miR-122, were not decreased in HBV-positive HCC tissues and cells compared to normal liver tissue and cells[25,34,35]. Therefore, the downregulation of mature miR-122 expression is thought to be the result of binding to a conserved sequence at the 3’ end of all HBV transcripts following degradation. Although the precise mechanisms remain to be clarified, viral non-coding RNAs may play a critical role modulating the turnover of host miRNAs through the degradation of target miRNAs[36-40].
miR-122 negatively regulates HBV replication. It has been reported that one possible mechanism mediating the negative regulation of HBV replication by miR-122 depends on the expression level of cyclin G1, a target of miR-122. Decreased expression or function of miR-122 would result in the suppression of p53 through upregulation of cyclin G1 expression, which further increases HBV transcription by blocking specific binding of p53 to HBV enhancer elements[33].
The miR-15 family is also reported to regulate HBV replication. For instance, HBV RNA can sequester miR-15a and miR-16-1, and overexpression of these miRNAs decreases viral replication. Although the direct molecular mechanism of miR-15 family members has not been fully elucidated, among the multiple targets of miR-15a and miR16-1, cyclin D1 is thought to be involved in the regulation. Specifically, the up-regulation of cyclin D1 was demonstrated to be required for HBV replication[26].
Yang et al[41] recently showed that HBV-encoded HBV-miR-3 was expressed in HBV-infected tissues and cells. The viral-derived miRNA targeted the 3.5-kb HBV transcript to reduce HBc protein and pgRNA/HBV-RI production. The inhibition of HBV replication was suggested to contribute to the development of persistent infection in chronic hepatitis B patients. However, there is insufficient direct evidence for this mechanism, and, therefore, further studies are warranted.
Liver fibrosis underlies the majority of chronic liver diseases and is a precursor to cirrhosis and HCC. The cycle of liver damage and repair leads to the deposition of extracellular matrix proteins and the development of fibrosis. Some miRNAs, such as miR-21, miR-221/222 and miR-181b, cause liver fibrosis through deregulation of the transforming growth factor-β (TGF-β) or nuclear factor-κB (NF-κB) pathways[42-44]. On the other hand, miR-29b, miR-101, miR-122, and miR-214-3p inhibit fibrosis by blocking collagen synthesis or the TGF-β pathway[45-48]. Among these miRNAs, miR-122 was reported to have complementary lesion(s) in HBV RNAs.
As previously mentioned, miR-122 is highly expressed in the healthy liver, but is downregulated in HBV-infected livers via sequestration by HBV RNA. This change in miR-122 expression leads to the development of liver fibrosis through the activation of collagen synthesis via the TGF-β pathway[47].
HBV is the leading risk factor for the development of HCC worldwide. Many mechanisms have been reported to lead to the development of HCC, and one such mechanism involves the sequestration of host miRNAs by HBV RNA.
Decreased miR-122 levels resulted in increased pituitary tumor transforming gene 1 (PTTG1)-binding factor (PBF) expression, which enhanced the proliferation and invasiveness of HCC in vitro and tumorigenicity in vivo, through PBF-mediated activation of the PTTG1 transcription factor[25]. The possible contribution of these mechanisms to HBV-related carcinogenesis should be further examined in studies on human samples.
miRNAs in the let-7 family are classified as putative tumor suppressor miRNAs. The expression level of this family of miRNAs is often decreased in human cancers, including HCC, and promotes transformation by suppressing oncogenic targets, such as LIN28B, HMGA2 and c-Myc. Studies conducted by our group and others found that let-7 family miRNAs (e.g., let-7g and let-7a) could be sequestered by HBV-RNA[15,24]. Furthermore, we demonstrated that this functional downregulation could lead to the promotion of tumorigenesis.
miR-199a-3p is also involved in carcinogenesis and contributes to the malignant potential of HCC. Indeed, downregulation of miR-199a-3p correlated with poor HCC patient survival[49]. This miRNA targets mammalian target of rapamycin (mTOR) and c-Met in HCC cells. The restoration of miR-199a-3p levels in HCC cells resulted in G(1)-phase cell cycle arrest, decreased invasive capability, enhanced susceptibility to hypoxia, and increased sensitivity to doxorubicin-induced apoptosis.
miR-15a can be sponged off by HBV mRNAs. One of the proposed targets of miR-15a is Smad7, an inhibitor of the TGF-β pathway. Thus, HBV mRNA can interfere with TGF-β signaling by upregulating Smad7 expression, which obstructs TGF-β-induced apoptosis and promotes tumor development[50].
HBV DNA can integrate into host chromosomes at various locations. Integrated HBV DNA lacks the ability to transcribe pgRNA because HBV double-stranded linear DNA is only ~16 nt longer than the length of the genome, making it too short to transcribe pgRNA. Despite this, integrated HBV DNA levels correlate with the development of HCC. Indeed, the majority of HBV-related HCCs contain at least one HBV genome integration site[51]. While the mechanism of carcinogenesis induced by the integration of the HBV genome has been explained in several ways, virus-related RNAs from the integration sites are definitely involved.
HBV DNA integration often occurs within or near repetitive, non-coding sequences, such as long interspersed nuclear element 1 (LINEs) and short interspersed nuclear elements (SINEs)[52]. By applying Viral-Fusion-Seq to detect possible fusions between viral and human sequences[53], a viral-human hybrid RNA transcript called HBx-LINE1 was identified in HBV-related HCCs[54]. The presence of this long non-coding RNA, a fusion of the human LINE1 and HBx genes, was correlated with poor prognosis in HCC patients[54].
HBx-LINE1 contains six binding sites for miR-122, which enable the chimeric HBx-LINE1 transcript to act as a molecular sponge for miR-122. This sequestration leads to an increase in hepatic cell β-catenin signaling, a decrease in E-cadherin levels, increased cell migration, and significant mouse liver injury, leading to HCC[35]. Therefore, HBx-LINE1 is a potential therapeutic target and prognostic biomarker for HCC. While this is an interesting result, further studies are needed to uncover the precise mechanism of oncogenesis.
Cyclin A2 (CCNA2) is a cell cycle regulatory protein that acts as a regulatory subunit of cyclin-dependent kinase[55]. Integration of HBV into the CCNA2 gene has been observed in HBV-positive HCCs[56]. The integration site is intron 2 of CCNA2, which results in the formation of a new splice site in the pre-mRNA. This new splice site leads to the formation of a 177-bp in-frame pseudo-exon and produces a novel and recurrent HBV-CCNA2 fusion transcript, A2S[56]. Disruption of the destruction box of A2S causes A2S to become non-degradable; however, the function enhancing cell cycle progression of CCNA2 is retained, which demonstrates its potential role in hepatocarcinogenesis.
In this review, we summarized current knowledge on the roles of HBV RNAs, including viral replication, promotion of liver fibrosis, and carcinogenesis, in HBV-related pathogenesis. Specifically, we discussed how HBV RNAs deregulate miRNA function and lead to the synthesis of host-viral fusion RNA from integration sites. However, HBV RNAs may still have other, as yet unknown biological functions, such as deregulating host protein function or long non-coding RNA function through direct interactions or associations. Therefore, further studies are needed to fully elucidate the biological roles of HBV RNAs.
Certainly, anti-HBV therapeutics must focus on the elimination of HBV RNAs; however, no such therapeutic is currently available. The ultimate therapeutic goal is to destroy cccDNA. While gene-editing approaches, such as those focused on the CRISPR/Cas9 system, may be reasonable for directly targeting cccDNA, further studies are necessary to identify strategies to maximize positive effects and minimize toxicity[17,57]. In the meantime, transcriptional silencing of cccDNA may be a practical approach to attenuate HBV-related pathogenesis. For this purpose, a full understanding of HBV transcriptional control and HBV RNA-mediated pathogenesis is urgently needed.
HBV RNAs are not only templates for protein synthesis and viral DNA replication but also exhibit biological functions that play a role in pathogenesis. Because current therapies are unable to solve this problem, novel therapeutic agents that target the cccDNA itself, or inhibit its transcription, are strongly warranted.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Japan
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Parvez MK S- Editor: Gong ZM L- Editor: Filipodia E- Editor: Huang Y
| 1. | Patterson Ross Z, Klunk J, Fornaciari G, Giuffra V, Duchêne S, Duggan AT, Poinar D, Douglas MW, Eden JS, Holmes EC. The paradox of HBV evolution as revealed from a 16th century mummy. PLoS Pathog. 2018;14:e1006750. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 48] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 2. | Alter HJ, Blumberg BS. Further studies on a “new” human isoprecipitin system (Australia antigen). Blood. 1966;27:297-309. [PubMed] |
| 3. | Dane DS, Cameron CH, Briggs M. Virus-like particles in serum of patients with Australia-antigen-associated hepatitis. Lancet. 1970;1:695-698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 648] [Cited by in RCA: 560] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 4. | Kim GA, Lim YS, An J, Lee D, Shim JH, Kim KM, Lee HC, Chung YH, Lee YS, Suh DJ. HBsAg seroclearance after nucleoside analogue therapy in patients with chronic hepatitis B: clinical outcomes and durability. Gut. 2014;63:1325-1332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 329] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 5. | Gao Y, Feng J, Yang G, Zhang S, Liu Y, Bu Y, Sun M, Zhao M, Chen F, Zhang W. Hepatitis B virus X protein-elevated MSL2 modulates hepatitis B virus covalently closed circular DNA by inducing degradation of APOBEC3B to enhance hepatocarcinogenesis. Hepatology. 2017;66:1413-1429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 53] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 6. | Ono A, Suzuki F, Kawamura Y, Sezaki H, Hosaka T, Akuta N, Kobayashi M, Suzuki Y, Saitou S, Arase Y. Long-term continuous entecavir therapy in nucleos(t)ide-naïve chronic hepatitis B patients. J Hepatol. 2012;57:508-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 132] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 7. | Chang TT, Lai CL, Kew Yoon S, Lee SS, Coelho HS, Carrilho FJ, Poordad F, Halota W, Horsmans Y, Tsai N. Entecavir treatment for up to 5 years in patients with hepatitis B e antigen-positive chronic hepatitis B. Hepatology. 2010;51:422-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 447] [Cited by in RCA: 466] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 8. | Heathcote EJ, Marcellin P, Buti M, Gane E, De Man RA, Krastev Z, Germanidis G, Lee SS, Flisiak R, Kaita K. Three-year efficacy and safety of tenofovir disoproxil fumarate treatment for chronic hepatitis B. Gastroenterology. 2011;140:132-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 364] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 9. | Buti M, Gane E, Seto WK, Chan HL, Chuang WL, Stepanova T, Hui AJ, Lim YS, Mehta R, Janssen HL. Tenofovir alafenamide versus tenofovir disoproxil fumarate for the treatment of patients with HBeAg-negative chronic hepatitis B virus infection: a randomised, double-blind, phase 3, non-inferiority trial. Lancet Gastroenterol Hepatol. 2016;1:196-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 386] [Cited by in RCA: 361] [Article Influence: 40.1] [Reference Citation Analysis (0)] |
| 10. | Fisman DN, Agrawal D, Leder K. The effect of age on immunologic response to recombinant hepatitis B vaccine: a meta-analysis. Clin Infect Dis. 2002;35:1368-1375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 144] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 11. | Nelson PK, Mathers BM, Cowie B, Hagan H, Des Jarlais D, Horyniak D, Degenhardt L. Global epidemiology of hepatitis B and hepatitis C in people who inject drugs: results of systematic reviews. Lancet. 2011;378:571-583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 949] [Cited by in RCA: 973] [Article Influence: 69.5] [Reference Citation Analysis (0)] |
| 12. | Degenhardt L, Charlson F, Stanaway J, Larney S, Alexander LT, Hickman M, Cowie B, Hall WD, Strang J, Whiteford H. Estimating the burden of disease attributable to injecting drug use as a risk factor for HIV, hepatitis C, and hepatitis B: findings from the Global Burden of Disease Study 2013. Lancet Infect Dis. 2016;16:1385-1398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 221] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 13. | World Health Organization. Global hepatitis report, 2017. Geneva. 2017;1-68. |
| 14. | Otsuka M, Kishikawa T, Yoshikawa T, Yamagami M, Ohno M, Takata A, Shibata C, Ishibashi R, Koike K. MicroRNAs and liver disease. J Hum Genet. 2017;62:75-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 15. | Takata A, Otsuka M, Ohno M, Kishikawa T, Yoshikawa T, Koike K. Mutual antagonism between hepatitis B viral mRNA and host microRNA let-7. Sci Rep. 2016;6:23237. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 16. | Lamontagne J, Steel LF, Bouchard MJ. Hepatitis B virus and microRNAs: Complex interactions affecting hepatitis B virus replication and hepatitis B virus-associated diseases. World J Gastroenterol. 2015;21:7375-7399. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 49] [Cited by in RCA: 57] [Article Influence: 5.7] [Reference Citation Analysis (1)] |
| 17. | Ohno M, Otsuka M, Kishikawa T, Yoshikawa T, Takata A, Koike K. Novel therapeutic approaches for hepatitis B virus covalently closed circular DNA. World J Gastroenterol. 2015;21:7084-7088. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 13] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 18. | Doitsh G, Shaul Y. Enhancer I predominance in hepatitis B virus gene expression. Mol Cell Biol. 2004;24:1799-1808. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 75] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 19. | Murphy CM, Xu Y, Li F, Nio K, Reszka-Blanco N, Li X, Wu Y, Yu Y, Xiong Y, Su L. Hepatitis B Virus X Protein Promotes Degradation of SMC5/6 to Enhance HBV Replication. Cell Rep. 2016;16:2846-2854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 241] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 20. | Decorsière A, Mueller H, van Breugel PC, Abdul F, Gerossier L, Beran RK, Livingston CM, Niu C, Fletcher SP, Hantz O. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature. 2016;531:386-389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 309] [Cited by in RCA: 414] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 21. | Niu C, Livingston CM, Li L, Beran RK, Daffis S, Ramakrishnan D, Burdette D, Peiser L, Salas E, Ramos H. The Smc5/6 Complex Restricts HBV when Localized to ND10 without Inducing an Innate Immune Response and Is Counteracted by the HBV X Protein Shortly after Infection. PLoS One. 2017;12:e0169648. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 119] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 22. | Jeppsson K, Kanno T, Shirahige K, Sjögren C. The maintenance of chromosome structure: positioning and functioning of SMC complexes. Nat Rev Mol Cell Biol. 2014;15:601-614. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 171] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 23. | Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8847] [Cited by in RCA: 9285] [Article Influence: 464.3] [Reference Citation Analysis (0)] |
| 24. | Deng M, Hou J, Hu J, Wang S, Chen M, Chen L, Ju Y, Li C, Meng S. Hepatitis B virus mRNAs functionally sequester let-7a and enhance hepatocellular carcinoma. Cancer Lett. 2016;383:62-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 25. | Li C, Wang Y, Wang S, Wu B, Hao J, Fan H, Ju Y, Ding Y, Chen L, Chu X. Hepatitis B virus mRNA-mediated miR-122 inhibition upregulates PTTG1-binding protein, which promotes hepatocellular carcinoma tumor growth and cell invasion. J Virol. 2013;87:2193-2205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 120] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 26. | Wang Y, Jiang L, Ji X, Yang B, Zhang Y, Fu XD. Hepatitis B viral RNA directly mediates down-regulation of the tumor suppressor microRNA miR-15a/miR-16-1 in hepatocytes. J Biol Chem. 2013;288:18484-18493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 88] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 27. | Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1612] [Cited by in RCA: 1638] [Article Influence: 86.2] [Reference Citation Analysis (0)] |
| 28. | Castoldi M, Vujic Spasic M, Altamura S, Elmén J, Lindow M, Kiss J, Stolte J, Sparla R, D’Alessandro LA, Klingmüller U. The liver-specific microRNA miR-122 controls systemic iron homeostasis in mice. J Clin Invest. 2011;121:1386-1396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 197] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 29. | Gatfield D, Le Martelot G, Vejnar CE, Gerlach D, Schaad O, Fleury-Olela F, Ruskeepää AL, Oresic M, Esau CC, Zdobnov EM. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev. 2009;23:1313-1326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 310] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 30. | Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, Yu L, Bai S, La Perle K, Chivukula RR. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122:2871-2883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 583] [Cited by in RCA: 628] [Article Influence: 48.3] [Reference Citation Analysis (0)] |
| 31. | Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, Huang Y, Chen HC, Lee CH, Tsai TF. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884-2897. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 612] [Cited by in RCA: 669] [Article Influence: 51.5] [Reference Citation Analysis (0)] |
| 32. | Chen Y, Shen A, Rider PJ, Yu Y, Wu K, Mu Y, Hao Q, Liu Y, Gong H, Zhu Y. A liver-specific microRNA binds to a highly conserved RNA sequence of hepatitis B virus and negatively regulates viral gene expression and replication. FASEB J. 2011;25:4511-4521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 149] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 33. | Wang S, Qiu L, Yan X, Jin W, Wang Y, Chen L, Wu E, Ye X, Gao GF, Wang F. Loss of microRNA 122 expression in patients with hepatitis B enhances hepatitis B virus replication through cyclin G(1) -modulated P53 activity. Hepatology. 2012;55:730-741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 209] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 34. | Zeisel MB, Pfeffer S, Baumert TF. miR-122 acts as a tumor suppressor in hepatocarcinogenesis in vivo. J Hepatol. 2013;58:821-823. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 35. | Liang HW, Wang N, Wang Y, Wang F, Fu Z, Yan X, Zhu H, Diao W, Ding Y, Chen X. Hepatitis B virus-human chimeric transcript HBx-LINE1 promotes hepatic injury via sequestering cellular microRNA-122. J Hepatol. 2016;64:278-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 101] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 36. | McCaskill J, Praihirunkit P, Sharp PM, Buck AH. RNA-mediated degradation of microRNAs: A widespread viral strategy? RNA Biol. 2015;12:579-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 37. | Cazalla D, Yario T, Steitz JA. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328:1563-1566. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 421] [Cited by in RCA: 391] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 38. | Libri V, Helwak A, Miesen P, Santhakumar D, Borger JG, Kudla G, Grey F, Tollervey D, Buck AH. Murine cytomegalovirus encodes a miR-27 inhibitor disguised as a target. Proc Natl Acad Sci U S A. 2012;109:279-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 119] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 39. | Marcinowski L, Tanguy M, Krmpotic A, Rädle B, Lisnić VJ, Tuddenham L, Chane-Woon-Ming B, Ruzsics Z, Erhard F, Benkartek C. Degradation of cellular mir-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo. PLoS Pathog. 2012;8:e1002510. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 168] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 40. | Lee S, Song J, Kim S, Kim J, Hong Y, Kim Y, Kim D, Baek D, Ahn K. Selective degradation of host MicroRNAs by an intergenic HCMV noncoding RNA accelerates virus production. Cell Host Microbe. 2013;13:678-690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 97] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 41. | Yang X, Li H, Sun H, Fan H, Hu Y, Liu M, Li X, Tang H. Hepatitis B Virus-Encoded MicroRNA Controls Viral Replication. J Virol. 2017;91:pii: e01919-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 85] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 42. | Zhang J, Jiao J, Cermelli S, Muir K, Jung KH, Zou R, Rashid A, Gagea M, Zabludoff S, Kalluri R. miR-21 Inhibition Reduces Liver Fibrosis and Prevents Tumor Development by Inducing Apoptosis of CD24+ Progenitor Cells. Cancer Res. 2015;75:1859-1867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 82] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 43. | Ogawa T, Enomoto M, Fujii H, Sekiya Y, Yoshizato K, Ikeda K, Kawada N. MicroRNA-221/222 upregulation indicates the activation of stellate cells and the progression of liver fibrosis. Gut. 2012;61:1600-1609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 191] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 44. | Wang B, Li W, Guo K, Xiao Y, Wang Y, Fan J. miR-181b promotes hepatic stellate cells proliferation by targeting p27 and is elevated in the serum of cirrhosis patients. Biochem Biophys Res Commun. 2012;421:4-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 86] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 45. | Wang J, Chu ES, Chen HY, Man K, Go MY, Huang XR, Lan HY, Sung JJ, Yu J. microRNA-29b prevents liver fibrosis by attenuating hepatic stellate cell activation and inducing apoptosis through targeting PI3K/AKT pathway. Oncotarget. 2015;6:7325-7338. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 149] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 46. | Xie Y, Yao Q, Butt AM, Guo J, Tian Z, Bao X, Li H, Meng Q, Lu J. Expression profiling of serum microRNA-101 in HBV-associated chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. Cancer Biol Ther. 2014;15:1248-1255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 70] [Article Influence: 6.4] [Reference Citation Analysis (1)] |
| 47. | Zeng C, Wang YL, Xie C, Sang Y, Li TJ, Zhang M, Wang R, Zhang Q, Zheng L, Zhuang SM. Identification of a novel TGF-β-miR-122-fibronectin 1/serum response factor signaling cascade and its implication in hepatic fibrogenesis. Oncotarget. 2015;6:12224-12233. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 62] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 48. | Chen R, Wu JC, Liu T, Qu Y, Lu LG, Xu MY. MicroRNA profile analysis in the liver fibrotic tissues of chronic hepatitis B patients. J Dig Dis. 2017;18:115-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 49. | Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong Q, Qin L, Wu X, Zheng Y, Yang Y. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19:232-243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 567] [Cited by in RCA: 588] [Article Influence: 42.0] [Reference Citation Analysis (0)] |
| 50. | Liu N, Jiao T, Huang Y, Liu W, Li Z, Ye X. Hepatitis B virus regulates apoptosis and tumorigenesis through the microRNA-15a-Smad7-transforming growth factor beta pathway. J Virol. 2015;89:2739-2749. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 40] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 51. | Matsubara K, Tokino T. Integration of hepatitis B virus DNA and its implications for hepatocarcinogenesis. Mol Biol Med. 1990;7:243-260. [PubMed] |
| 52. | Ding D, Lou X, Hua D, Yu W, Li L, Wang J, Gao F, Zhao N, Ren G, Li L. Recurrent targeted genes of hepatitis B virus in the liver cancer genomes identified by a next-generation sequencing-based approach. PLoS Genet. 2012;8:e1003065. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 135] [Cited by in RCA: 153] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 53. | Li JW, Wan R, Yu CS, Co NN, Wong N, Chan TF. ViralFusionSeq: accurately discover viral integration events and reconstruct fusion transcripts at single-base resolution. Bioinformatics. 2013;29:649-651. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 58] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 54. | Lau CC, Sun T, Ching AK, He M, Li JW, Wong AM, Co NN, Chan AW, Li PS, Lung RW. Viral-human chimeric transcript predisposes risk to liver cancer development and progression. Cancer Cell. 2014;25:335-349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 227] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 55. | Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. EMBO J. 1992;11:961-971. [PubMed] |
| 56. | Chiu YT, Wong JK, Choi SW, Sze KM, Ho DW, Chan LK, Lee JM, Man K, Cherny S, Yang WL. Novel pre-mRNA splicing of intronically integrated HBV generates oncogenic chimera in hepatocellular carcinoma. J Hepatol. 2016;64:1256-1264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 57. | Li H, Sheng C, Wang S, Yang L, Liang Y, Huang Y, Liu H, Li P, Yang C, Yang X. Removal of Integrated Hepatitis B Virus DNA Using CRISPR-Cas9. Front Cell Infect Microbiol. 2017;7:91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 97] [Article Influence: 12.1] [Reference Citation Analysis (0)] |