Published online Jul 14, 2017. doi: 10.3748/wjg.v23.i26.4744
Peer-review started: February 11, 2017
First decision: March 16, 2017
Revised: April 23, 2017
Accepted: May 19, 2017
Article in press: May 19, 2017
Published online: July 14, 2017
Processing time: 154 Days and 7.3 Hours
To investigate toll-like receptor 2 (TLR2) and TLR4 expression, following bifidobacteria and low-dose EPEC endotoxin treatment, and intestinal barrier function in rat intestinal epithelial cell18 (IEC18).
Six experimental groups were established - normal control, EPEC, Bifidobacteria infantis (B. infantis), B. longum, B. bifidum, and B. youth groups. Optimal EPEC endotoxin concentration, bifidobacteria fold dilution, and treatment duration were determined. Quantitative real-time polymerase chain reaction and western blot, respectively, were conducted to detect TLR2 and TLR4 mRNA and protein expression in IEC-18 cells. Transepithelial electrical resistance (TEER) was measured by the EVOM chopstick voltohmmeter in each group. All experiments were conducted in triplicate and data were analyzed on SPSS 16.
TLR2 and TLR4 mRNA and protein expression in the EPEC group were significantly higher than in the control group (P < 0.05). TLR2 mRNA and protein expression in the B. infantis, B. longum and B. youth groups were significantly lower than in the normal control group (P < 0.05). TLR4 mRNA and protein expression in the B. bifidum and B. youth groups were significantly lower than in normal controls (P < 0.05). In addition, the TEER in B. infantis, B. longum, B. bifidum, and B. youth groups were decreased by 19%, 18%, 23% and 23%, respectively, after 120 min of intervention, as compared to the control group. However, the TEER in the EPEC group was significantly decreased by 67% in comparison to the normal control group (P < 0.05).
Bifidobacteria protect IEC-18 cells against injury by down-regulating TLR2 and TLR4 expression and enhance intestinal barrier function to protect the intestinal epithelial cells from pathogenic invasion.
Core tip: Toll-like receptor (TLRs) are key components of the innate immune system that trigger antimicrobial host defense responses. EPEC promoted the expression of toll-like receptor 2 (TLR2) and TLR4 and increased cell membrane permeability, where as bifidobacteria inhibited the expression of TLR2 and TLR4 and prevented TLR-mediated inflammation. Therefore, bifidobacteria can potentially play a protective role by inhibiting inflammation and preventing penetration of pathogenic bacteria in patients with inflammatory bowel disease.
- Citation: Yang X, Gao XC, Liu J, Ren HY. Effect of EPEC endotoxin and bifidobacteria on intestinal barrier function through modulation of toll-like receptor 2 and toll-like receptor 4 expression in intestinal epithelial cell-18. World J Gastroenterol 2017; 23(26): 4744-4751
- URL: https://www.wjgnet.com/1007-9327/full/v23/i26/4744.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i26.4744
The incidence of inflammatory bowel disease (IBD) is high in North America and Nordic countries, but lower in Asia. However, increasing incidence of IBD has been observed over the past few decades in parallel with better living standards and advances in diagnostics for IBD[1]. Current knowledge on the specific etiopathogenesis of IBD is still poorly understood. In general, however, IBD is considered to have several risk factors, including environmental, genetic, infectious and immune contributors[2]. The increasing incidence of IBD in developing countries indicates that progression of IBD epidemiology is associated with westernization and industrialization[3]. IBD is not only a polygenic disease, but also displays genetic heterogeneity. A recent study found that intraintestinal interaction of symbiotic gut bacteria is genetically determined and associated with epithelial dysfunction, which further induces IBD[4]. Thus far, no specific pathogens have been found to be associated with IBD. However, in an immunodeficient animal model of IBD, researchers found that intestinal inflammation did not occur in a sterile intestinal environment[5], suggesting that infective factors may play a key role in IBD pathogenesis.
More than 200 species in approximately one hundred trillion numbers constitute the characteristic distribution of commensal bacteria in a healthy human; of these, 99.9% of the bacteria grow on the intestinal mucosal surface, and along with those in feces, comprise three main species-bifidobacteria, Escherichia coli, and Bacteroidesfragilis[6]. Low-grade chronic inflammation in the intestinal lamina propria of healthy patients confers immune tolerance through various antibodies. However, such intestinal immune tolerance may be absent in patients with IBD, with resultant activation of intestinal mucosal immune response in the absence of a self-limiting mucosal immune response. Deficits in intestinal immune tolerance may majorly be attributed to intestinal flora imbalance[7]. Gut bacteria compete for nutrients and space with foreign pathogens in the intestinal environment, thereby effecting biological antagonism through inhibiting foreign pathogens from adhering to the intestinal mucosa[8]. Moreover, normal flora can promote the development of immune pathways and activation of macrophages to resist damage caused by foreign pathogens[9].
The intestinal epithelial barrier forms the first line of defense against invading pathogens. Several types of pattern-recognition receptors (PRRs), including a group of signaling toll-like receptors (TLRs) found in epithelial cells, play an important role in the intestinal mucosal immune system[10]. They facilitate recognition of microorganisms; for instance, TLR2 recognizes Gram-positive bacterial peptidoglycan, TLR4 recognizes lipopolysaccharide of Gram-negative bacteria, and TLR9 recognizes CpG DNA sequences of microorganisms[11]. Pathogen-associated molecular patterns (PAMPs) are recognized by TLRs and other PRRs[12]. Distinguishing between intestinal commensal bacteria and pathogens by intestinal epithelial cells (IECs) is a prerequisite for immune surveillance, although their mechanism of action is unclear.
Bifidobacterium, a bacterial commensal, reduces release of endotoxins through reduction of superfluous Gram-negative bacilli to normal levels, thereby functioning as a detoxifying agent[13]. Acidic products produced by protective bifidobacterium are essential to maintain normal intestinal conditions and improve mediation of intestinal defense by epithelial cells[14]. Furthermore, cholesterol in the intestine is converted to cholestane and fecal alkyls by acidic products generated by bifidobacterium[15]. In recent years, increasing attention has been paid to bifidobacterium-based therapy due to its stable efficacy and fewer side effects. Bifidobacterium has been used to treat diarrhea, IBD and other disorders of digestion[16-19].
This study was conducted to investigate the role of intestinal commensal bacteria in inducing immune tolerance and IBD pathogenesis. To evaluate intestinal barrier function, four different subtypes of bifidobacteria and the EPEC endotoxin were used to induce effects in rat normal small IEC-18, and their transepithelial electrical resistance (TEER) was detected after intervention. Changes of TLR2 and TLR4 expression levels were investigated to evaluate the effect of bifidobacteria on immune signal recognition and transduction in IECs.
Rat IEC-18 cells [Catalog No. CRL-1589; American Tissue Culture Collection (ATCC), Manassas, VA, United States] were purchased from the ATCC and cultured in 95% Dulbecco’s modified Eagle’s medium supplemented with 5% fetal bovine serum and 0.1 U/mL bovine insulin (Gibco, Carlsbad, CA, United States). Bifidobacteria infantis (B. infantis), B. longum, B. bifidum and B. youth were provided by the Institute of Light Industry (Wuhan, China). EPEC (serotype O127: B8) endotoxin was purchased from Sigma-Aldrich (St Louis, MO, United States) and constituted for use at a concentration of 1 mg/mL.
Six groups were established in this experiment, including normal control, EPEC, B. infantis, B. longum, B. bifidum, and B. youth. The EPEC group received intervention with the EPEC endotoxin, whereas the bifidobacteria groups underwent treatment with B. infantis, B. longum, B. bifidum and B. youth separately. Normal controls did not undergo any intervention.
In order to obtain optimal conditions for the intervention, IEC-18 cells were cultured in 6-well plates and treated with EPEC endotoxin at three different concentrations (0.5, 1 and 5 mg/mL) and with four different kinds of bifidobacteria (B. infantis, B. longum, B. bifidum and B. youth), with each subspecies diluted 100-fold and 300-fold, respectively. Then the cells were cultivated in a 37 °C humidified atmosphere, consisting of 95% air and 5% CO2. After intervention for 4, 8, 12 and 16 h, levels of TLR2 and TLR4 mRNA were detected in each intervention group by using qRT-PCR. Briefly, total mRNA was extracted from each intervention group using TRIzol reagent (Invitrogen Corp, Carlsbad, CA, United States) according to the manufacturer’s instruction. One microgram mRNA was used for cDNA synthesis in the PrimeScrip RT Reagent Kit (Takara Biotechnology, Dalian, China) under the following conditions: reverse transcription at 37 °C for 25 min, followed by incubation at 85 °C for 5 s in 20 μL reaction volume. Then, real-time PCR based on SYBR premix EX TAQ was conducted to quantify mRNA levels on the ABI StepOnePlus Real-Time PCR System (Applied Biosystems Inc, Foster City, CA, United States) under the following conditions: denaturation at 95 °C for 30 s, followed by 40 cycles of amplification (95 °C for 5 s, and 60 °C for 30 s). Primers were designed using NCBI Primer-BLAST as follows: TLR2 (forward, 5’-TGGAGGTCTCCAGGTCAAATC-3’; reverse, 5’-ACAGAGATGCCTGGGCAGAAT-3’); TLR4 (forward, 5’-TCCCTGCATAGAGGTACTTC-3’; reverse, 5’-CACACCTGGATAAATCCAGC-3’), GAPDH (forward, 5’-TCTTCCAGGAGCGAGATCCC-3’; reverse, 5’-TTCAGGTGAGCCCCAGCCTT-3’). Cycle threshold (CT) values were standardized to CT values of GAPDH, and 2-ΔΔCT was used to determine fold-change differences between intervention groups.
Based on the results obtained from qRT-PCR of TLR2 and TLR4 mRNA expression post-intervention in all the study groups, we obtained the optimal values of EPEC endotoxin concentration, bifidobacteria dilution concentrations, and duration of action. Thereafter, we conducted tests with these optimal concentrations of EPEC endotoxin and four different strains of bifidobacteria at the optimal duration of action, and expression levels of TLR2 and TLR4 proteins were detected in each group using western blotting.
Briefly, total protein was extracted from each group using RIPA buffer (Sigma-Aldrich) in accordance with the manufacturer’s instructions. Protein concentration was determined with the BCA Protein Assay Kit (BosterBio, Wuhan, China). For this, protein samples were resolved on a 10% SDS-PAGE (Bio-Rad, Hercules, CA, United States) and transferred to polyvinylene difluoride membranes (Millipore Corporation, Billerica, MA, United States). These blotted membranes were blocked with TBST buffer supplemented with 5% fat-free milk for 2 h at room temperature, and incubated with primary rabbit polyclonal antibody TLR2 and TLR4 (1:2000 dilution; Abcam Inc, Cambridge, MA, United States) overnight. GAPDH (BosterBio, Wuhan, China) was used as a loading control. Thereafter, these membranes were washed and incubated with peroxidase-conjugated secondary antibodies (BosterBio, Wuhan, China) for 2 h at room temperature according to the manufacturer’s instructions. Then the membranes were washed thrice with TBST for 10 min each at room temperature to terminate the reaction. Immunoreactive bands were visualized by enhanced chemiluminescence using SuperSignal West Pico Trail Kit (ThermoFisher, Rockford, IL, United States), and the intensity of the detected bands was analyzed using Image J software.
IEC-18 cells were cultured in Transwell chambers with 3.0-mm polycarbonate filters (Corning Costar, Corning, NY,United States) at 37 °C in a humidified atmosphere of 95% air and 5% CO2. Optimal concentrations of the EPEC endotoxin and four different types of bifidobacteria were added to the upper compartment of the Transwell plates in five intervention groups.An epithelial cell transmembrane resistance meter was used to test TEER changes in IEC-18 cells at 30, 60, 90 and 120 min in all of the five intervention groups and the normal control group. The TEER was calculated as: TEER (1 cm2) = Measured value of the resistance (Ω) × Effective membrane area (cm2).
All data were analyzed using SPSS 16.0 for Windows (SPSS Inc, Chicago, IL, United States). Between-group comparisons were conducted by two-way ANOVA. Three replicate wells were tested per assay, and each experiment was performed in triplicate. All statistical tests were two-tailed, and statistical significance was assumed for values with P < 0.05.
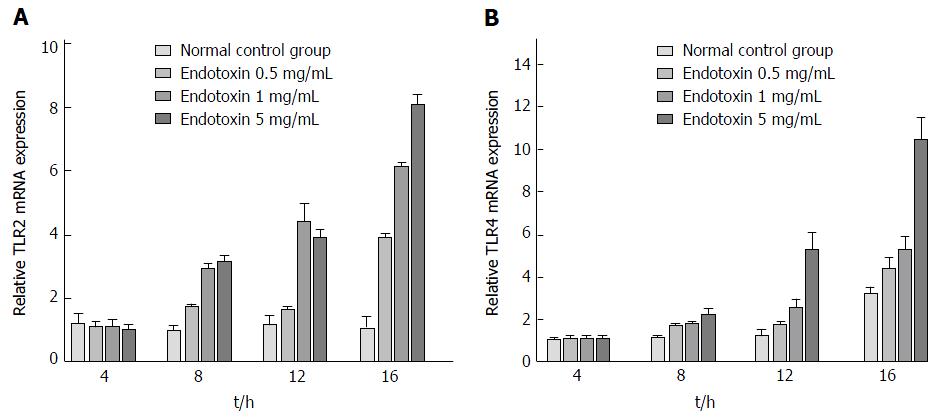
TLR2 mRNA expression was significantly increased after intervention with EPEC endotoxin at 12 h and 16 h, as compared with the control group (P < 0.05; Figure 1A). However, TLR2 mRNA expression did not change markedly in the intervention groups treated for 4 h and 8 h (P > 0.05; Figure 1A). Moreover, TLR4 mRNA expression was highest in the 16-h intervention groups (P < 0.05; Figure 1B). After treatment for 16 h, TLR2 mRNA expression was significantly higher in the 1 mg/mL and 5 mg/mL EPEC endotoxin groups, as compared to the 0.5 mg/mL EPEC and control groups (P < 0.05; Figure 1A). However, TLR2 mRNA expression did not differ considerably between the 1 mg/mL and 5 mg/mL EPEC groups (P > 0.05; Figure 1A). After treatment for 8 h and 16 h, TLR4 mRNA expression was significantly up-regulated in the 5 mg/mL EPEC group, as compared with that in the 0.5 mg/mL and 1 mg/mL EPEC groups and normal controls (P < 0.05; Figure 1B). However, there were no remarkable between-group differences in TLR4 mRNA expression among the different EPEC groups following intervention for 4 h and 12 h (P > 0.05; Figure 1B). We determined that the optimal EPEC endotoxin concentration was 5 mg/mL and optimal treatment duration was 16 h. qRT-PCR of IEC-18 cells post-treatment with EPEC endotoxin (0.5, 1 and 5 mg/mL) and with four different types of bifidobacteria diluted 100-fold and 300-fold at different time points, revealed that the optimal bifidobacteria dilution concentration was 300-fold.
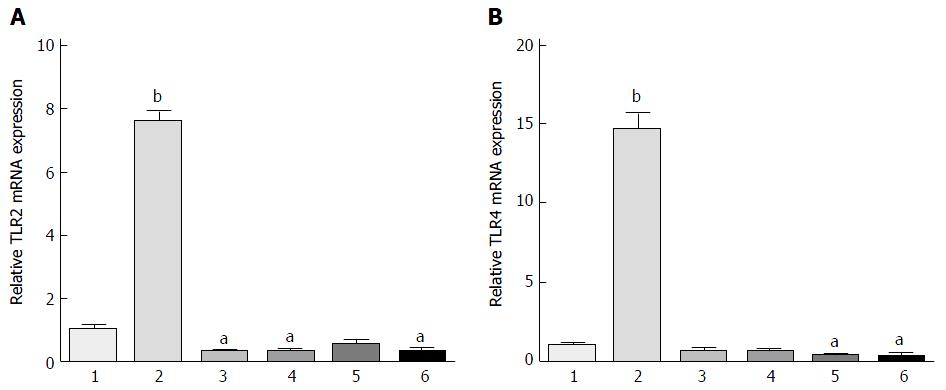
Next, we applied these optimal parameters (EPEC endotoxin 5 mg/mL, four different strains of bifidobacteria diluted 300-fold, for 16 h) and evaluated TLR2 and TLR4 protein expression (Figure 2). TLR2 and TLR4 mRNA expression was significantly up-regulated in the 5 mg/mL EPEC group, as compared with the control group (P < 0.001; Figure 2). TLR2 mRNA expression in the B. infant, B. longum and B. youth groups were significantly lower than in the control group (P < 0.05), although reduction in expression levels in the bifidum group did not reach significance (Figure 2A). Moreover, TLR4 mRNA expression in the B. bifidum and B. youth groups were significantly lower than in the control group (P < 0.05; Figure 2B).
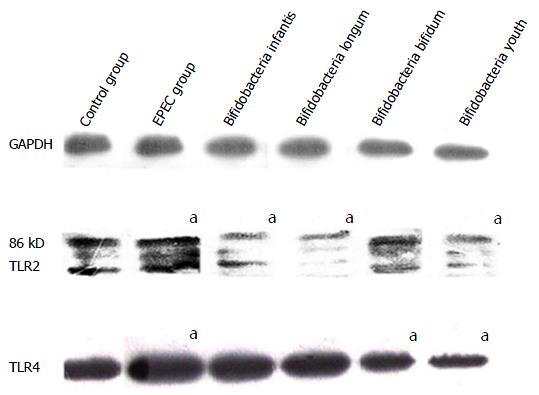
After treatment with EPEC endotoxin (5 mg/mL) and four different bifidobacteria strains (diluted 300-fold) for 16 h, TLR2 and TLR4 protein expression in the EPEC groups were significantly higher than in controls (P < 0.05; Figure 3). TLR2 protein expression in the B. infant, B. longum and B. youth groups were lower than those in the control group (P < 0.05; Figure 3). TLR4 protein expression in the B. bifidum and B. youth groups were down-regulated in comparison with that in the control groups (P < 0.05; Figure 3).
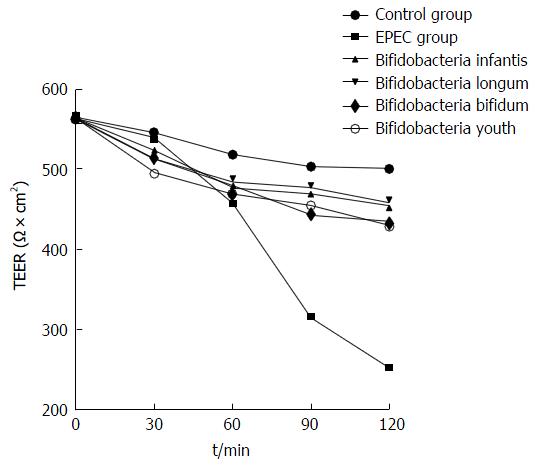
The TEER in the B. infantis, B. longum, B. bifidum and B. youth groups decreased by only 19%, 18%, 23% and 23%, respectively, post-intervention for 120 min compared with the control group. However, the TEER in the EPEC endotoxin intervention groups reduced significantly by 67%, as compared with the control group (P < 0.05; Figure 4).
Recent studies have shown that infective factors may play a key role in the pathogenesis of IBD, and that changes in the composition of intestinal microbiota are closely correlated with IBD progression[17]. In addition, it has been suggested that probiotics can potentially influence equilibrium of commensal and pathogenic bacteria, and thereby destroy homeostasis in inflammatory conditions[13]. Moreover, probiotics modulate intestinal mucosal immune function, which plays a protective role in maintaining the intestinal barrier function[20]. Therefore, we focus on understanding the role of probiotics in the induction and maintenance of IBD remission and discuss their influence on gut microbiota.
Besides bifidobacteria, lactobacilli, and other probiotic organisms, a large number of opportunistic pathogens exist as intestinal commensals. Immune stimulation from microbial antigens, food antigens, and allergens occur in intestinal epithelial tissue all the time, leading to different immune responses. Although there are various pathogenic bacteria in the intestinal epithelial tissue of the healthy population, these people seldom fall sick, which is attributed to immune tolerance and homeostasis. Moreover, macrobiotic imbalance will induce loss of intestinal immune tolerance and immune homeostasis and induce a series of immune responses, which eventually damages normal tissue. To investigate the effect of bacteria and probiotic interventions on intestinal immune barrier function and the signal transduction pathway,we established an in vitro normal intestinal immune system experimental model using the probiotic bacterial species bifidobacterium and toxins from EPEC.
Probiotics are defined as “living microorganisms which, when administered in adequate amounts, confer a health benefit on the host”[17]. Probiotics, as a food additive, are being adopted extensively and can help improve imbalances of intestinal flora induced by antibiotic use or by pathologic conditions, particularly IBD[2,20]. Probiotics can prevent or improve diarrheal symptoms, alleviate symptoms of lactose intolerance, enhance the immune system, and promote intestinal digestion and absorption of nutrients[7]. Therefore, research on the mechanism of action of probiotics will be beneficial to understand the etiopathogenesis and treatment of IBD, and could potentially be applied as a preventive measure against IBD. In addition to important applications in the treatment of gastrointestinal diseases, probiotics may have beneficial therapeutic effects on other systemic diseases, such as diabetes and disorders of the reproductive system.
Bifidobacterium is an anaerobic Gram-positive bacteria, with 32 subspecies, of which 28 subspecies have been detected in the human gut[13]. Intestinal bifidobacteria appear within 3 to 4 d postnatally, and bifidobacteria comprise approximately 25% of total intestinal bacteria[20]. The number of bifidobacteria reduce gradually with increasing age, and decrease to 7.9% in individuals over age 65[20]. Reduction in the number of bifidobacteria is suggested to be an indicator of lack of health[21].
EPEC was first discovered as a causative agent of diarrhea in epidemiological studies. Considerable EPEC proliferation occurs on mucosal surfaces of the duodenum, jejunum and ileum. The adhesion of EPEC to microvilli can lead to brush border damage, atrophy of microvilli, epithelial cell disorder and dysfunction, and, eventually, diarrhea[12]. EPEC and opportunistic pathogens grow in the normal intestine; however, when immune function is impaired, these bacteria can cause disease.
Due to a complicated interaction between the pathogen and the host immune system in microbial infection, microorganisms express a large amount of special molecules, such as the viral nucleic acid sequences known as the PAMPs[10]. PAMPs are recognized by a number of receptors, including PRRs. Following PAMP recognition by PRRs, downstream signaling pathways activate innate immunity and cell functions such as phagocytosis and release of inflammatory cytokines, which are important for a balanced adaptive immune response against pathogens[22,23]. TLRs appear to be the most important members of the PRR family; furthermore, TLR2 and TLR4, two main subtypes of the TLR family, play a key role in the maintenance of intestinal epithelial homeostasis[24]. Previous studies have shown that high expression of TLR2 and TLR4 is significantly associated with increased risk of IBD[25,26]. Activation of TLR4 by pathogenic bacteria damages the mucosal barrier function, facilitating pathogen translocation from the gut and release of proinflammatory factors, such as interleukin (IL)-1, IL-6, and tumor necrosis factor-alpha[15,24,27]. Therefore, TLR2 and TLR4 were selected in this research to validate the effect of intervention on intestinal immune barrier function and the signal transduction pathway, as well as to further study the mechanism of immune responses in the gut.
EPEC is highly pathogenic, and culturing IEC cells with live EPEC induces cell death. EPEC endotoxin, the main PAMP, is present in the cell wall of Gram-negative bacteria, whereas exopolysaccharide, a secretory product expressed by bifidobacterium during proliferation and death, is found in Gram-positive bacteria. Therefore, EPEC endotoxin and live bifidobacterium were selected as interventional agents for use in IEC-18 cells. Due to the uncertain effects of EPEC endotoxin and bifidobacteria in IEC-18 cells, it was necessary to treat the IEC-18 cells with different concentrations of EPEC endotoxin (0.5, 1 and 5 mg/mL) and four different kinds of bifidobacteria, diluted 100-fold and 300-fold, at different time points to identify the optimal EPEC endotoxin treatment concentration, bifidobacteria diluted concentration, and duration of action.
Following intervention with the optimal concentration of EPEC endotoxin (5 mg/mL) and four different strains of bifidobacteria(diluted 300-fold) for 16 h, TLR2 and TLR4 mRNA and protein expressions were up-regulated in the EPEC group but down-regulated in some bifidobacterium groups, as compared with those in the normal control group. From these results, we infer that after IECs are stimulated by pathogenic bacteria, bifidobacteria can induce immunologic tolerance to those bacteria through down-regulation of TLR2 and TLR4 expression or compete with pathogens for binding TLRs to prevent TLR-mediated inflammation. Therefore, bifidobacteria can provide a protective role or benefit through inhibition of inflammation in patients with IBD.
In our study, the TEERs in the B. longum, B. infantis, B. bifidum and B. youth groups were decreased only by 18%, 19%, 23% and 23%, respectively, after 120 min of intervention, as compared to the normal control group. However, the TEER in the EPEC group was reduced significantly by 67%. The TEER is inversely proportional to cell membrane permeability, and decreased TEER would lead to gap junction damage, increase cell osmotic pressure, and increase cell permeability; this could further result in pathogen penetration of the cell barrier and consequent inflammation[28,29]. These results indicate that EPEC may increase cell membrane permeability, thus playing a pathogenic role in patients with IBD. However, bifidobacteria exert a protective role in preventing penetration of pathogenic bacteria by the natural barrier function of the IEC. The lack of animal experimental model could be considered a major limitation of this study. Therefore, additional in vivo experiments should be conducted to further explore the mechanism of IBD in future research endeavors.
In conclusion, the EPEC endotoxin can promote TLR2 and TLR4 expression and increase cell membrane permeability in the IEC. In contrast, bifidobacteria can inhibit TLR2 and TLR4 expression and prevent TLR-mediated inflammation. Individualized use of a suitable probiotic may be helpful to design effective therapeutic strategies for symptom improvement in IBD.
This manuscript was edited by the Medjaden Academy and Research Foundation.
The incidence of inflammatory bowel disease (IBD) has increased in the past few decades, making it a global healthcare problem and an interesting research area. Current knowledge about the specific mechanism of IBD is deficient.
IBD has been considered to be caused by a variety of risk factors, including environmental, genetic, infectious and immune triggers. Toll-like receptors (TLRs) are key components of the innate immune system that trigger antimicrobial host defense responses. This study aimed to investigate alterations of intestinal barrier function and the TLR2 and TLR4 expression in intestinal epithelial cell (IEC)-18 cells and to further elucidate the mechanism of IEC immune defense function following induction by low-dose EPEC endotoxin and bifidobacteria.
This study analyzed the potential protective role of bifidobacteria on IECs through regulation of TLR2 and TLR4 expression and intestinal cell membrane permeability. The findings revealed that the EPEC endotoxin promoted TLR2 and TLR4 expression and increased intestinal cell membrane permeability. In contrast, bifidobacteria inhibited TLR2 and TLR4 expression and prevented TLR-mediated inflammation.
Bifidobacteria can provide a protective role through inhibiting inflammation and preventing penetration of pathogenic bacteria across the intestinal cell barrier in patients with IBD. Individualized use of a suitable probiotic may be helpful to design an effective and specific therapy to improve disease symptoms in IBD.
This is a very well designed, performed and written experimental study for investigation of the effect of enteropathogenic Escherichia coli endotoxin and the effect of bifidobacteria on the expression of TLR2 and TLR4 mRNA and protein in IECs and possible influence of intestinal barrier function of cells and its role in pathogenesis of IBD.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Caboclo JLF, Vorobjova T S- Editor: Ma YJ L- Editor: Filipodia E- Editor: Li D
| 1. | Baumgart DC, Bernstein CN, Abbas Z, Colombel JF, Day AS, D’Haens G, Dotan I, Goh KL, Hibi T, Kozarek RA. IBD Around the world: comparing the epidemiology, diagnosis, and treatment: proceedings of the World Digestive Health Day 2010--Inflammatory Bowel Disease Task Force meeting. Inflamm Bowel Dis. 2011;17:639-644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 59] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 2. | Mady R, Grover W, Butrus S. Ocular complications of inflammatory bowel disease. ScientificWorldJournal. 2015;2015:438402. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 3. | Ng SC, Bernstein CN, Vatn MH, Lakatos PL, Loftus EV, Tysk C, O’Morain C, Moum B, Colombel JF, Epidemiology . Geographical variability and environmental risk factors in inflammatory bowel disease. Gut. 2013;62:630-649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 470] [Cited by in RCA: 442] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 4. | Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119-124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3979] [Cited by in RCA: 3603] [Article Influence: 277.2] [Reference Citation Analysis (0)] |
| 5. | Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745-757. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1659] [Cited by in RCA: 1578] [Article Influence: 112.7] [Reference Citation Analysis (0)] |
| 6. | Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM. Enterotypes of the human gut microbiome. Nature. 2011;473:174-180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5822] [Cited by in RCA: 5038] [Article Influence: 359.9] [Reference Citation Analysis (2)] |
| 7. | Carbonero F, Benefiel AC, Alizadeh-Ghamsari AH, Gaskins HR. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front Physiol. 2012;3:448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 291] [Cited by in RCA: 380] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 8. | Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, McCoy KD, Macpherson AJ. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794-806. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 599] [Cited by in RCA: 656] [Article Influence: 46.9] [Reference Citation Analysis (0)] |
| 9. | Donohoe DR, Garge N, Zhang X, Sun W, O’Connell TM, Bunger MK, Bultman SJ. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13:517-526. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1507] [Cited by in RCA: 1355] [Article Influence: 96.8] [Reference Citation Analysis (0)] |
| 10. | Chang JH, McCluskey PJ, Wakefield D. Recent advances in Toll-like receptors and anterior uveitis. Clin Exp Ophthalmol. 2012;40:821-828. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 11. | Beutler BA. TLRs and innate immunity. Blood. 2009;113:1399-1407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 588] [Cited by in RCA: 642] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
| 12. | Zeuthen LH, Fink LN, Frøkiaer H. Toll-like receptor 2 and nucleotide-binding oligomerization domain-2 play divergent roles in the recognition of gut-derived lactobacilli and bifidobacteria in dendritic cells. Immunology. 2008;124:489-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 140] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 13. | Moratalla A, Caparrós E, Juanola O, Portune K, Puig-Kröger A, Estrada-Capetillo L, Bellot P, Gómez-Hurtado I, Piñero P, Zapater P. Bifidobacterium pseudocatenulatum CECT7765 induces an M2 anti-inflammatory transition in macrophages from patients with cirrhosis. J Hepatol. 2016;64:135-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 14. | Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543-547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1444] [Cited by in RCA: 1693] [Article Influence: 120.9] [Reference Citation Analysis (0)] |
| 15. | Ruiz PA, Hoffmann M, Szcesny S, Blaut M, Haller D. Innate mechanisms for Bifidobacterium lactis to activate transient pro-inflammatory host responses in intestinal epithelial cells after the colonization of germ-free rats. Immunology. 2005;115:441-450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 84] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 16. | Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084-1089. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1979] [Cited by in RCA: 2845] [Article Influence: 284.5] [Reference Citation Analysis (2)] |
| 17. | Saez-Lara MJ, Gomez-Llorente C, Plaza-Diaz J, Gil A. The role of probiotic lactic acid bacteria and bifidobacteria in the prevention and treatment of inflammatory bowel disease and other related diseases: a systematic review of randomized human clinical trials. Biomed Res Int. 2015;2015:505878. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 244] [Cited by in RCA: 239] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 18. | Martinez FA, Balciunas EM, Converti A, Cotter PD, de Souza Oliveira RP. Bacteriocin production by Bifidobacterium spp. A review. Biotechnol Adv. 2013;31:482-488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 147] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 19. | O’Mahony L, McCarthy J, Kelly P, Hurley G, Luo F, Chen K, O’Sullivan GC, Kiely B, Collins JK, Shanahan F. Lactobacillus and bifidobacterium in irritable bowel syndrome: symptom responses and relationship to cytokine profiles. Gastroenterology. 2005;128:541-551. [PubMed] |
| 20. | Fontana L, Bermudez-Brito M, Plaza-Diaz J, Muñoz-Quezada S, Gil A. Sources, isolation, characterisation and evaluation of probiotics. Br J Nutr. 2013;109 Suppl 2:S35-S50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 244] [Cited by in RCA: 193] [Article Influence: 16.1] [Reference Citation Analysis (1)] |
| 21. | Hodges K, Hecht G. Bacterial infections of the small intestine. Curr Opin Gastroenterol. 2013;29:159-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 22. | Robinson JA, Moehle K. Structural aspects of molecular recognition in the immune system. Part II: Pattern recognition receptors. Pure Appl Chem. 2014;10:1483-1538. [RCA] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 23. | Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. 2011;30:16-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1380] [Cited by in RCA: 1545] [Article Influence: 110.4] [Reference Citation Analysis (0)] |
| 24. | Cario E. Toll-like receptors in inflammatory bowel diseases: a decade later. Inflamm Bowel Dis. 2010;16:1583-1597. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 242] [Cited by in RCA: 259] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 25. | Cheng Y, Zhu Y, Huang X, Zhang W, Han Z, Liu S. Association between TLR2 and TLR4 Gene Polymorphisms and the Susceptibility to Inflammatory Bowel Disease: A Meta-Analysis. PLoS One. 2015;10:e0126803. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 26. | Bermudez-Brito M, Plaza-Díaz J, Muñoz-Quezada S, Gómez-Llorente C, Gil A. Probiotic mechanisms of action. Ann Nutr Metab. 2012;61:160-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 921] [Cited by in RCA: 692] [Article Influence: 53.2] [Reference Citation Analysis (0)] |
| 27. | Mann ER, Li X. Intestinal antigen-presenting cells in mucosal immune homeostasis: crosstalk between dendritic cells, macrophages and B-cells. World J Gastroenterol. 2014;20:9653-9664. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 76] [Cited by in RCA: 81] [Article Influence: 7.4] [Reference Citation Analysis (1)] |
| 28. | Lodemann U, Einspanier R, Scharfen F, Martens H, Bondzio A. Effects of zinc on epithelial barrier properties and viability in a human and a porcine intestinal cell culture model. Toxicol In Vitro. 2013;27:834-843. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 29. | Petrof EO. Probiotics and Gastrointestinal Disease: Clinical Evidence and Basic Science. Antiinflamm Antiallergy Agents Med Chem. 2009;8:260-269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 37] [Article Influence: 2.3] [Reference Citation Analysis (0)] |