Published online Feb 14, 2016. doi: 10.3748/wjg.v22.i6.2060
Peer-review started: June 26, 2015
First decision: August 26, 2015
Revised: September 10, 2015
Accepted: November 13, 2015
Article in press: November 13, 2015
Published online: February 14, 2016
Processing time: 214 Days and 13.7 Hours
AIM: To investigate the expression profiles of hsa-miR-29c and hsa-miR-135b in gastric mucosal samples and their values as gastric carcinogenesis biomarkers.
METHODS: The expression levels of hsa-miR-29c and hsa-miR-135b in normal gastric mucosa, non-atrophic chronic gastritis, intestinal metaplasia and intestinal-type gastric adenocarcinoma were analysed using quantitative real-time PCR. The difference between hsa-miR-29c and hsa-miR-135b expression profiles in the grouped samples was evaluated by ANOVA and Student’s t-test tests. The results were adjusted for multiple testing by using Bonferroni’s correction. P values ≤ 0.05 were considered statistically significant. To evaluate hsa-miR-29c and hsa-miR-135b expressions as potential biomarkers of gastric carcinogenesis, we performed a receiver operating characteristic curve analysis and the derived area under the curve, and a Categorical Principal Components Analysis. In silico identification of the genetic targets of hsa-miR-29c and hsa-miR-135b was performed using different prediction tools, in order to identify possible genes involved in gastric carcinogenesis.
RESULTS: The expression levels of hsa-miR-29c were higher in normal gastric mucosal samples, and decreased progressively in non-atrophic chronic gastritis samples, intestinal metaplasia samples and intestinal-type gastric adenocarcinoma samples. The expression of hsa-miR-29c in the gastric lesions showed that non-atrophic gastritis have an intermediate profile to gastric normal mucosa and intestinal-type gastric adenocarcinoma, and that intestinal metaplasia samples presented an expression pattern similar to that in intestinal-type gastric adenocarcinoma. This microRNA (miRNA) has a good discriminatory accuracy between normal gastric samples and (1) intestinal-type gastric adenocarcinoma; and (2) intestinal metaplasia, and regulates the DMNT3A oncogene. hsa-miR-135b is up-regulated in non-atrophic chronic gastritis and intestinal metaplasia samples and down-regulated in normal gastric mucosa and intestinal-type gastric adenocarcinoma samples. Non-atrophic chronic gastritis and intestinal metaplasia are significantly different from normal gastric mucosa samples. hsa-miR-135b expression presented a greater discriminatory accuracy between normal samples and gastric lesions. This miRNA was associated with Helicobacter pylori presence in non-atrophic chronic gastritis samples and regulates the APC and KLF4 tumour suppressor genes.
CONCLUSION: Our results provide evidence of epigenetic alterations in non-atrophic chronic gastritis and intestinal metaplasia and suggest that hsa-miR-29c and hsa-miR-135b are promising biomarkers of gastric carcinogenesis.
Core tip: The miRNAs hsa-miR-29c and hsa-miR-135b were reported as potential biomarkers of intestinal-type gastric adenocarcinoma. We evaluated and compared the expression profile of these miRNAs in gastric mucosal samples, including normal gastric mucosa, non-atrophic chronic gastritis, intestinal metaplasia and intestinal-type gastric adenocarcinoma. Our results provided evidence of epigenetic alterations in non-atrophic chronic gastritis and intestinal metaplasia and suggest that hsa-miR-29c and hsa-miR-135b are promising biomarkers of gastric carcinogenesis.
- Citation: Vidal AF, Cruz AM, Magalhães L, Pereira AL, Anaissi AK, Alves NC, Albuquerque PJ, Burbano RM, Demachki S, Ribeiro-dos-Santos Â. hsa-miR-29c and hsa-miR-135b differential expression as potential biomarker of gastric carcinogenesis. World J Gastroenterol 2016; 22(6): 2060-2070
- URL: https://www.wjgnet.com/1007-9327/full/v22/i6/2060.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i6.2060
Since the middle of the last century, the histological classification of adenocarcinomas has been largely based on the criteria proposed by Lauren[1]. According to this classification, there are three gastric adenocarcinoma types: intestinal, diffuse and undifferentiated, which is also classified as indeterminate[1]. Intestinal-type and diffuse-type gastric adenocarcinomas have their own characteristics and specific risk factors[2].
In 1988, Pelayo Correa proposed a paradigm for intestinal-type gastric adenocarcinoma carcinogenesis, which became known as the Correa cascade. According to this cascade, a subset of patients who develop intestinal-type gastric adenocarcinomas undergo a multi-stage and complex process of carcinogenesis, initiated by (1) chronic superficial gastritis, also called non-atrophic chronic gastritis; followed by (2) chronic atrophic gastritis; then (3) intestinal metaplasia; and finally (4) dysplasia[3,4].
Helicobacter pylori (H. pylori) infection is the major risk factor among all the main risk factors involved in chronic gastritis, intestinal metaplasia, dysplasia and intestinal-type gastric adenocarcinoma. For this reason, in 1994, this bacterium was classified as a type 1 carcinogen by the World Health Organization[5].
Despite the fact that the different inflammatory stages of the Correa cascade are pathologically well defined, the molecular signatures of these stages have not been well explored and the mechanisms that lead to carcinogenic progression are still unknown[6].
The pre-cancerous or pre-malignant lesions are defined as those that precede invasive cancers which many of the molecular changes and phenotypic characteristics of invasive cancer are present but not fully expressed[7]. Thus, it is assumed that the changes found in intestinal-type gastric adenocarcinomas may also be present in different stages of gastric lesions, such as chronic gastritis, intestinal metaplasia and dysplasia.
Several studies have shown that gastric cancer is a complex disease involving changes in oncogenes, tumour suppressor genes, DNA repair regulatory genes, cell cycle and cell adhesion, as well as numerous epigenetic changes[8]. A class of small non-coding RNAs, called microRNAs (miRNAs), have emerged as key agents in these epigenetic changes[9].
MiRNAs are short (approximately 22 nucleotides in length), endogenous, noncoding RNAs that regulate the expression of target mRNAs at a post-transcriptional level[10]. Based on the results obtained in studies by Ribeiro-dos-Santos et al[11], Moreira et al[12], Gomes et al[13] and Darnet et al[14], hsa-miR-29c and hsa-miR-135b were reported as potential biomarkers of intestinal-type gastric adenocarcinoma. However, more studies are needed to confirm and validate hsa-miR-29c and hsa-miR-135b as potential biomarkers.
The objective of this study was to investigate the expression profiles of hsa-miR-29c and hsa-miR-135b in gastric mucosal samples, including normal gastric mucosa, non-atrophic chronic gastritis, intestinal metaplasia and intestinal-type gastric adenocarcinoma, and their values as gastric carcinogenesis biomarkers. Additionally, in silico prediction was performed to identify potential driver genes involved in the carcinogenic mechanism[15] regulated by these miRNAs.
This study comprised randomly selected frozen tissue samples of normal gastric mucosa (n = 20), FFPE samples of non-atrophic chronic gastritis (n = 20) and of intestinal metaplasia (n = 10) from patients undergoing endoscopic gastric biopsy samples, and gastric intestinal adenocarcinoma frozen tissue samples obtained from patients undergoing gastrectomies (n = 14). All cases investigated in this study were reviewed and confirmed by a pathologist.
Histological processing was performed using glass slide-mounted 3 μm-thick rotary microtome slices (Leica 2125RT). These preparations were deparaffinised, stained with haematoxylin-eosin (HE) and analysed by light microscopy. After histological processing, manual microdissection was performed to increase the accuracy of histopathological characterisation.
Non-atrophic chronic gastritis samples were defined by the presence of lymphocytes and plasmocytes in the lamina propria. The presence or absence of neutrophils permeating the glandular and the surface epithelia, and the presence or absence of lymphoid follicles were also evaluated[16].
Samples of intestinal metaplasia were histopathologically diagnosed by the replacement of the surface and glandular gastric columnar epithelial cells by metaplastic cells of intestinal morphology, such as absorptive and goblet cells.
Fresh tissue samples were immediately stored in RNAlater Solution (Ambion) at -80 °C until total RNA extraction. Only samples with a pure tumour area occupying at least 80% of the slide were used. Pathological TNM staging was evaluated according to the 2010 criteria of The American Joint Committee on Cancer.
The histological sections of gastric mucosal biopsy were stained with HE and cresyl fast violet to perform the H. pylori detection.
Samples were obtained from the Hospital Universitário João de Barros Barreto - Federal University of Pará (Belém, Pará, Brazil) and from the Hospital São Camilo e São Luís (Macapá, Amapá, Brazil). Informed consent was obtained from all individual participants included and the study protocol was approved by the Local Ethics Committee (Protocol number: 657 666) in accordance with the Helsinki Declaration of 1964.
The total RNA was extracted using the High Pure Kit miRNA Isolation Kit (Roche Diagnostics) according to manufacturer’s protocol, and stored at -80 °C to avoid degradation. The total RNA concentrations were determined by the Qubit® 2.0 Fluorometer (Life Technologies) using the Qubit RNA HS Assay kit (Life Technologies). The samples were diluted to the final concentration of 4 ng/μL.
Assays for measuring the miRNAs expression were performed using TaqMan MicroRNA Assays (Applied Biosystems) according to the manufacturer’s instructions. Initially, 10 ng of total RNA was subjected to reverse transcription polymerase chain reaction using the TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems) according to manufacturer’s protocol. The thermocycling conditions were: 30 min at 16 °C, followed by 30 min at 42 °C, 5 min at 85 °C and 5 min at 4 °C.
The quantitative real-time polymerase chain reaction (qRT-PCR) was performed using TaqMan Universal PCR Master Mix Kit (Applied Biosystems) according to the manufacturer’s protocol and the equipment 7500 Real-Time PCR System (Applied Biosystems). The reactions were performed in triplicate and incubated in optical 96-well reaction plates. The thermocycling conditions were: 95 °C for 10 min, and 40 cycles of 15 s at 95 °C, followed by 1 min at 60 °C.
After finalization of the qRT-PCR experiments, the average values of the cycle threshold (Ct) of the reactions in triplicate were determined. The comparative Ct method was adopted, and Z30 was used as an endogenous control. The relative amount of miRNA expression was normalized by the average values of CtZ30 and calculated by the equation 2-ΔCt, where ΔCt = CtmiRNA - CtZ30.
In order to identify genes that may be involved in gastric carcinogenesis process, we compared the driver genes ranked by Vogelstein et al[15] with the target genes of hsa-miR-29c and hsa-miR-135b.
In silico identification of the target genes was performed using miRecords (http://mirecords.biolead.org) (which integrates 11 prediction tools), TargetCompare (http://54.187.40.156:8080/targetcompare/), miRTarBase (http://mirtarbase.mbc.nctu.edu.tw), MicroCosm/miRBase (http://ebi.ac.uk), miRDB (http://mirdb.org), miRo (http://ferrolab.dmi.unict.it) and miRNAMap (http://mirnamap.mbc.nctu.edu.tw). We considered target genes the ones that were observed in no less than 10 tools.
We also used the miRTarBase database to check which miRNA target genes have already been validated experimentally. The mRNA sequences of target genes were obtained from NCBI.
The pattern of distribution of the data was determined by the Shapiro-Wilk test. The difference between hsa-miR-29c and hsa-miR-135b expression profiles in the grouped samples was evaluated by ANOVA and Student’s t-test tests. The results were adjusted for multiple testing by using Bonferroni’s correction. P values ≤ 0.05 were considered statistically significant.
To evaluate hsa-miR-29c and hsa-miR-135b as potential biomarkers of gastric carcinogenesis, we performed a receiver operating characteristic (ROC) curve analysis and the derived area under the curve (AUC), and a Categorical Principal Components Analysis (CATPCA). Statistical tests and graphics were performed using IBM SPSS Statistics software (version 20), GraphPad Prism (GraphPad Software), MATLAB® 8.3 (Release 2014a) and RStudio (version 0.98.1103).
To determine and compare the expression levels of hsa-miR-29c and hsa-miR-135b in gastric mucosal samples, we performed qRT-PCR.
We used the ANOVA test to compare the miRNAs expression levels between normal gastric mucosa, non-atrophic chronic gastritis, intestinal metaplasia and intestinal-type gastric adenocarcinoma samples. The results were adjusted for multiple testing by using Bonferroni’s correction.
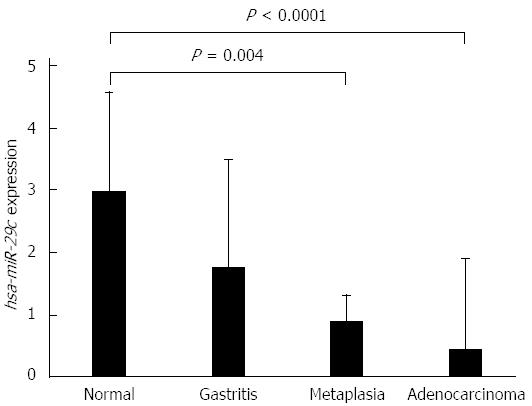
The expression levels of hsa-miR-29c were higher in normal gastric mucosa samples, and decreased progressively in non-atrophic chronic gastritis, intestinal metaplasia and intestinal-type gastric adenocarcinoma samples (Figure 1). The decrease in the expression of hsa-miR-29c as the Correa cascade stages progress is molecular evidence for the Correa cascade pathogenesis.
ANOVA test showed that non-atrophic chronic gastritis samples have an intermediate hsa-miR-29c profile to gastric normal mucosa and intestinal-type gastric adenocarcinoma. Furthermore, intestinal metaplasia hsa-miR-29c expression is significantly different to its in normal gastric mucosa samples (P = 0.004) but is similar to its in intestinal-type gastric adenocarcinoma. There is no difference between non-atrophic chronic gastritis and intestinal metaplasia expression profiles (Figure 1).
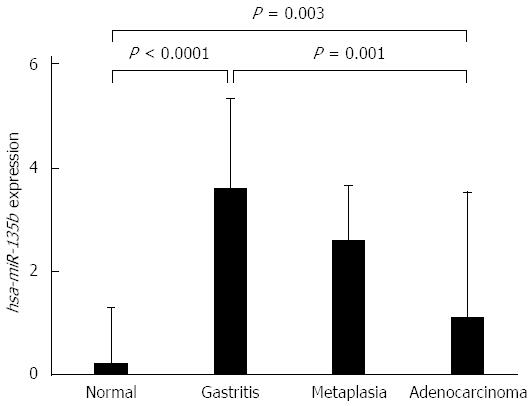
The expression levels of hsa-miR-135b showed higher values in the non-atrophic chronic gastritis and the intestinal metaplasia samples and lower values in the normal gastric mucosa and intestinal-type gastric adenocarcinoma. ANOVA test showed non-atrophic chronic gastritis (P < 0.0001) and the intestinal metaplasia (P = 0.003) are different to normal gastric mucosa, but similar to each other. Non-atrophic chronic gastritis showed a significant difference in expression of hsa-miR-135b in comparison to intestinal-type gastric adenocarcinoma (P = 0.001) (Figure 2).
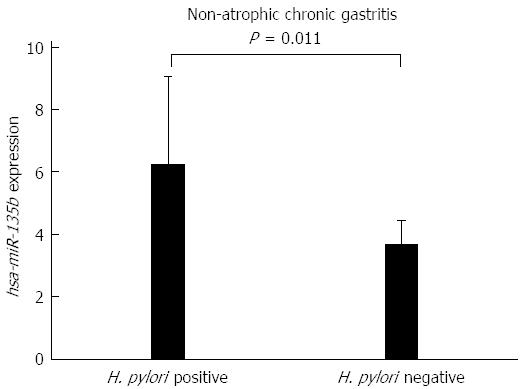
To determine whether the presence of H. pylori affects the expression of hsa-miR-29c and hsa-miR-135b, a Student’s t-test was used to compare the H. pylori-positive and H. pylori-negative non-atrophic chronic gastritis samples. No significant difference was found in the expression of hsa-miR-29c (P = 0.0939) between those groups, however, for hsa-miR-135b, there was a significant difference (P = 0.011). Samples of H. pylori-positive non-atrophic chronic gastritis have a higher expression of hsa-miR-135b than the H. pylori-negative non-atrophic chronic gastritis samples (Figure 3), indicating that this miRNA may be involved in the immune response modulation in association with H. pylori infection.
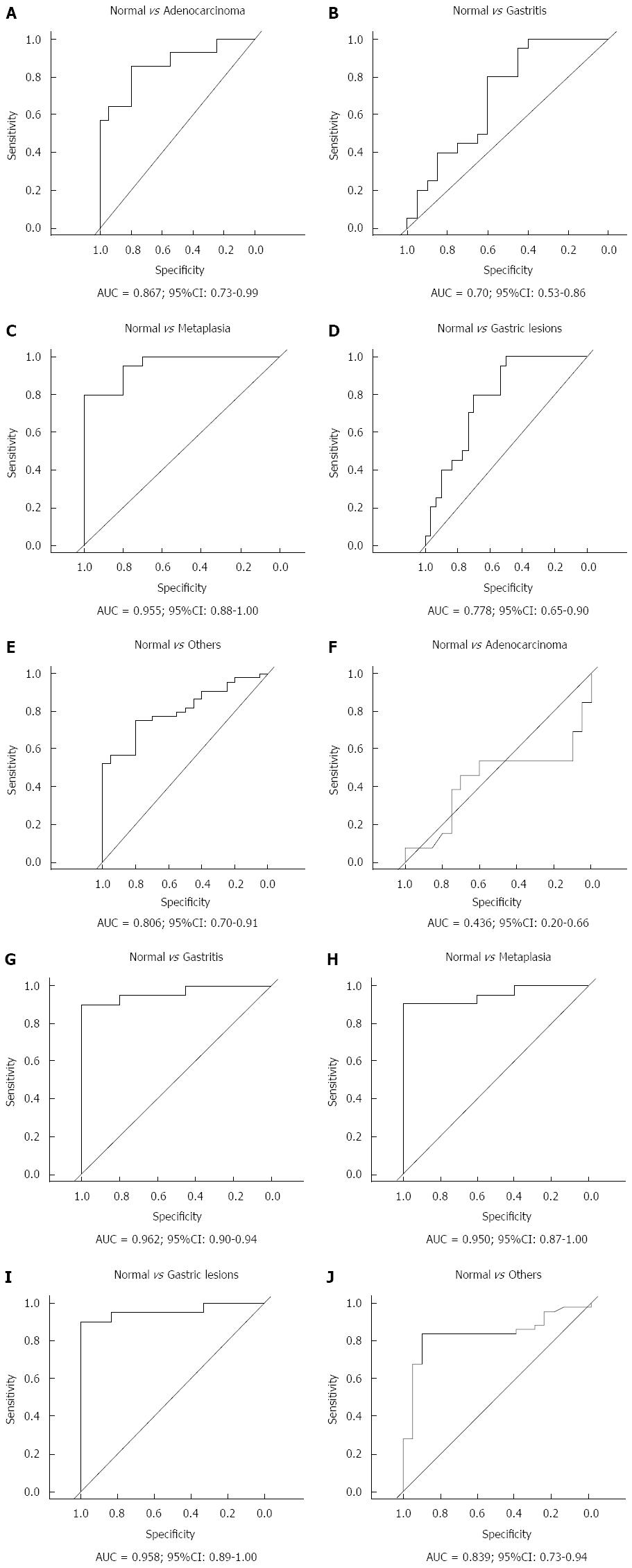
To evaluate hsa-miR-29c and hsa-miR-135b expression as potential biomarker of gastric carcinogenesis, ROC curve analysis and the discriminatory accuracy by AUC values were performed. As shown in Figure 4, hsa-miR-29c expression presented a greater discriminatory accuracy between normal gastric samples and (1) intestinal-type gastric adenocarcinoma (Figure 4A); and (2) intestinal metaplasia (Figure 4C). Otherwise, hsa-miR-135b expression presented a greater discriminatory accuracy between normal samples and gastric lesions (Figure 4G-I).
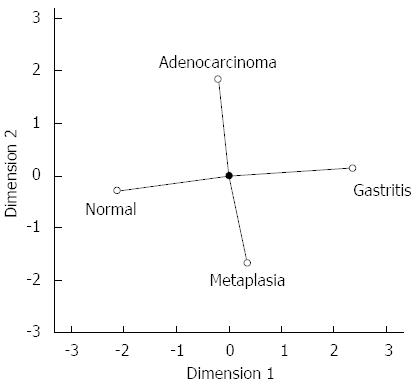
To provide a global view of hsa-miR-29c and hsa-miR-135b expression in all samples groups studied simultaneously, CATPCA analysis was performed. The CATPCA analysis of hsa-miR-29c expression resulted in two dimensions (first component: Cronbach’s alpha = 0.778 and eigenvalue = 2.401; second component: Cronbach’s alpha = 0.422 and eigenvalue = 1.463). According to the angles between the vectors (Figure 5), the hsa-miR-29c expression was able to distinguish each group of samples. It was not possible to construct a three-dimensional graphic due to the negative value of the third component’s Cronbach’s alpha.
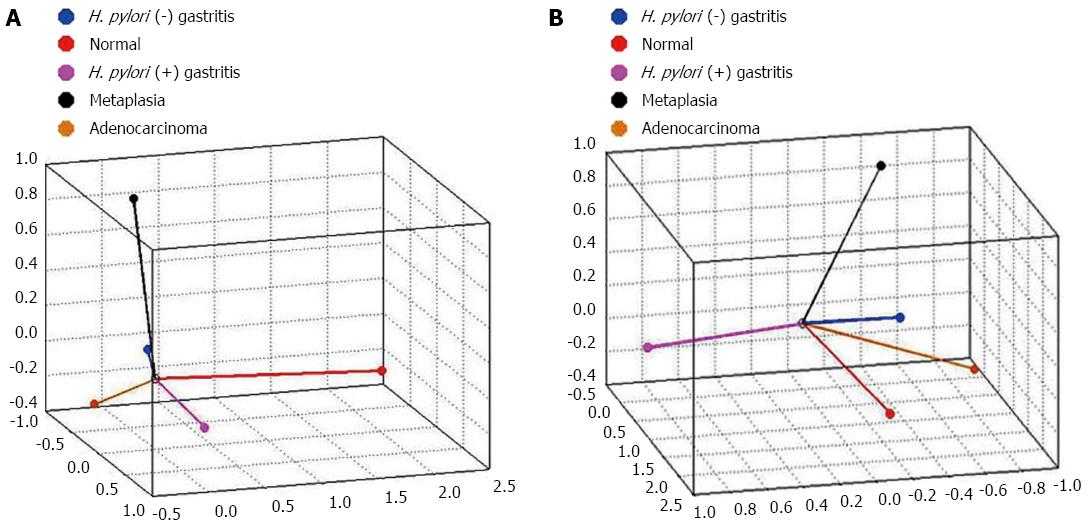
The CATPCA analysis for hsa-miR-135b expression resulted in three dimensions (first component: Cronbach’s alpha = 0.938 and eigenvalue = 8.051; second component: Cronbach’s alpha = 0.829 and eigenvalue = 4.424; third component: Cronbach’s alpha = 0.81 and eigenvalue = 4.101). Figure 6 shows the same CATPCA in a three-dimensional space in two different angles (Figure 6). According to the angles between the vectors, hsa-miR-135b expression was able to distinguish the gastric normal mucosa samples to H. pylori-positive non-atrophic chronic gastritis and intestinal metaplasia samples.
To predict the target genes of hsa-miR-29c and hsa-miR-135b and identify possible genes involved in gastric carcinogenesis, we used 17 different tools and compared the results with the driver genes list ranked by Vogelstein et al[15]. The DNMT3A driver gene is a validated target of hsa-miR-29c, and the APC and KLF4 driver genes are validated targets of hsa-miR-135b (Table 1).
| MicroRNA | Target genes | Ref. Seq. RNA | Validation methods | No. of papers | ||||
| Reporter assay | Western blot | qPCR | Microarray | Other | ||||
| hsa-miR-29c | DNMT3A | NM_022552 | × | × | × | × | × | 3 |
| NM_153759 | ||||||||
| NM_175629 | ||||||||
| NM_175630 | ||||||||
| hsa-miR-135b | APC | NM_000038 | × | × | × | 1 | ||
| NM_001127510 | ||||||||
| NM_001127511 | ||||||||
| KLF4 | NM_004235 | × | × | × | × | 1 | ||
This study compared the expression levels of two miRNA candidates for gastric cancer biomarkers (hsa-miR-29c and hsa-miR-135b) between normal gastric mucosa, gastric lesions (non-atrophic chronic gastritis and intestinal metaplasia) and intestinal-type gastric adenocarcinoma.
The results showed a progressive down-regulation of hsa-miR-29c in normal gastric mucosa, non-atrophic chronic gastritis, intestinal metaplasia and intestinal-type gastric adenocarcinoma, providing evidence for the pathogenesis of the Correa cascade. The expression of this miRNA in the gastric lesions showed that non-atrophic gastritis have a intermediate profile to gastric normal mucosa and intestinal-type gastric adenocarcinoma samples, and that intestinal metaplasia samples presented an expression pattern similar to that in intestinal-type gastric adenocarcinoma.
Furthermore, we observed a significantly difference between hsa-miR-29c profiles in normal gastric mucosa and intestinal-type gastric adenocarcinoma. Different studies analysed the expression of hsa-miR-29c in gastric cancer and found results consistent with those of this investigation[17-23].
The down-regulation of hsa-miR-29c has been reported in several human malignancies, including nasopharyngeal carcinoma[24], bladder carcinoma cells[25], lung cancer[26], esophaeal cancer[27,28], chronic lymphocytic leukaemia[29,30] and melanoma[31].
Considering the expression profile of hsa-miR-29c in gastric cancer, it is suggested that hsa-miR-29c acts as a TS-miR. Therefore, down-regulation of this miRNA can lead to overexpression of oncogenes, such as DNMT3A.
The DNMT3A gene encodes the DNA methyltransferase 3A, an enzyme responsible for the dynamics of DNA methylation during embryogenesis and pathogenesis. The de-regulation of DNA methylation patterns can shut-down or cripple the normal transcriptional activity and is considered an early event in tumour development[32]. This gene was validated to be a target of hsa-miR-29c in three different papers[32-34].
According to these papers, in normal conditions, highly expressed hsa-miR-29c may control DNMT3A through a conserved function. Therefore, expression of this miRNA in tumour cells can lead to reduced global DNA methylation, restoring expression of tumour suppressor genes and inhibiting tumourigenicity both in vivo and in vitro[32-34].
Indeed, overexpression of DNMT3A was reported not only in gastric cancer itself but also in gastric lesions[35-37]. Hyper-methylation of the hMLH1, P16, DAP-kinase, THBS1 and TIMP-3 genes was detected in chronic gastritis and intestinal metaplasia samples[38].
It is possible that hsa-miR-29c inhibits tumourigenicity both in vivo and in vitro[32] by restoring expression of tumour suppression genes involved in the control of cell proliferation. Indeed, Matsuo et al[18] demonstrated that hsa-miR-29c is involved in regulating the S phase of the cell cycle in gastric cancer. Furthermore, Wang et al[23] find that hsa-miR-29c act as metastasis suppressor in gastric cancer. These findings suggested that this miRNA not only functioned as TS-miR in gastric cancer but also might serve as effective predictors for gastric cancer prevention[23].
Up-regulated expression of hsa-miR-135b was observed in gastric lesions compared to normal gastric mucosa and intestinal-type gastric adenocarcinoma samples. Non-atrophic chronic gastritis and intestinal metaplasia are significantly different from normal gastric mucosa samples.
To date, the expression of hsa-miR-135b has been analysed in gastric cancer in a few studies[17,21]. This miRNA has been most extensively studied in other types of human cancer, such as colon[39], breast[40], cutaneous squamous cell carcinoma[41] and lung cancer[42,43]. In all of these cases, its overexpression points to the hypothesis that this miRNA acts as oncomiR in the process of carcinogenesis.
In silico analysis showed that hsa-miR-135b has two validated target genes, KLF4[44] and APC[45], which are both tumour suppressor genes.
The KLF4 gene encodes a zinc-finger transcription factor, which is involved in mediating pro-inflammatory responses and regulating cell proliferation and differentiation[46,47]. Down-regulation of this gene is reported in gastric cancer, indicating its participation in the regulation of homeostasis and maintenance of the gastric mucosa. In addition, restoration of KLF4 expression was able to inhibit tumour growth in vivo and in vitro by inducing apoptosis in gastric cancer cells. Thus, altering KLF4 expression plays a critical role in gastric cancer development and progression[48].
The APC gene encodes a protein that binds to the transcription factor β-catenin and results in degradation of β-catenin. The loss of function of this gene causes the nuclear accumulation of APC-free β-catenin, which stimulates the Wnt signalling pathway and leads to de-regulated cell growth and adhesion[45]. Mutations in APC have been identified in patients with gastric adenocarcinoma, especially in those with the intestinal type[49-52]. These results suggest that the loss of APC expression plays an important role during gastric carcinogenesis.
Several studies have suggested that miRNAs represent a bridge between chronic gastritis and gastric cancer development[53-56]. This study showed that hsa-miR-29c expression has a good discriminatory accuracy between normal gastric samples and (1) intestinal-type gastric adenocarcinoma; and (2) intestinal metaplasia; hsa-miR-135b expression presented a greater discriminatory accuracy between normal samples and gastric lesions.
In conclusion, our results suggest that hsa-miR-29c and hsa-miR-135b are promising biomarkers of gastric carcinogenesis and provide evidence of epigenetic alterations in non-atrophic chronic gastritis and intestinal metaplasia, indicating that better understanding of these gastric lesions is required for the prevention of gastric cancer.
We thank Dr. Paulo Assumpção, PhD. Sidney Santos, MsC. André Santos, PhD. Fabiano Moreira and MsC. Pablo Pinto for their comments, ideas and help.
The molecular signatures of the gastric pre-cancerous lesions, such as non-atrophic chronic gastritis and intestinal metaplasia, have not been well explored and the mechanisms that lead to carcinogenic progression are still unknown. microRNAs (miRNAs) were reported as potential biomarkers of intestinal-type gastric adenocarcinoma. hsa-miR-29c and hsa-miR-135b are promising biomarkers of gastric carcinogenesis and provide evidence of epigenetic alterations in non-atrophic chronic gastritis and intestinal metaplasia.
A subset of patients who develop intestinal-type gastric adenocarcinomas undergo a multi-stage and complex process of carcinogenesis, initiated by chronic gastritis and intestinal metaplasia. Hsa-miR-29c and hsa-miR-135b expressions are altered in non-atrophic chronic gastritis and intestinal metaplasia, indicating that better understanding of these gastric lesions is required for the prevention of gastric cancer.
Previous studies have shown that hsa-miR-29c and hsa-miR-135b are potential biomarkers of intestinal-type gastric adenocarcinoma. This study investigated the expression profiles of hsa-miR-29c and hsa-miR-135b in gastric mucosal samples, including normal gastric mucosa, non-atrophic chronic gastritis, intestinal metaplasia and intestinal-type gastric adenocarcinoma. The expression of hsa-miR-29c in the gastric lesions showed that non-atrophic gastritis have a intermediate profile to gastric normal mucosa and intestinal-type gastric adenocarcinoma, and that intestinal metaplasia samples presented an expression pattern similar to that in intestinal-type gastric adenocarcinoma. This miRNA regulates the DMNT3A oncogene. Hsa-miR-135b is up-regulated in non-atrophic chronic gastritis and intestinal metaplasia samples and down-regulated in normal gastric mucosa and intestinal-type gastric adenocarcinoma samples. Non-atrophic chronic gastritis and intestinal metaplasia are significantly different from normal gastric mucosa samples. Hsa-miR-135b expression presented a greater discriminatory accuracy between normal samples and gastric lesions. This miRNA was associated with Helicobacter pylori presence in non-atrophic chronic gastritis samples and regulates the APC and KLF4 tumour suppressor genes.
This study showed that hsa-miR-29c expression has a good discriminatory accuracy between normal gastric samples and (1) intestinal-type gastric adenocarcinoma; and (2) intestinal metaplasia; hsa-miR-135b expression presented a greater discriminatory accuracy between normal samples and gastric lesions. These results suggest that hsa-miR-29c and hsa-miR-135b are promising biomarkers of gastric carcinogenesis, indicating that better understanding of the gastric lesions is required for the prevention of gastric cancer.
According to the Correa cascade, a subset of patients who develop intestinal-type gastric adenocarcinomas undergo a multi-stage and complex process of carcinogenesis, initiated by (1) non-atrophic chronic gastritis; followed by (2) chronic atrophic gastritis; then (3) intestinal metaplasia; and finally (4) dysplasia. hsa-miR-29c and hsa-miR-135b are miRNAs, a class of noncoding RNAs, that regulate the expression of target mRNAs at a post-transcriptional level. These miRNAs are related to the gastric carcinogenesis and may be potential biomarkers.
The authors used gastric tissues derived from normal, gastritis, metaplasia, and carcinoma to investigate expression of 2 miRNAs and evaluate their potential to be biomarkers to distinguish these 4 groups. The concept is clear and the flow of data presentation is straightforward.
P- Reviewer: Liang Y S- Editor: Gong ZM L- Editor: A E- Editor: Ma S
| 1. | Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. an attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31-49. [PubMed] |
| 2. | Hu B, El Hajj N, Sittler S, Lammert N, Barnes R, Meloni-Ehrig A. Gastric cancer: Classification, histology and application of molecular pathology. J Gastrointest Oncol. 2012;3:251-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 256] [Reference Citation Analysis (0)] |
| 3. | Correa P. A human model of gastric carcinogenesis. Cancer Res. 1988;48:3554-3560. [PubMed] |
| 4. | Correa P. Human gastric carcinogenesis: a multistep and multifactorial process--First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992;52:6735-6740. [PubMed] |
| 5. | Correa P; IARC. Schistosomes, liver flukes and Helicobacter pylori. Lyon: IARC Monogr. Eval. Carcinog. Risks Hum 1994; 177-240. |
| 6. | Zabaleta J. MicroRNA: A Bridge from H. pylori Infection to Gastritis and Gastric Cancer Development. Front Genet. 2012;3:294. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 7. | Kim HS, Lee JS, Freund JN, Min KW, Lee JS, Kim W, Juhng SW, Park CS. CDX-2 homeobox gene expression in human gastric carcinoma and precursor lesions. J Gastroenterol Hepatol. 2006;21:438-442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 8. | Wu HH, Lin WC, Tsai KW. Advances in molecular biomarkers for gastric cancer: miRNAs as emerging novel cancer markers. Expert Rev Mol Med. 2014;16:e1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 133] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 9. | Ross SA, Davis CD. MicroRNA, nutrition, and cancer prevention. Adv Nutr. 2011;2:472-485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 92] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 10. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25833] [Cited by in RCA: 27822] [Article Influence: 1324.9] [Reference Citation Analysis (0)] |
| 11. | Ribeiro-dos-Santos Â, Khayat AS, Silva A, Alencar DO, Lobato J, Luz L, Pinheiro DG, Varuzza L, Assumpção M, Assumpção P. Ultra-deep sequencing reveals the microRNA expression pattern of the human stomach. PLoS One. 2010;5:e13205. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 58] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 12. | Moreira FC, Assumpção M, Hamoy IG, Darnet S, Burbano R, Khayat A, Gonçalves AN, Alencar DO, Cruz A, Magalhães L. MiRNA expression profile for the human gastric antrum region using ultra-deep sequencing. PLoS One. 2014;9:e92300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 13. | Gomes LL, Moreira FC, Hamoy IG, Santos S, Assumpção P, Santana AL, Ribeiro-Dos-Santos A. Identification of miRNAs Expression Profile in Gastric Cancer Using Self-Organizing Maps (SOM). Bioinformation. 2014;10:246-250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 14. | Darnet S, Moreira FC, Hamoy IG, Burbano R, Khayat A, Cruz A, Magalhães L, Silva A, Santos S, Demachki S. High-Throughput Sequencing of miRNAs Reveals a Tissue Signature in Gastric Cancer and Suggests Novel Potential Biomarkers. Bioinform Biol Insights. 2015;9:1-8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 15. | Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546-1558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5906] [Cited by in RCA: 5591] [Article Influence: 465.9] [Reference Citation Analysis (0)] |
| 16. | Kumar V, Abbas AK, Aster JC, Robbins SL, Cotran RS. Robbins and Cotran Pathologic Basis of Disease. 8th ed. Philadelphia: Elsevier Saunders 2005; 768-773. |
| 17. | Lim JY, Yoon SO, Seol SY, Hong SW, Kim JW, Choi SH, Lee JS, Cho JY. Overexpression of miR-196b and HOXA10 characterize a poor-prognosis gastric cancer subtype. World J Gastroenterol. 2013;19:7078-7088. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 47] [Cited by in RCA: 58] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 18. | Matsuo M, Nakada C, Tsukamoto Y, Noguchi T, Uchida T, Hijiya N, Matsuura K, Moriyama M. MiR-29c is downregulated in gastric carcinomas and regulates cell proliferation by targeting RCC2. Mol Cancer. 2013;12:15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 71] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 19. | Saito Y, Suzuki H, Imaeda H, Matsuzaki J, Hirata K, Tsugawa H, Hibino S, Kanai Y, Saito H, Hibi T. The tumor suppressor microRNA-29c is downregulated and restored by celecoxib in human gastric cancer cells. Int J Cancer. 2013;132:1751-1760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 59] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 20. | Tsukamoto Y, Nakada C, Noguchi T, Tanigawa M, Nguyen LT, Uchida T, Hijiya N, Matsuura K, Fujioka T, Seto M. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res. 2010;70:2339-2349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 320] [Cited by in RCA: 346] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 21. | Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 644] [Cited by in RCA: 676] [Article Influence: 45.1] [Reference Citation Analysis (0)] |
| 22. | Gong J, Li J, Wang Y, Liu C, Jia H, Jiang C, Wang Y, Luo M, Zhao H, Dong L. Characterization of microRNA-29 family expression and investigation of their mechanistic roles in gastric cancer. Carcinogenesis. 2014;35:497-506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 74] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 23. | Wang Y, Liu C, Luo M, Zhang Z, Gong J, Li J, You L, Dong L, Su R, Lin H. Chemotherapy-Induced miRNA-29c/Catenin-δ Signaling Suppresses Metastasis in Gastric Cancer. Cancer Res. 2015;75:1332-1344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 24. | Sengupta S, den Boon JA, Chen IH, Newton MA, Stanhope SA, Cheng YJ, Chen CJ, Hildesheim A, Sugden B, Ahlquist P. MicroRNA 29c is down-regulated in nasopharyngeal carcinomas, up-regulating mRNAs encoding extracellular matrix proteins. Proc Natl Acad Sci USA. 2008;105:5874-5878. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 322] [Cited by in RCA: 342] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 25. | Friedman JM, Liang G, Liu CC, Wolff EM, Tsai YC, Ye W, Zhou X, Jones PA. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009;69:2623-2629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 321] [Cited by in RCA: 314] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 26. | Wang H, Zhu Y, Zhao M, Wu C, Zhang P, Tang L, Zhang H, Chen X, Yang Y, Liu G. miRNA-29c suppresses lung cancer cell adhesion to extracellular matrix and metastasis by targeting integrin β1 and matrix metalloproteinase2 (MMP2). PLoS One. 2013;8:e70192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 105] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 27. | Ding DP, Chen ZL, Zhao XH, Wang JW, Sun J, Wang Z, Tan FW, Tan XG, Li BZ, Zhou F. miR-29c induces cell cycle arrest in esophageal squamous cell carcinoma by modulating cyclin E expression. Carcinogenesis. 2011;32:1025-1032. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 76] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 28. | Guo Y, Chen Z, Zhang L, Zhou F, Shi S, Feng X, Li B, Meng X, Ma X, Luo M. Distinctive microRNA profiles relating to patient survival in esophageal squamous cell carcinoma. Cancer Res. 2008;68:26-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 268] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 29. | Mraz M, Malinova K, Kotaskova J, Pavlova S, Tichy B, Malcikova J, Stano Kozubik K, Smardova J, Brychtova Y, Doubek M. miR-34a, miR-29c and miR-17-5p are downregulated in CLL patients with TP53 abnormalities. Leukemia. 2009;23:1159-1163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 134] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 30. | Stamatopoulos B, Meuleman N, Haibe-Kains B, Saussoy P, Van Den Neste E, Michaux L, Heimann P, Martiat P, Bron D, Lagneaux L. microRNA-29c and microRNA-223 down-regulation has in vivo significance in chronic lymphocytic leukemia and improves disease risk stratification. Blood. 2009;113:5237-5245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 181] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 31. | Nguyen T, Kuo C, Nicholl MB, Sim MS, Turner RR, Morton DL, Hoon DS. Downregulation of microRNA-29c is associated with hypermethylation of tumor-related genes and disease outcome in cutaneous melanoma. Epigenetics. 2011;6:388-394. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 163] [Cited by in RCA: 146] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 32. | Pandi G, Nakka VP, Dharap A, Roopra A, Vemuganti R. MicroRNA miR-29c down-regulation leading to de-repression of its target DNA methyltransferase 3a promotes ischemic brain damage. PLoS One. 2013;8:e58039. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 84] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 33. | Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805-15810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1300] [Cited by in RCA: 1279] [Article Influence: 71.1] [Reference Citation Analysis (0)] |
| 34. | Kovalchuk O, Zemp FJ, Filkowski JN, Altamirano AM, Dickey JS, Jenkins-Baker G, Marino SA, Brenner DJ, Bonner WM, Sedelnikova OA. microRNAome changes in bystander three-dimensional human tissue models suggest priming of apoptotic pathways. Carcinogenesis. 2010;31:1882-1888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 49] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 35. | Fan H, Liu D, Qiu X, Qiao F, Wu Q, Su X, Zhang F, Song Y, Zhao Z, Xie W. A functional polymorphism in the DNA methyltransferase-3A promoter modifies the susceptibility in gastric cancer but not in esophageal carcinoma. BMC Med. 2010;8:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 71] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 36. | Hino R, Uozaki H, Murakami N, Ushiku T, Shinozaki A, Ishikawa S, Morikawa T, Nakaya T, Sakatani T, Takada K. Activation of DNA methyltransferase 1 by EBV latent membrane protein 2A leads to promoter hypermethylation of PTEN gene in gastric carcinoma. Cancer Res. 2009;69:2766-2774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 258] [Cited by in RCA: 288] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 37. | Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A, MacLeod AR. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat Genet. 2003;33:61-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 456] [Cited by in RCA: 470] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 38. | Kang GH, Shim YH, Jung HY, Kim WH, Ro JY, Rhyu MG. CpG island methylation in premalignant stages of gastric carcinoma. Cancer Res. 2001;61:2847-2851. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 90] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 39. | Khatri R, Subramanian S. MicroRNA-135b and Its Circuitry Networks as Potential Therapeutic Targets in Colon Cancer. Front Oncol. 2013;3:268. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 30] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 40. | Arigoni M, Barutello G, Riccardo F, Ercole E, Cantarella D, Orso F, Conti L, Lanzardo S, Taverna D, Merighi I. miR-135b coordinates progression of ErbB2-driven mammary carcinomas through suppression of MID1 and MTCH2. Am J Pathol. 2013;182:2058-2070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 49] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 41. | Olasz EB, Seline LN, Schock AM, Duncan NE, Lopez A, Lazar J, Flister MJ, Lu Y, Liu P, Sokumbi O. MicroRNA-135b Regulates Leucine Zipper Tumor Suppressor 1 in Cutaneous Squamous Cell Carcinoma. PLoS One. 2015;10:e0125412. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 42. | Halappanavar S, Nikota J, Wu D, Williams A, Yauk CL, Stampfli M. IL-1 receptor regulates microRNA-135b expression in a negative feedback mechanism during cigarette smoke-induced inflammation. J Immunol. 2013;190:3679-3686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 57] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 43. | Lin CW, Chang YL, Chang YC, Lin JC, Chen CC, Pan SH, Wu CT, Chen HY, Yang SC, Hong TM. MicroRNA-135b promotes lung cancer metastasis by regulating multiple targets in the Hippo pathway and LZTS1. Nat Commun. 2013;4:1877. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 239] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 44. | Lutherborrow M, Bryant A, Jayaswal V, Agapiou D, Palma C, Yang YH, Ma DD. Expression profiling of cytogenetically normal acute myeloid leukemia identifies microRNAs that target genes involved in monocytic differentiation. Am J Hematol. 2011;86:2-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 40] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 45. | Nagel R, le Sage C, Diosdado B, van der Waal M, Oude Vrielink JA, Bolijn A, Meijer GA, Agami R. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008;68:5795-5802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 376] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 46. | Wei D, Kanai M, Huang S, Xie K. Emerging role of KLF4 in human gastrointestinal cancer. Carcinogenesis. 2006;27:23-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 177] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 47. | Li Q, Jia Z, Wang L, Kong X, Li Q, Guo K, Tan D, Le X, Wei D, Huang S. Disruption of Klf4 in villin-positive gastric progenitor cells promotes formation and progression of tumors of the antrum in mice. Gastroenterology. 2012;142:531-542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 69] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 48. | Wei D, Gong W, Kanai M, Schlunk C, Wang L, Yao JC, Wu TT, Huang S, Xie K. Drastic down-regulation of Krüppel-like factor 4 expression is critical in human gastric cancer development and progression. Cancer Res. 2005;65:2746-2754. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 240] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 49. | Ebert MP, Fei G, Kahmann S, Müller O, Yu J, Sung JJ, Malfertheiner P. Increased beta-catenin mRNA levels and mutational alterations of the APC and beta-catenin gene are present in intestinal-type gastric cancer. Carcinogenesis. 2002;23:87-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 115] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 50. | Tsuchiya T, Tamura G, Sato K, Endoh Y, Sakata K, Jin Z, Motoyama T, Usuba O, Kimura W, Nishizuka S. Distinct methylation patterns of two APC gene promoters in normal and cancerous gastric epithelia. Oncogene. 2000;19:3642-3646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 141] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 51. | Park WS, Oh RR, Park JY, Lee SH, Shin MS, Kim YS, Kim SY, Lee HK, Kim PJ, Oh ST. Frequent somatic mutations of the beta-catenin gene in intestinal-type gastric cancer. Cancer Res. 1999;59:4257-4260. [PubMed] |
| 52. | Fang DC, Jass JR, Wang DX, Zhou XD, Luo YH, Young J. Infrequent loss of heterozygosity of APC/MCC and DCC genes in gastric cancer showing DNA microsatellite instability. J Clin Pathol. 1999;52:504-508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 53. | Zhu YM, Zhong ZX, Liu ZM. Relationship between let-7a and gastric mucosa cancerization and its significance. World J Gastroenterol. 2010;16:3325-3329. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 14] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 54. | Zhang Z, Li Z, Gao C, Chen P, Chen J, Liu W, Xiao S, Lu H. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Invest. 2008;88:1358-1366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 339] [Cited by in RCA: 375] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 55. | Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 678] [Cited by in RCA: 693] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 56. | Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105-D110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1025] [Cited by in RCA: 1116] [Article Influence: 65.6] [Reference Citation Analysis (0)] |