Published online Mar 14, 2017. doi: 10.3748/wjg.v23.i10.1816
Peer-review started: October 1, 2016
First decision: December 1, 2016
Revised: December 21, 2016
Accepted: February 7, 2017
Article in press: February 8, 2017
Published online: March 14, 2017
Processing time: 165 Days and 2.5 Hours
To investigate whether microRNA (miR)-34a mediates oxaliplatin (OXA) resistance of colorectal cancer (CRC) cells by inhibiting macroautophagy via the transforming growth factor (TGF)-β/Smad4 pathway.
miR-34a expression levels were detected in CRC tissues and CRC cell lines by quantitative real-time polymerase chain reaction. Computational search, functional luciferase assay and western blotting were used to demonstrate the downstream target of miR-34a in CRC cells. Cell viability was measured with Cell Counting Kit-8. Apoptosis and macroautophagy of CRC cells were analyzed by flow cytometry and transmission electron microscopy, and expression of beclin I and LC3-II was detected by western blotting.
Expression of miR-34a was significantly reduced while expression of TGF-β and Smad4 was increased in CRC patients treated with OXA-based chemotherapy. OXA treatment also resulted in decreased miR-34a levels and increased TGF-β and Smad4 levels in both parental cells and the OXA-resistant CRC cells. Activation of macroautophagy contributed to OXA resistance in CRC cells. Expression levels of Smad4 and miR-34a in CRC patients had a significant inverse correlation and overexpressing miR-34a inhibited macroautophagy activation by directly targeting Smad4 through the TGF-β/Smad4 pathway. OXA-induced downregulation of miR-34a and increased drug resistance by activating macroautophagy in CRC cells.
miR-34a mediates OXA resistance of CRC by inhibiting macroautophagy via the TGF-β/Smad4 pathway.
Core tip: This study demonstrated a significant association between microRNA (miR)-34a expression and the acquired chemoresistance to oxaliplatin (OXA) in colorectal cancer (CRC). miR-34a mediates OXA resistance through its inhibitory effects on macroautophagy by the transforming growth factor (TGF)-β/Smad4 pathway. Our findings suggest that miR-34a is a potential therapeutic target for improving the chemotherapeutic effect of OXA in CRC.
- Citation: Sun C, Wang FJ, Zhang HG, Xu XZ, Jia RC, Yao L, Qiao PF. miR-34a mediates oxaliplatin resistance of colorectal cancer cells by inhibiting macroautophagy via transforming growth factor-β/Smad4 pathway. World J Gastroenterol 2017; 23(10): 1816-1827
- URL: https://www.wjgnet.com/1007-9327/full/v23/i10/1816.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i10.1816
Colorectal cancer (CRC) is one of the most common causes of cancer-related deaths, leading to approximately 600000 deaths worldwide annually[1,2]. Current standard treatments for CRC include surgical resection, systematic chemotherapy, radiotherapy or a combination of these methods. Oxaliplatin (OXA), a third-generation platinum compound, is the first platinum-based compound to show efficacy in the treatment of CRC[3]. Despite this demonstrated efficacy, the advances in chemotherapy for CRC have been limited because of its resistance to OXA. Therefore, revealing the underlying mechanism for the development of chemoresistance is necessary for developing effective chemotherapeutic agents.
MicroRNAs (miRNAs) are a group of noncoding RNAs that have been highly conserved during evolution and have emerged recently as potent regulators of gene expression, cell proliferation, apoptosis and tumorigenesis[4,5]. Our previous study demonstrated that miR-34a inhibits epithelial mesenchymal transition in human cholangiocarcinoma by targeting Smad4 through the transforming growth factor (TGF)-β/Smad pathway[6]. Recent research has found that miR-34a is one of the well-characterized miRNAs and is consistently downregulated in various types of malignancies, including CRC[7-9]. More importantly, miR-34a has been reported to be involved in drug resistance and regulating chemosensitivity in CRC[10].
Macroautophagy is a catabolic degradation process that is required to maintain cellular homeostasis[11]. Some researchers have shown that macroautophagy is involved in the regulation of chemoresistance. He et al[12] found that miR-21 mediates sorafenib resistance of hepatocellular carcinoma cells via inhibition of macroautophagy through the PTEN/Akt pathway. Pan et al[13] have reported that miR-200b regulates macroautophagy associated with chemoresistance in human lung adenocarcinoma. Whether miR-34a is involved in mediating the chemoresistance of OXA by affecting macroautophagy activity remains unclear.
In the present study, we found that expression of miR-34a was significantly reduced while expression of TGF-β and Smad4 was increased in CRC patients treated with OXA-based chemotherapy. OXA treatment also decreased miR-34a levels and increased TGF-β and Smad4 levels in both parental cells and OXA-resistant CRC cells. Activation of macroautophagy contributed to OXA resistance in CRC cells. In addition, our data indicated that expression of Smad4 and miR-34a in CRC patients had a significant inverse correlation and overexpression of miR-34a inhibited macroautophagy activation by directly targeting Smad4 through the TGF-β/Smad4 pathway. We further demonstrated that OXA-induced downregulation of miR-34a increased drug resistance by activating macroautophagy in CRC cells. We identified a novel mechanism by which miR-34a mediates OXA resistance of CRC by inhibiting autophagy via the TGF-β/Smad4 pathway.
We included 30 CRC patients (median age: 61 years, range: 48-75 years) who underwent potentially curative surgery between 2013 and 2015 at the Second Affiliated Hospital of Harbin Medical University (Harbin, China). CRC was verified by a pathologist and patients received OXA-based combination chemotherapy. Blood samples were taken after obtaining informed consent according to institutional guidelines and the study was approved by the Institutional Review Board of Harbin Medical University, Harbin, China. The protocols used in the study were approved by the Committee of Human Subjects Protection of our hospital.
To establish the chemoresistance model in vitro, the human CRC cell line HT29 obtained from the Cell Bank of Chinese Academy of Sciences (Shanghai, China) was seeded in a six-well plate at 104 cells/well and 2 μmol/L OXA was added to the medium. The medium was changed after 48 h. The concentration of OXA was increased by 1 μmol/L for each subculture. After subculture and incubation with OXA solution was repeated 10 times, OXA-resistant cell lines were obtained, termed as HT29-OXA, and continuously maintained by culturing them in complete culture medium containing 2 μmol/L OXA[14,15].
Real-time polymerase chain reaction (RT-PCR) was used to confirm the expression levels of mRNAs and miRNAs as previously described[6]. For mRNA detection, total RNA from cultured cells and blood samples was extracted using TRIzol (Invitrogen, Carlsbad, CA, United States). Reverse transcription was performed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, United States). Total miRNA from cultured cells and fresh surgical tissues was extracted using the mirVana miRNA Isolation Kit (Ambion, Austin, TX, United States). cDNA was synthesized from 2 μg total RNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Expression of miRNA and mRNA was assessed with qRT-PCR using Power SYBR Green Master Mix in an Applied Biosystems 7500 Sequence Detection System. The expression level of mRNA and miRNA was defined based on the threshold cycle (Ct), and relative expression levels were calculated using the 2−ΔΔCt method, with the expression level of β-actin mRNA and U6 small nuclear RNA as a reference gene. The names of the genes and the primers are listed in Supplementary Table 1.
Protein lysates were separated using SDS-PAGE and transferred to nitrocellulose membranes (Amersham Pharmacia Biotech, Piscataway, NJ, United States). Membranes were probed with the appropriate primary antibodies (Supplementary Table 2). Alexa Fluor 680 donkey anti-mouse IgG (H + L) or Alexa Fluor 680 donkey anti-rabbit IgG (H + L) were used as secondary antibodies (1:5000; Invitrogen). Fluorophores were detected using the Odyssey Infrared Imaging System (Li-Cor, Lincoln, NE, United States).
Transfection of pre-miR-34a was conducted as previously described[6]. A synthetic miR-34a mimic (GenePharma, Shanghai, China) was used to increase miR-34a expression. A scrambled oligonucleotide (GenePharma) was used as a control. Transfection was performed using Lipofectamine 2000 transfection reagent (Invitrogen). A mixture of Lipofectamine 2000 and RNA was added to CRC cells, which were 70% confluent, for 4-6 h, and the cells were then incubated for 24 h in fresh medium. The cells were harvested using lysis buffer for luciferase assay. Total RNA and protein were prepared 48 h after transfection and used for qRT-PCR or western blot analysis.
The luciferase reporter assay was conducted as described previously[6]. The fragment containing miR-34a-binding sites in the Smad4 3’-untranslated region (UTR) was amplified by PCR and inserted downstream of the firefly luciferase gene in a pGL3-promoter vector (Promega, Madison, WI, United States). The mutant reporter plasmids were constructed using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA, United States). These constructed plasmids were all sequenced to confirm their orientation. Luciferase activity was measured with the Dual-Luciferase Reporter Assay System (Promega) as described previously[16]. Promoter activities were expressed as the ratio between firefly luciferase and Renilla luciferase activities.
The cells were seeded into a 96-well plate (3 × 103/well) in triplicate and cultured overnight. The culture medium was replaced with fresh fetal-calf-serum-free medium containing vehicle or testing reagents for 24 h. Cell viability was measured with the Cell Counting Kit-8 (CCK-8) (Boster Bio, Wuhan, China). Untreated cells served as controls. Cell viability (%) was calculated according to the formula: experimental OD value/control OD value × 100%.
The evaluation of macroautophagy and apoptosis by flow cytometry was performed as previously described[17]. Apoptosis was examined using the Annexin V-FITC/PI Kit (Becton Dickinson, Franklin Lakes, NJ, United States). Cells were collected in 400 mL medium. Following resuspension, approximately 10 mL Annexin V solution and 5 mL propidium iodide were added and incubated for 30 min at room temperature in the dark. The cell suspension was analyzed by flow cytometry (Becton Dickinson). Cell Quest software was used to analyze 104 cells.
Cell macroautophagy was detected by monodansylcadaverine (MDC; Sigma-Aldrich, St Louis, MO, United States) staining. Cells were washed with phosphate-buffered saline (PBS) and then incubated with 0.05 mmol/L MDC in PBS at 37 °C for 45 min. After incubation, the cells were washed three times with PBS and analyzed by flow cytometry immediately. Cell Quest software was used to analyze 104 cells.
All the presented data were expressed as the mean ± SD and representative results were from at least three independent experiments. Statistical comparisons were calculated by Student’s two-tailed t test. When multiple comparisons were possible, ANOVA coupled with Tukey correction was used. Correlation analysis between relative expressions of Smad4 and miR-34a was examined by logistic regression analysis. P < 0.05 was considered statistically significant. Statistical analysis was carried out using SPSS version 21 (IBM Corporation, Armonk, NY, United States) or GraphPad Prism version 5.0 (La Jolla, CA, United States).
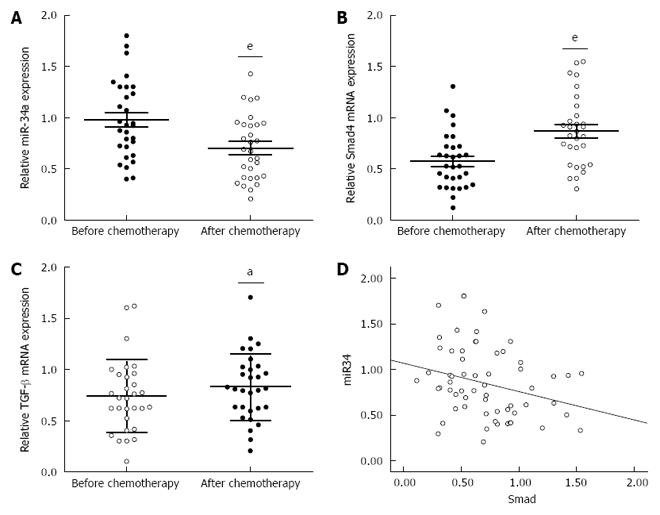
To investigate the underlying mechanism for the resistance of CRC patients to OXA treatment, we focused on identifying the changes in miR-34a and TGF-β/Smad4 pathway expression after OXA-based combination chemotherapy. miR-34a expression was significantly decreased while TGF-β and Smad4 mRNA expression was significantly increased in the blood samples of CRC patients after chemotherapy (Figure 1A-C). Spearman correlation analysis was applied to compare the relative expression levels of Smad4 and miR-34a in CRC patients. We obtained a statistically significant inverse correlation (R = -0.283, P = 0.029) in 30 CRC patients (Figure 1D). These data suggest that downregulation of miR-34a and upregulation of TGF-β/Smad4 may be related to OXA-based combination chemotherapy.
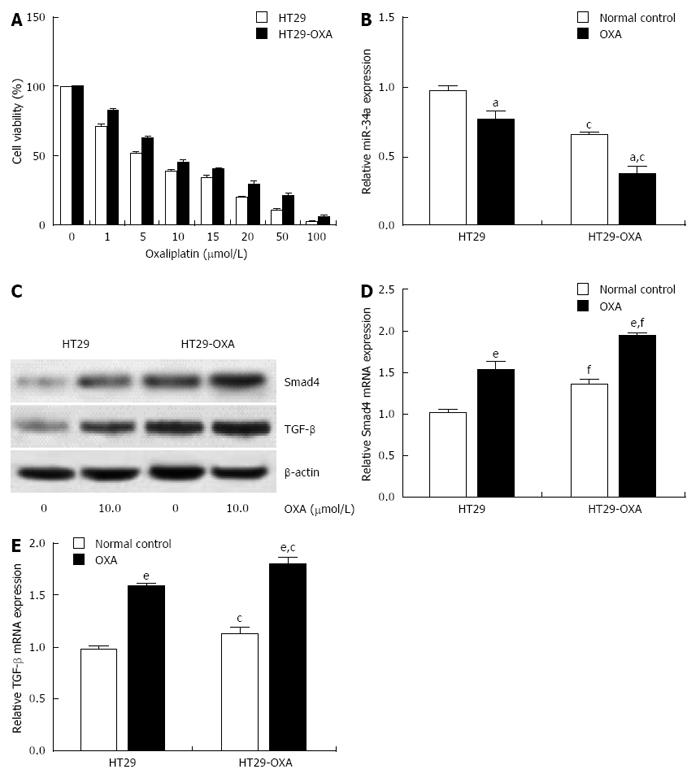
To determine further whether miR-34a levels are correlated with chemotherapeutic response, the OXA-resistant cell line, HT29-OXA, was established by chronic exposure of HT29 cells to increasing concentrations of OXA. Incubation of OXA with HT29 cells reduced their viability in a concentration-dependent manner. However, HT29-OXA cells became resistant to OXA as their viability was significantly higher than that of respective parental cells when exposed to the same concentration of OXA (Figure 2A). Expression of miR-34a in HT29 and HT29-OXA cells was downregulated when exposed to 10 μmol/L OXA for 48 h, although HT29-OXA cells expressed lower levels of miR-34a than HT29 cells with or without OXA treatment (Figure 2B).
Smad4 is a known miR-34a target gene in some types of tumor cells. We examined expression of TGF-β and Smad4 in HT29 and HT29-OXA cells in the presence or absence of OXA treatment. Incubation of OXA induced increased expression of TGF-β and Smad4 in HT29 and HT29-OXA cells. HT29-OXA cells expressed higher levels of TGF-β, resulting in upregulation of Smad4 compared with HT29 cells (Figure 2C-E). These results indicate that sustained exposure of CRC cells to OXA could inhibit miR-34a expression and activate the TGF-β/Smad4 pathway.
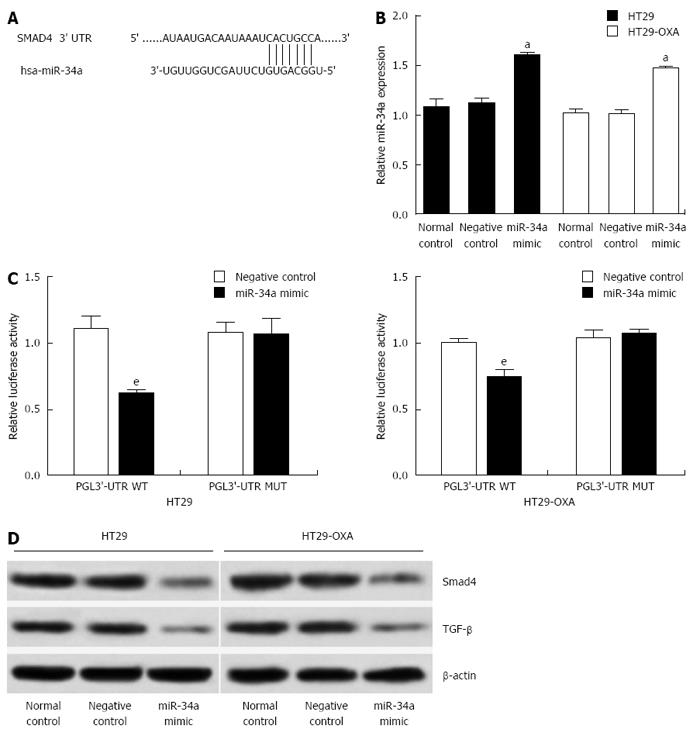
To investigate the underlying mechanism of miR-34a in CRC, targetscan (http://www.targetscan.org) and miRanda (http://www.microrna.org) were used to search for potential targets of miR-34a. Consistent with our previous study[6], the result showed that the 3’-UTR of Smad4 contained the complementary site for the seed region of miR-34a (Figure 3A). To verify that Smad4 is one of the direct targets of miR-34a in OXA-resistant and parental CRC cells, we constructed a reporter vector consisting of the luciferase coding sequence followed by the 3’-UTR of Smad4. A dual luciferase reporter assay was performed in HT29 and HT29-OXA cells. miR-34a significantly decreased the luciferase activity of the wild-type Smad4 3’-UTR compared with the vector-only control (Figure 3B). Additionally, partial mutation of the perfectly complementary sites in the 3’-UTR of Smad4 abolished the suppressive effect due to the disruption of the interaction between miR-34a and Smad4.
miR-34a mimics were transfected into HT29 and HT29-OXA cells to increase the endogenous level of miR-34a to investigate further the biological function of miR-34a in CRC cells (Figure 3C). Compared to the control groups, transfecting with miR-34a mimics downregulated Smad4 and TGF-β expression in HT29 and HT29-OXA cells (Figure 3D). These data suggest that Smad4 is a direct target of miR-34a and miR-34a mediated TGF-β/Smad4 pathway in CRC cell lines.
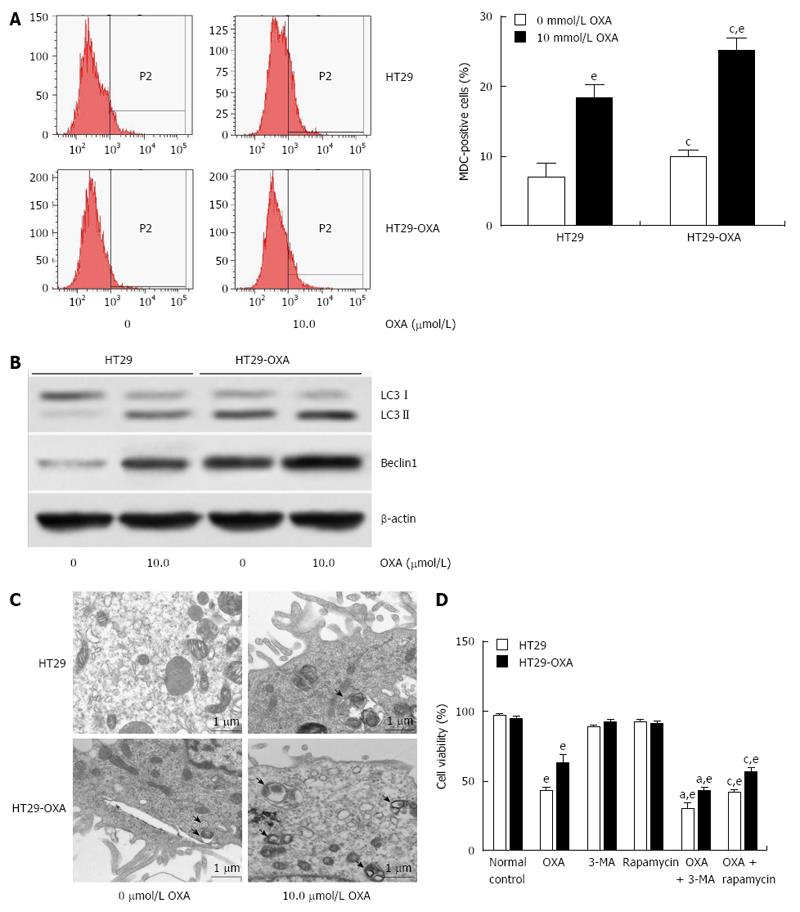
Macroautophagy is a key regulator of resistance to OXA[18]. We investigated whether activation of macroautophagy contributes to the acquired resistance to OXA. Incubation with OXA led to more MDC-positive HT29 and HT29-OXA cells, but there were more MDC-positive HT29-OXA than HT29 cells in the presence or absence of OXA treatment (Figure 4A). Western blotting confirmed that the HT29-OXA cells had significantly lower LC3-II and beclin 1 intensity than TH29 cells had in the presence or absence of OXA treatment (Figure 4B). Representative electron micrographs demonstrated less autophagic vacuole formation in the HT29-OXA cells than HT29 cells in the presence or absence of OXA treatment (Figure 4C). Suppression of macroautophagy by 3-methyladenine (3-MA) increased OXA-induced cell death. In contrast, rapamycin, an inhibitor of mammalian target of rapamycin, and positive regulator of macroautophagy, protected OXA-resistant cells against OXA-induced reduction in cell viability (Figure 4D).
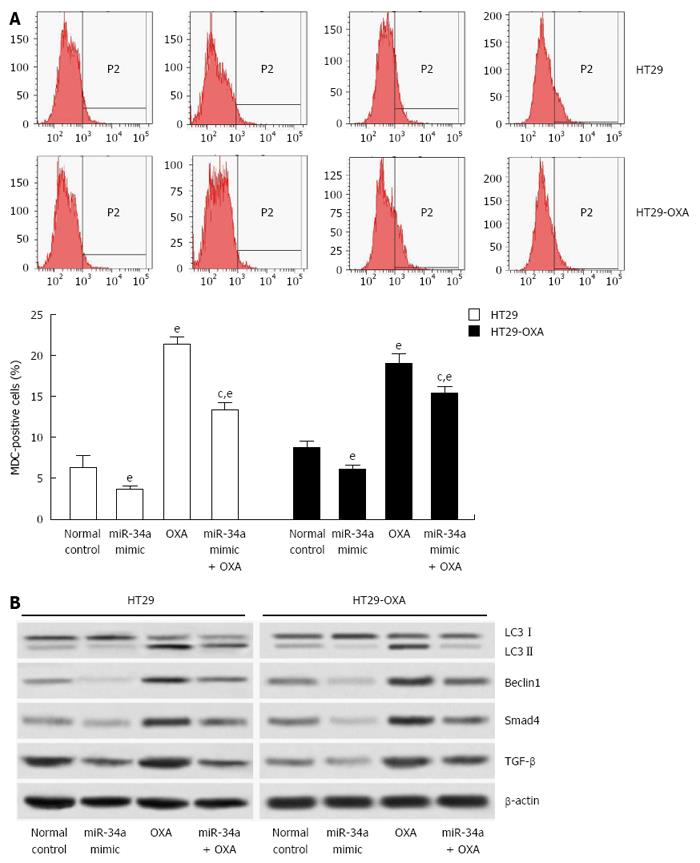
Since macroautophagy participates in OXA resistance in CRC cells[19], we investigated the effects of miR-34a on macroautophagy of CRC cells. miR-34a mimics inhibited macroautophagy in parental and OXA-resistant cells, and fewer MDC-stained cells were observed in cells transfected with miR-34a mimics than in normal control cells. OXA treatment increased autophagy of parental and OXA-resistant cells, as shown by the presence of more MDC-stained cells. However, induction of macroautophagy was inhibited by treatment with miR-34a mimics in parental and OXA-resistant cells (Figure 5A). miR-34a mimics decreased expression of LC3-II, beclin-1, Smad4 and TGF-β, while OXA treatment increased their expression, and in agreement with quantitative flow cytometry, induction was impaired by treatment with miR-34a mimics mixed with OXA in parental and OXA-resistant cells (Figure 5B). These results indicate that miR-34a participates in the regulation of OXA-induced macroautophagy in CRC cells through downregulation of the TGF-β/Smad4 signaling pathway.
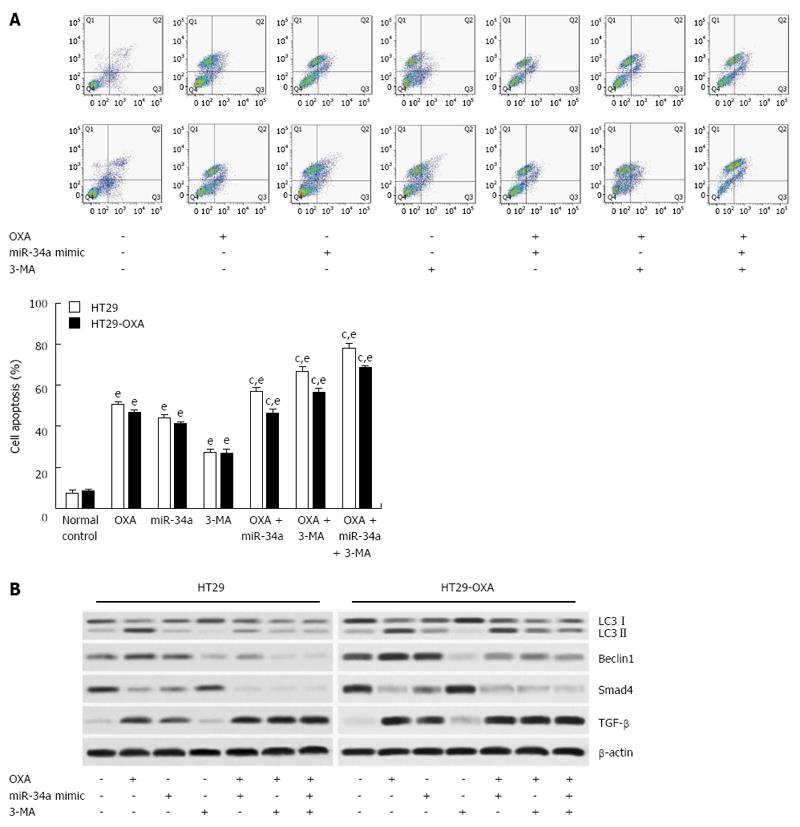
To determine the mechanism of macroautophagy in OXA-induced apoptosis in CRC cells, we exposed cells to OXA for 48 h with miR-34a mimics or 3-MA and assessed apoptotic rate. 3-MA attenuated the activation of macroautophagy and the anti-apoptotic capacity in HT29 and HT29-OXA cells treated with OXA, as shown by a higher apoptotic rate (Figure 6A). In addition, lower expression of LC3-II, beclin 1, Bax, Smad4 and TGF-β and higher expression of Bcl-2 were found compared with the control group and the OXA treatment group (Figure 6B). Moreover, when treated with OXA, miR-34a mimics and 3-MA, more significantly inhibitory effects on autophagic activity and the anti-apoptotic rate in HT29 and HT29-OXA cells were achieved. These results indicated that the activation of macroautophagy may serve as a protective mechanism against OXA-induced apoptosis by inhibiting miR-34a through the TGF-β/Smad4 pathway in CRC cells.
In the present study, we investigated whether miR-34a mediates OXA resistance of colorectal cancer cells by inhibiting macroautophagy via the TGF-β/Smad4 pathway. We found that miR-34a levels were significantly decreased in CRC patients after systematic chemotherapy by OXA and in CRC cells treated with OXA. We further demonstrated that miR-34a downregulation increased macroautophagy activation by targeting Smad4. More important, macroautophagy inhibition abolished miR-34a downregulation-induced apoptosis. These data suggest that OXA-induced downregulation of miR-34a increases drug resistance by activating macroautophagy in CRC cells. The TGF-β/Smad4 pathway plays an important role in regulating macroautophagy response[20]. Further investigation found that OXA treatment enhanced macroautophagy via inhibiting the expression of miR-34a and activating the TGF-β/Smad4 pathway, which at least partly enacted the protective role of macroautophagy on CRC cell apoptosis induced by OXA.
CRC is a major cause of cancer-related mortality worldwide and its incidence is increasing. Chemotherapy can be used in addition to surgery in certain cases as an adjuvant therapy. However, drug resistance represents a major obstacle for chemotherapy of CRC. OXA is a promising drug for the treatment of advanced CRC. Despite a rapid shrinkage in tumor mass following chemotherapy, the resistance of cancer cells to OXA frequently results in the subsequent recurrence and metastasis of cancer, and the exact mechanism for this effect is still not well understood. Therefore, there is an urgent need to investigate these adverse factors that might help us better understand the mechanism of drug resistance in CRC patients.
Macroautophagy is considered to play a dual role in the regulation of cell fate in the context of cancer treatment[21,22]. Numerous studies have focused on the association between the cytoprotective effects of autophagy and the development of chemoresistance. Sustained drug exposure can induce an imbalance in apoptotic pathways and lead to resistance to apoptosis[23]. Modest macroautophagy is implicated in promoting chemoresistance of cancer cells and attenuating the efficacy of chemotherapy. Lv et al[24] have demonstrated that upregulation of CD44v6 contributes to acquired chemoresistance via the modulation of macroautophagy in CRC cells. Yang et al[14] have reported that macroautophagy contributes to the enrichment and survival of CRC stem cells under OXA treatment. These observations suggest that macroautophagy serves as a pro-survival mechanism that promotes chemoresistance, and selective inhibition of macroautophagy regulators has the potential to improve chemotherapeutic regimens. Here, we found that OXA treatment promoted macroautophagy activation in CRC cells, which protected the cells from OXA-induced apoptosis.
miRNAs are frequently dysregulated in chemoresistant cancers, and they have been shown to target macroautophagy-related genes or modulators[12,13]. It has been well established that miR-34a is an important anti-oncogene by regulation of different downstream targets in various types of cancer[25]. miR-34a has also been identified as one of the molecular species associated with drug resistance in CRC[10]. Here, we reported that miR-34a levels were significantly decreased in CRC patients after systematic chemotherapy by OXA and in CRC cells treated with OXA. Expression of miR-34a was downregulated in OXA-resistant cells. Forced expression of miR-34a promoted OXA-induced apoptosis in parental and OXA-resistant cells.
Among the miR-34a targets, Smad4 is a key regulator of the TGF-β/Smad pathway, which is well characterized as a positive mediator of macroautophagy[6]. Based on our previous study, here we found that mRNA expression of TGF-β and Smad4 was upregulated in CRC patients after OXA chemotherapy. Moreover, expression levels of miR-34a and Smad4 were inversely correlated in human blood samples from CRC patients. Our results showed forced upregulation of miR-34a significantly inhibited protein expression of Smad4 and TGF-β. Contrasting results were observed when the CRC cells were treated with OXA. We have also applied bioinformatic methods to demonstrate that Smad4 is a potential target of miR-34a[6]. Our data reveal that miR-34a directly targets the 3’-UTR of Smad4 and that ectopic expression of miR-34a represses Smad4 and TGF-β protein levels in parental and OXA-resistant CRC cells. Our results demonstrate that the TGF-β/Smad4 pathway at least partly is involved in regulating miR-34a-inhibited macroautophagy and contributes to OXA resistance in CRC.
Our data demonstrate that transfection of miR-34a mimics enhances the therapeutic effect of OXA by inhibition of cell macroautophagy and improves the efficacy of OXA against OXA-resistant CRC cells. The macroautophagy inhibitor, 3-MA, enhances the pro-apoptotic effect in cells treated with OXA or exogenous miR-34a. These data suggest that activation of macroautophagy protects CRC cells from OXA-induced apoptosis by inhibiting miR-34a expression.
In summary, the results of this study demonstrate a significant association between miR-34a expression and the acquired chemoresistance of OXA in CRC. The miR-34a-mediated OXA resistance occurs through its inhibitory effects on macroautophagy via regulation of the TGF-β/Smad4 pathway. Our findings suggest that miR-34a could be a potentially therapeutic target for improving the chemotherapeutic effect of OXA in CRC.
Colorectal cancer (CRC) is one of the most common cancers worldwide. Oxaliplatin (OXA)-based systemic chemotherapy combined with surgical resection plays an important role in the treatment of CRC. However, chemoresistance to OXA is a major limitation in the clinic, and the underlying mechanism is not clear.
miRNAs have been conserved during evolution and have emerged recently as potent regulators of gene expression, cell proliferation, apoptosis and tumorigenesis. The authors, in a previous study, have demonstrated that miRNA (miR)-34a inhibits epithelial mesenchymal transition in human cholangiocarcinoma by targeting Smad4 through the transforming growth factor (TGF)-β/Smad pathway. More importantly, miR-34a is involved in drug resistance and regulating chemosensitivity in CRC. Some researchers have shown that autophagy is involved in the regulation of chemoresistance. However, whether miR-34a is involved in mediating the chemoresistance of OXA by affecting autophagic activity remained unclear.
This study found that expression of miR-34a was significantly reduced while expression of TGF-β and Smad4 was increased in CRC patients treated with OXA-based chemotherapy. OXA treatment also decreased miR-34a and increased TGF-β and Smad4 levels in parental cells and OXA-resistant CRC cells. Activation of autophagy contributed to OXA resistance in CRC cells. In addition, the data in this study indicated that the relative expression levels of Smad4 and miR-34a in CRC patients had a significant inverse correlation, and overexpression of miR-34a inhibited autophagy activation by directly targeting Smad4 through the TGF-β/Smad4 pathway. The authors further demonstrated that OXA-induced downregulation of miR-34a increased drug resistance by activating autophagy in CRC cells. Moreover, they identified that miR-34a mediated TGF-β/Smad4 pathway induced macroautophagy, which may represent a novel mechanism regulating chemoresistance in CRC.
The findings in this study suggest that miR-34a could be a potentially therapeutic target for improving the chemotherapeutic effect of OXA in CRC.
OXA is a third-generation platinum compound, and is the first platinum-based compound to show efficacy in the treatment of CRC. miRNAs are a group of noncoding RNAs that have been highly conserved during evolution and have emerged recently as potent regulators of gene expression, cell proliferation, apoptosis and tumorigenesis. Macroautophagy is a catabolic degradation process that is required to maintain cellular homeostasis.
In the original article of Sun et al the authors demonstrated that a significant association exists between miR-34a expression and the acquired chemoresistance of OXA in CRC. The miR-34a mediated OXA resistance was found to be mediated by the inhibitory effects of miR34a on autophagy by regulating the TGF-β/Smad4 pathway. Their findings suggest that miR-34a could be a potentially therapeutic target for improving the OXA-based chemotherapeutic effect in CRC. The study is well designed, and moderately presented.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Sipos F S- Editor: Gong ZM L- Editor: Filipodia E- Editor: Wang CH
| 1. | Ung L, Lam AK, Morris DL, Chua TC. Tissue-based biomarkers predicting outcomes in metastatic colorectal cancer: a review. Clin Transl Oncol. 2014;16:425-435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 2. | Sostres C, Gargallo CJ, Lanas A. Aspirin, cyclooxygenase inhibition and colorectal cancer. World J Gastrointest Pharmacol Ther. 2014;5:40-49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 55] [Cited by in RCA: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 3. | Meyerhardt JA, Mayer RJ. Systemic therapy for colorectal cancer. N Engl J Med. 2005;352:476-487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 859] [Cited by in RCA: 859] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 4. | Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14460] [Cited by in RCA: 16079] [Article Influence: 1004.9] [Reference Citation Analysis (2)] |
| 5. | Matamala N, Vargas MT, González-Cámpora R, Miñambres R, Arias JI, Menéndez P, Andrés-León E, Gómez-López G, Yanowsky K, Calvete-Candenas J. Tumor microRNA expression profiling identifies circulating microRNAs for early breast cancer detection. Clin Chem. 2015;61:1098-1106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 156] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 6. | Qiao P, Li G, Bi W, Yang L, Yao L, Wu D. microRNA-34a inhibits epithelial mesenchymal transition in human cholangiocarcinoma by targeting Smad4 through transforming growth factor-beta/Smad pathway. BMC Cancer. 2015;15:469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 81] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 7. | Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017-5022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 593] [Cited by in RCA: 601] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 8. | Zhou JY, Chen X, Zhao J, Bao Z, Chen X, Zhang P, Liu ZF, Zhou JY. MicroRNA-34a overcomes HGF-mediated gefitinib resistance in EGFR mutant lung cancer cells partly by targeting MET. Cancer Lett. 2014;351:265-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 73] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 9. | Ranadive IN, Sussman DA. Prevention of Colorectal Cancer: The Future Is Now. Curr Colorectal Cancer Rep. 2014;10:1-10. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 10. | Toden S, Tran HM, Tovar-Camargo OA, Okugawa Y, Goel A. Epigallocatechin-3-gallate targets cancer stem-like cells and enhances 5-fluorouracil chemosensitivity in colorectal cancer. Oncotarget. 2016;7:16158-16171. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 134] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 11. | Duncan C, Papanikolaou T, Ellerby LM. Autophagy: polyQ toxic fragment turnover. Autophagy. 2010;6:312-314. [PubMed] |
| 12. | He C, Dong X, Zhai B, Jiang X, Dong D, Li B, Jiang H, Xu S, Sun X. MiR-21 mediates sorafenib resistance of hepatocellular carcinoma cells by inhibiting autophagy via the PTEN/Akt pathway. Oncotarget. 2015;6:28867-28881. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 171] [Cited by in RCA: 184] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 13. | Pan B, Feng B, Chen Y, Huang G, Wang R, Chen L, Song H. MiR-200b regulates autophagy associated with chemoresistance in human lung adenocarcinoma. Oncotarget. 2015;6:32805-32820. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 63] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 14. | Yang HZ, Ma Y, Zhou Y, Xu LM, Chen XJ, Ding WB, Zou HB. Autophagy contributes to the enrichment and survival of colorectal cancer stem cells under oxaliplatin treatment. Cancer Lett. 2015;361:128-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 56] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 15. | Zhou Y, Wan G, Spizzo R, Ivan C, Mathur R, Hu X, Ye X, Lu J, Fan F, Xia L. miR-203 induces oxaliplatin resistance in colorectal cancer cells by negatively regulating ATM kinase. Mol Oncol. 2014;8:83-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 150] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 16. | Li G, Thomas AM, Williams JA, Kong B, Liu J, Inaba Y, Xie W, Guo GL. Farnesoid X receptor induces murine scavenger receptor Class B type I via intron binding. PLoS One. 2012;7:e35895. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 17. | Qiao PF, Yao L, Zhang XC, Li GD, Wu DQ. Heat shock pretreatment improves stem cell repair following ischemia-reperfusion injury via autophagy. World J Gastroenterol. 2015;21:12822-12834. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 32] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (1)] |
| 18. | Yang AD, Fan F, Camp ER, van Buren G, Liu W, Somcio R, Gray MJ, Cheng H, Hoff PM, Ellis LM. Chronic oxaliplatin resistance induces epithelial-to-mesenchymal transition in colorectal cancer cell lines. Clin Cancer Res. 2006;12:4147-4153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 366] [Cited by in RCA: 435] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 19. | Burada F, Nicoli ER, Ciurea ME, Uscatu DC, Ioana M, Gheonea DI. Autophagy in colorectal cancer: An important switch from physiology to pathology. World J Gastrointest Oncol. 2015;7:271-284. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 123] [Cited by in RCA: 122] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 20. | Suzuki HI, Kiyono K, Miyazono K. Regulation of autophagy by transforming growth factor-β (TGF-β) signaling. Autophagy. 2010;6:645-647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 118] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 21. | Kenific CM, Debnath J. Cellular and metabolic functions for autophagy in cancer cells. Trends Cell Biol. 2015;25:37-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 207] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 22. | White E. The role for autophagy in cancer. J Clin Invest. 2015;125:42-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 802] [Cited by in RCA: 1033] [Article Influence: 103.3] [Reference Citation Analysis (0)] |
| 23. | Lubner SJ, Loconte NK, Holen KD, Schelman W, Thomas JP, Jumonville A, Eickhoff JC, Seo S, Mulkerin DL. A phase II study of oxaliplatin, 5-fluorouracil, leucovorin, and high-dose capecitabine in patients with metastatic colorectal cancer. Clin Colorectal Cancer. 2010;9:157-161. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 24. | Lv L, Liu HG, Dong SY, Yang F, Wang QX, Guo GL, Pan YF, Zhang XH. Upregulation of CD44v6 contributes to acquired chemoresistance via the modulation of autophagy in colon cancer SW480 cells. Tumor Biology. 2016;37:8811-8824. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 65] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 25. | Li H, Rokavec M, Hermeking H. Soluble IL6R represents a miR-34a target: potential implications for the recently identified IL-6R/STAT3/miR-34a feed-back loop. Oncotarget. 2015;6:14026-14032. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |