Published online Oct 14, 2016. doi: 10.3748/wjg.v22.i38.8489
Peer-review started: May 2, 2016
First decision: June 20, 2016
Revised: July 19, 2016
Accepted: August 5, 2016
Article in press: August 5, 2016
Published online: October 14, 2016
Processing time: 164 Days and 12.4 Hours
Currently, hepatitis B virus (HBV), upon attaching to human hepatocytes, is considered to interact first with heparan sulfate proteoglycan (HSPG) via an antigenic loop of HBV envelope S protein. Then, it is promptly transferred to the sodium taurocholate cotransporting polypeptide (NTCP) via the myristoylated N-terminal sequence of pre-S1 region (from Gly-2 to Gly-48, HBV genotype D), and it finally enters the cell by endocytosis. However, it is not clear how HSPG passes HBV to NTCP and how NTCP contributes to the cellular entry of HBV. Owing to the poor availability and the difficulty of manipulations, including fluorophore encapsulation, it has been nearly impossible to perform biochemical and cytochemical analyses using a substantial amount of HBV. A bio-nanocapsule (BNC), which is a hollow nanoparticle consisting of HBV envelope L protein, was efficiently synthesized in Saccharomyces cerevisiae. Since BNC could encapsulate payloads (drugs, genes, proteins) and specifically enter human hepatic cells utilizing HBV-derived infection machinery, it could be used as a model of HBV infection to elucidate the early infection machinery. Recently, it was demonstrated that the N-terminal sequence of pre-S1 region (from Asn-9 to Gly-24) possesses low pH-dependent fusogenic activity, which might play a crucial role in the endosomal escape of BNC payloads and in the uncoating process of HBV. In this minireview, we describe a model in which each domain of the HBV L protein contributes to attachment onto human hepatic cells through HSPG, initiation of endocytosis, interaction with NTCP in endosomes, and consequent provocation of membrane fusion followed by endosomal escape.
Core tip: Owing to the poor availability and the difficulty of manipulations of hepatitis B virus (HBV), it has been difficult to analyze its early infection events in human hepatocytes. Using a bio-nanocapsule, a unique model of HBV, we could study these events by biochemical and cytochemical methods, and finally identify a low pH-dependent fusogenic domain in HBV pre-S1 region, which might play a pivotal role in the endosomal escape of HBV. We hereby postulate a model in which each domain in HBV envelope L protein participates in cell attachment, endocytosis, membrane fusion, and consequent endosomal escape (i.e., uncoating process of HBV).
- Citation: Liu Q, Somiya M, Kuroda S. Elucidation of the early infection machinery of hepatitis B virus by using bio-nanocapsule. World J Gastroenterol 2016; 22(38): 8489-8496
- URL: https://www.wjgnet.com/1007-9327/full/v22/i38/8489.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i38.8489
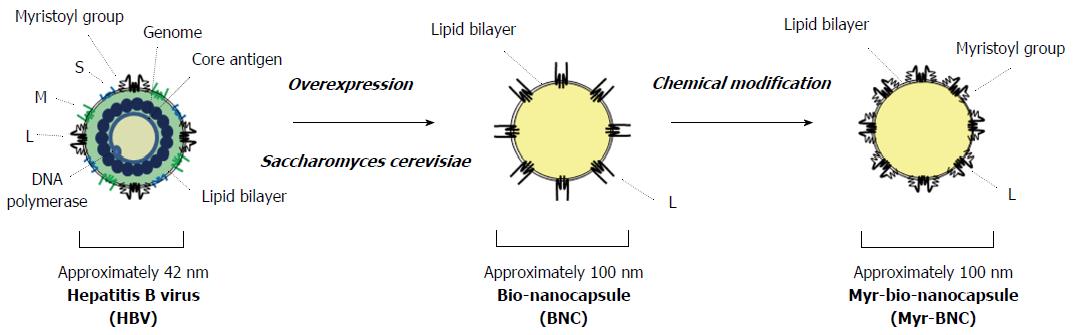
Hepatitis B virus (HBV) is an approximately 42 nm envelope virus containing a nucleocapsid with an approximately 3.2 kilobase double-stranded DNA genome (Figure 1). There are three types of envelope proteins on the surface of HBV virion: The small (S) protein, middle (M, pre-S2 + S region) protein, and large (L, pre-S1 + pre-S2 + S region) protein[1] (Figure 2). Although nearly half a century has passed since the discovery of HBV, it has been difficult to obtain HBV in substantial amounts for biochemical analyses and establish an efficient in vitro system covering infection, replication, and virion release. As a first generation system, HBV was purified from chronically infected patient’s plasma, and primary human hepatocytes (PHH) were used as target cells. Since this system completely depends on clinical specimens, many researchers have reluctantly utilized human hepatoma-derived cell lines for a long time, but these often neither accept HBV infection nor reproduce it. Next, as a second generation system, primary Tupaia hepatocytes (PTH) were found to accept HBV infection efficiently in the presence of dimethyl sulfoxide (DMSO) and allow HBV reproduction[2]. Furthermore, as a third generation system, one human hepatoma cell line, HepaRG, was recently found to accept HBV infection in the presence of DMSO as well as PHH and PTH[3], providing a new possibility for studying HBV early infection. Although the aforementioned progress in HBV target cells has been made for the long time, it is still necessary to obtain a substantial amount of HBV exclusively from patient’s plasma. These situations have hampered the comprehensive elucidation of early infection machinery of HBV.
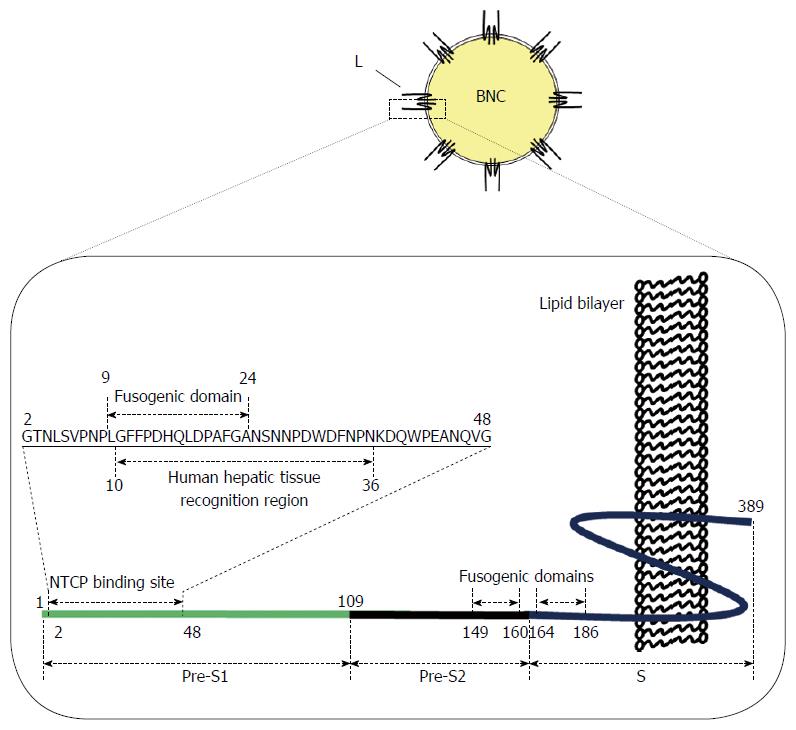
To elucidate the mechanism how HBV recognizes human hepatocytes, many parts of the HBV envelope L protein have been proposed to be indispensable for early infection, especially for the initial attachment. In 1986, the N-terminal part of pre-S1 region (from Pro-10 to Pro-36; see Figure 2) was shown to interact with PHH[4]. Monoclonal antibody against the pre-S1 region, MA18/7, could neutralize the in vitro infectivity of HBV to PTH, whereas antibodies against pre-S2 and S regions could not[5,6]. N-terminal myristoylation of pre-S1 region is also indispensable for HBV to infect PHH[7]. Furthermore, since the addition of heparin could efficiently inhibit in vitro infection with HBV, attachment to heparan sulfate proteoglycan (HSPG), a protein abundant in the extracellular matrix, is a prerequisite for infection of HepaRG cells[8] and PTH[9]. Highly conserved residues (Gly-282, Pro-283, Cys-284, Arg-285, Cys-287, and Lys-312; see Figure 2) in the antigenic loop (AGL) of S region could directly contact with HSPG[10-12]. It is interesting that other viruses, including herpes simplex virus-1 and human immunodeficiency virus-1 (HIV-1), also require interaction with HSPG for their entry[13]. These results strongly suggest that HSPG is a low-affinity receptor for HBV entry. In case of HIV-1, following attachment to HSPG, the virus interacts with CD4 receptor, leading to conformational changes in the viral envelope protein, and subsequently enters the cell using the CCR5 co-receptor[14]. However, it remains unclear what events occur during infection and how HBV exhibits such stringent specificity to human hepatocytes.
In 2012, a transmembrane protein, sodium taurocholate cotransporting polypeptide (NTCP), also known as SLC10A1[15], was indicated as a functional receptor responsible for HBV infection[16]. NTCP is predominantly expressed on the sinusoidal membrane of hepatocytes and is responsible for the majority of sodium-dependent bile acid translocation, playing an essential role in the enterohepatic cycle of bile acids[17]. Knockdown or overexpressed NTCP in human hepatocytes prevented or facilitated HBV infection, respectively[18]. Most importantly, an N-terminally myristoylated pre-S1 (2-47) peptide could inhibit both the transporter function and HBV interaction of NTCP efficiently[19] (Figure 2). These results indicate that NTCP is a high-affinity human liver-specific HBV receptor and would open up new avenues for understanding the early infection machinery of HBV.
Large fraction of the world’s population suffers from HBV infection. Since HBV can be transmitted through blood and body fluids, and there has not been any effective anti-HBV drug, vaccination against hepatitis B (HB) is essential for protection from blood-borne infection. Initially, subviral particles consisting of HBV envelope S protein [i.e., HBV surface antigen (HBsAg)] were purified from HBV e antigen negative patient’s plasma and were formulated as first generation HB vaccines. For eliminating the risk caused by contamination with HBV, HBV envelope S protein was expressed in yeast cells in a particle form, and used as the second generation HB vaccine[20]. However, even after repetitive injection with these vaccines, approx. Five percent of vaccinees could not be seroconverted (i.e., low and non-responders)[21]. The finding that pre-S2 region could elicit HBV-neutralizing antibodies in chimpanzees[22] led us to synthesize the HBV envelope M protein in particle form in yeast cells. This third generation HB vaccine candidate could effectively induce protective level of anti-pre-S2 antibodies even in low and non-responders[23]. Thereafter, owing to the recognition that pre-S1 region could elicit additional HBV-neutralizing antibodies[4], many researchers have attempted to synthesize the full-length HBV envelope protein (L protein) in particle form in eukaryotic cells, but the N-terminal part of pre-S1 region showed strong inhibitory effect on its synthesis. In 1992, fusion of the N-terminus with a chicken lysozyme-derived signal peptide could overcome the inhibitory effect and facilitated overexpression of L particles in yeast cells (up to approx. 42% of the total soluble protein)[24]. The particles could be purified by heat-treatment, affinity column chromatography, and size exclusion column chromatography[25]. Unlike the approx. 42-nm HBV virion, the particle exhibited approx. 100-nm spherical hollow structure, consisting of about 110 L proteins embedded in an yeast endoplasmic reticulum membrane-derived liposomal structure, whereas the stoichiometric ratio of L/M/S envelope proteins of HBV virion is approx. 1:1:4 (Figure 1)[26,27]. Furthermore, the density of particle is approx. 1.22 g/cm3[28], which is similar to that of HBV virion (approx. 1.17 g/cm3)[29]. The particle lacks the N-terminal myristoyl group and possesses additional sugar groups in the pre-S1+pre-S2 region. They could elicit anti-S, anti-pre-S1 and anti-pre-S2 antibodies effectively in mice[26]. According to proteinase protection assay, the pre-S1 region, pre-S2 region, and a part of S region (AGL) are deployed outwardly on the surface of L particle, similar to HBV[24]. This similarity in surface structure prompted us to utilize it as a bio-mimic of HBV in further studies. We therefore designated the HBV envelope L particle as a “bio-nanocapsule (BNC)”[30,31].
Since HBV specifically infects human hepatocytes and delivers its genetic material and associated proteins into the cytoplasm, we examined whether BNC also exhibited a similar function. Following the introduction of a fluorophore or enhanced green fluorescence protein -expression plasmid into the hollow space of BNC by electroporation, the human hepatic cells receiving BNCs were found to exhibit fluorescence in vitro[32]. Furthermore, after an intravenous injection of these BNCs, fluorescence was emitted exclusively from human hepatic cell-derived tumors of xenograft mice[32] and in normal human liver tissues under the kidney skin of severe combined immunodeficiency (SCID) mice[33]. These results strongly suggested that BNC target and enter human hepatic cells in vitro and in vivo using HBV-derived infection machinery, present in the L protein.
BNC was capable of fusing with liposomes (LPs), leading to the formation of BNC-LP complex[28]. Under high temperature (up to 70 °C) and acidic conditions, the complex was found to transform into a smooth and spherical structure (namely virosomes), in which L proteins translocated across the membrane in the correct topology[34]. Following incorporation of beads and genes, these virosomes could efficiently deliver into the cytoplasm of human hepatic cells in vitro[28] and specifically to human hepatic cells in vivo[28,34]. Furthermore, after incorporating doxorubicin by remote loading method, the virosomes were intravenously injected into xenograft mice harboring human hepatic cell-derived tumors. The virosomes could retard the tumor growth more effectively than LPs containing doxorubicin, strongly suggesting that virosome could delivery their payload into human hepatic cell-derived tumor efficiently utilizing HBV-derived infection machinery[34]. These results indicate that the virosome is a promising nanocarrier for cytoplasmic delivery of drugs and genes.
Intravenously injected nanoparticles were unexpectedly trapped by the reticuloendothelial system (RES) in liver, lung, spleen, and so on. However, some viruses can evade RES effectively and finally infect target cells and tissues in vivo, namely by virus-derived stealth activity. Meanwhile, it was demonstrated that HBV could associates with monomeric human serum albumin (HSA) and polymerized-HSA[35,36] through the polymerized-albumin receptor (PAR) domain in the pre-S2 region (120-129 aa)[37]. When LPs displaying the PAR domain-containing peptide were intravenously injected into mice, they could recruit albumins on their surface and evade RES effectively (Takagi et al personal communication). Indeed, in nude mice harboring human hepatic cell-derived tumors, intravenously injected BNC could target and enter the target tumors by evading RES[32]. Thus, BNC (presumably as well as HBV) could recruit albumin in blood stream by its PAR domain and then exhibit stealth activity.
As described in the above paragraph, it was demonstrated that BNC possesses HBV-derived infection machinery. Compared with HBV virion, BNC has the following advantages for the elucidation of early infection machinery of HBV. First, the substantial amount of purified BNCs could be easily obtained from recombinant yeast cells. Second, BNCs could be easily labeled with fluorophores and enzymes for cytochemical and biochemical analyses.
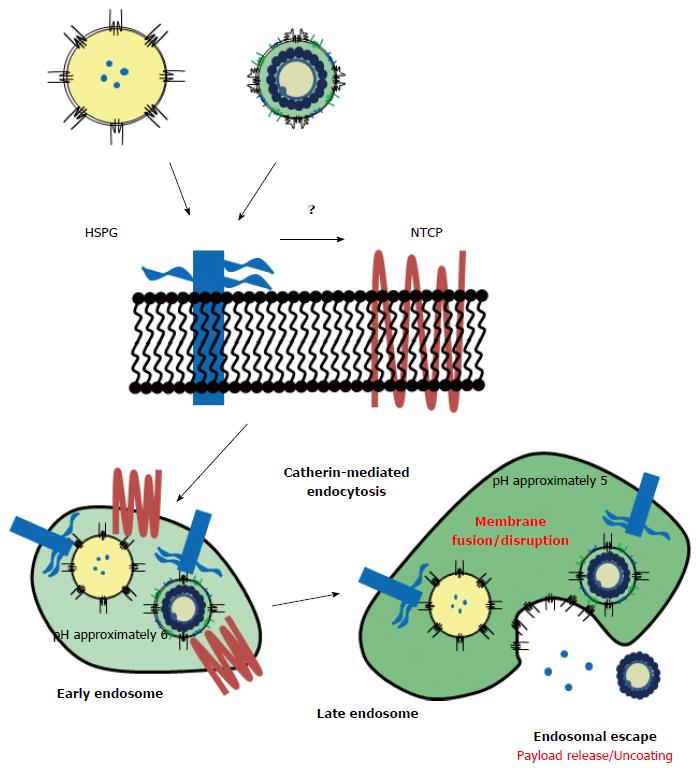
As described above, HBV is considered to interact primarily with HSPG (a low-affinity HBV receptor), change its own structure, and then interact with NTCP (a high-affinity HBV receptor)[18,19]. HSPG is abundantly expressed in the extracellular matrix of various tissues, which could interact with AGL of S region[8,11]. The binding of BNC to human hepatic cells[32] was efficiently suppressed by heparin[38]. Since the membrane topology of BNC is similar with HBV, HSPG might act as a low-affinity receptor for BNC. After BNC attached onto human hepatic cells, it was internalized mainly by clathrin-dependent endocytosis, with a rate very similar to HBV[39] (Figure 3). These results led us to assume that both BNC and HBV utilize the same machinery for cell attachment and endocytosis. The N-terminally myristoylated pre-S1 region (from Gly-2 to Val-47; see Figure 2) is essential for interaction of HBV with NTCP[40], but the original BNC lacks the myristoylated N-terminus owing to the addition of a signal peptide for expression in yeast cells[24]. After chemical modification with myristoyl group, the myristoylated BNC (Myr-BNC) was confirmed to interact with NTCP by immunoprecipitation assay by using lysate of NTCP-overexpressing HepG2 cells (HepG2/NTCP cells), and inhibit the infection of HepG2/NTCP cells with HBV in vitro, whereas BNC itself neither[41]. For evaluating the contribution of NTCP to the cell attachment and entry of HBV, fluorophore-labeled form of BNCs, Myr-BNCs, and HB patient plasma-derived HBsAg particles (containing native L protein with N-terminal myristoylation) was incubated with either HepG2 or HepG2/NTCP cells, and then analyzed by flow cytometry. Each of three particle was found to be equally associated with both cell types and be finally localized in late endosomes at comparable level (approx. 20% of each particle)[41]. Furthermore, overexpressed NTCP in HepG2 cells (HepG2/NTCP cells) could neither enhance the cell-surface interaction nor the internalization of fluorophore-labeled form of Myr-BNCs or HBsAg particles, strongly suggesting that cell surface NTCP is not involved in the interaction with HBV (Figure 3)[41]. It is likely that other receptors participate in the human hepatic cell-specific interaction and internalization of BNC and HBV.
For the majority of viruses, entry into the cell is via the endocytic pathway. Upon reaching late endosomes, the subsequent uncoating process assists in endosomal escape; otherwise, viruses would be degraded in lysosomes[42]. As is the case with other enveloped viruses, it was postulated that HBV also escapes from endosomes using the membrane fusion machinery[43]. The following fusogenic domains have been identified in L protein: C-terminal half of pre-S2 region (pH-independent; amino acid residues from 149 to 160)[44], N-terminal part of S region (low pH-dependent; amino acid residues from 164 to 186)[45], and the whole pre-S1 region (low pH-dependent)[46]. However, it has thus far remained controversial as to which domains are responsible for the uncoating process of HBV in endosomes. Recently, by lipid mixing assay using BNC or LPs displaying pre-S1-derived mutant peptide, we identified novel low pH-dependent fusogenic domain in the N-terminus of pre-S1 region (from Asn-9 to Gly-24; see Figure 2)[47]. When BNC lacking pre-S1 region was used for the lipid-mixing assay, the fusogenic activity was completely lost. Pre-incubation of BNC with anti-pre-S1 antibodies also inhibited fusion. These results indicated that the fusogenic activity of pre-S1 (9-24) peptide is dominant over those of other fusogenic domains (see above). Furthermore, upon mixing LPs containing fluorophore and quencher (model of endosomes) with BNC or LPs displaying pre-S1 (9-24) peptide, the fluorophore was immediately released at low pH. This suggested that the pre-S1 (9-24) peptide possesses low pH-dependent membrane disruption activity. When BNCs containing fluorophore were mixed with LPs, the fluorophore was released at low pH, suggesting that BNC as well as HBV rupture upon interaction with endosomal membrane at low pH. These results strongly suggested that the pre-S1 (9-24) peptide is essential for the endosomal escape of BNC payload as well as the uncoating process of HBV (Figure 3), which agreed well with Watashi et al[48]. More recently, the fusogenic activity of pre-S1 (9-24) peptide was shown to correlate with its hydrophobicity, which is significantly enhanced by the protonation of Asp-16 and Asp-20 under acidic conditions[49].
While many researchers have attempted to isolate functional HBV receptors since the last two decades, HSPG and NTCP were finally identified as low-affinity and high-affinity HBV receptors, respectively. However, it is still unclear whether both molecules by themselves support the stringent specificity of HBV to human hepatic cells. Since NTCP is not expressed in all HBV susceptible cells, other molecules may participate in the interaction of HBV with hepatic cells in a cell-specific manner. The mechanism of transfer of HSPG-bound HBV to NTCP, presumably in the endosomes, and the change in structure of HBV (pre-S1 region) at low pH for adaptation to NTCP remains to be elucidated (see Figure 3). Interestingly, the pre-S1 (9-24) peptide is well conserved among all HBV genotypes, and is located within the NTCP-binding site (from Gly-2 to Val-47) (see Figure 2)[16]. This positional relationship of the two domains implies that the uncoating process of HBV and the endosomal escape of BNC are initiated by interaction with NTCP in late endosomes. The elucidation of the molecular machinery of HBV early infection can open avenues for novel targets for anti-HBV drugs.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Japan
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Luo FL, McQuillan GM, Zhang YY S- Editor: Yu J L- Editor: A E- Editor: Zhang FF
| 1. | Tiollais P, Pourcel C, Dejean A. The hepatitis B virus. Nature. 1985;317:489-495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 934] [Cited by in RCA: 927] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 2. | Walter E, Keist R, Niederöst B, Pult I, Blum HE. Hepatitis B virus infection of tupaia hepatocytes in vitro and in vivo. Hepatology. 1996;24:1-5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 93] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 3. | Gripon P, Rumin S, Urban S, Le Seyec J, Glaise D, Cannie I, Guyomard C, Lucas J, Trepo C, Guguen-Guillouzo C. Infection of a human hepatoma cell line by hepatitis B virus. Proc Natl Acad Sci USA. 2002;99:15655-15660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 926] [Cited by in RCA: 990] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 4. | Neurath AR, Kent SB, Strick N, Parker K. Identification and chemical synthesis of a host cell receptor binding site on hepatitis B virus. Cell. 1986;46:429-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 412] [Cited by in RCA: 402] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 5. | Heermann KH, Goldmann U, Schwartz W, Seyffarth T, Baumgarten H, Gerlich WH. Large surface proteins of hepatitis B virus containing the pre-s sequence. J Virol. 1984;52:396-402. [PubMed] |
| 6. | Glebe D, Aliakbari M, Krass P, Knoop EV, Valerius KP, Gerlich WH. Pre-s1 antigen-dependent infection of Tupaia hepatocyte cultures with human hepatitis B virus. J Virol. 2003;77:9511-9521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 151] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 7. | Gripon P, Le Seyec J, Rumin S, Guguen-Guillouzo C. Myristylation of the hepatitis B virus large surface protein is essential for viral infectivity. Virology. 1995;213:292-299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 149] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 8. | Schulze A, Gripon P, Urban S. Hepatitis B virus infection initiates with a large surface protein-dependent binding to heparan sulfate proteoglycans. Hepatology. 2007;46:1759-1768. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 303] [Cited by in RCA: 351] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 9. | Leistner CM, Gruen-Bernhard S, Glebe D. Role of glycosaminoglycans for binding and infection of hepatitis B virus. Cell Microbiol. 2008;10:122-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 83] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 10. | Abou-Jaoudé G, Sureau C. Entry of hepatitis delta virus requires the conserved cysteine residues of the hepatitis B virus envelope protein antigenic loop and is blocked by inhibitors of thiol-disulfide exchange. J Virol. 2007;81:13057-13066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 87] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 11. | Salisse J, Sureau C. A function essential to viral entry underlies the hepatitis B virus “a” determinant. J Virol. 2009;83:9321-9328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 92] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 12. | Sureau C, Salisse J. A conformational heparan sulfate binding site essential to infectivity overlaps with the conserved hepatitis B virus a-determinant. Hepatology. 2013;57:985-994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 155] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 13. | Bernfield M, Götte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729-777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2114] [Cited by in RCA: 2115] [Article Influence: 84.6] [Reference Citation Analysis (0)] |
| 14. | Engelman A, Cherepanov P. The structural biology of HIV-1: mechanistic and therapeutic insights. Nat Rev Microbiol. 2012;10:279-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 252] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 15. | Hagenbuch B, Meier PJ. Molecular cloning, chromosomal localization, and functional characterization of a human liver Na+/bile acid cotransporter. J Clin Invest. 1994;93:1326-1331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 337] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 16. | Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife. 2012;1:e00049. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1280] [Cited by in RCA: 1593] [Article Influence: 122.5] [Reference Citation Analysis (1)] |
| 17. | Döring B, Lütteke T, Geyer J, Petzinger E. The SLC10 carrier family: transport functions and molecular structure. Curr Top Membr. 2012;70:105-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 100] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 18. | Ni Y, Lempp FA, Mehrle S, Nkongolo S, Kaufman C, Fälth M, Stindt J, Königer C, Nassal M, Kubitz R. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology. 2014;146:1070-1083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 540] [Cited by in RCA: 619] [Article Influence: 56.3] [Reference Citation Analysis (0)] |
| 19. | Urban S, Bartenschlager R, Kubitz R, Zoulim F. Strategies to inhibit entry of HBV and HDV into hepatocytes. Gastroenterology. 2014;147:48-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 256] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 20. | Valenzuela P, Medina A, Rutter WJ, Ammerer G, Hall BD. Synthesis and assembly of hepatitis B virus surface antigen particles in yeast. Nature. 1982;298:347-350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 629] [Cited by in RCA: 590] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 21. | Clemens R, Sänger R, Kruppenbacher J, Höbel W, Stanbury W, Bock HL, Jilg W. Booster immunization of low- and non-responders after a standard three dose hepatitis B vaccine schedule--results of a post-marketing surveillance. Vaccine. 1997;15:349-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 70] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 22. | Itoh Y, Takai E, Ohnuma H, Kitajima K, Tsuda F, Machida A, Mishiro S, Nakamura T, Miyakawa Y, Mayumi M. A synthetic peptide vaccine involving the product of the pre-S(2) region of hepatitis B virus DNA: protective efficacy in chimpanzees. Proc Natl Acad Sci USA. 1986;83:9174-9178. [PubMed] |
| 23. | Kuroda S, Fujisawa Y, Iino S, Akahane Y, Suzuki H. Induction of protection level of anti-pre-S2 antibodies in humans immunized with a novel hepatitis B vaccine consisting of M (pre-S2 + S) protein particles (a third generation vaccine). Vaccine. 1991;9:163-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 24. | Kuroda S, Otaka S, Miyazaki T, Nakao M, Fujisawa Y. Hepatitis B virus envelope L protein particles. Synthesis and assembly in Saccharomyces cerevisiae, purification and characterization. J Biol Chem. 1992;267:1953-1961. [PubMed] |
| 25. | Jung J, Iijima M, Yoshimoto N, Sasaki M, Niimi T, Tatematsu K, Jeong SY, Choi EK, Tanizawa K, Kuroda S. Efficient and rapid purification of drug- and gene-carrying bio-nanocapsules, hepatitis B virus surface antigen L particles, from Saccharomyces cerevisiae. Protein Expr Purif. 2011;78:149-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 26. | Yamada T, Iwabuki H, Kanno T, Tanaka H, Kawai T, Fukuda H, Kondo A, Seno M, Tanizawa K, Kuroda S. Physicochemical and immunological characterization of hepatitis B virus envelope particles exclusively consisting of the entire L (pre-S1 + pre-S2 + S) protein. Vaccine. 2001;19:3154-3163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 64] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 27. | Bruss V. Hepatitis B virus morphogenesis. World J Gastroenterol. 2007;13:65-73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 198] [Cited by in RCA: 213] [Article Influence: 11.8] [Reference Citation Analysis (1)] |
| 28. | Jung J, Matsuzaki T, Tatematsu K, Okajima T, Tanizawa K, Kuroda S. Bio-nanocapsule conjugated with liposomes for in vivo pinpoint delivery of various materials. J Control Release. 2008;126:255-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 62] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 29. | Bremer CM, Bung C, Kott N, Hardt M, Glebe D. Hepatitis B virus infection is dependent on cholesterol in the viral envelope. Cell Microbiol. 2009;11:249-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 74] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 30. | Somiya M, Kuroda S. Development of a virus-mimicking nanocarrier for drug delivery systems: The bio-nanocapsule. Adv Drug Deliv Rev. 2015;95:77-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 31. | Kasuya T, Jung J, Kinoshita R, Goh Y, Matsuzaki T, Iijima M, Yoshimoto N, Tanizawa K, Kuroda S. Chapter 8 - Bio-nanocapsule-liposome conjugates for in vivo pinpoint drug and gene delivery. Methods Enzymol. 2009;464:147-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 32. | Yamada T, Iwasaki Y, Tada H, Iwabuki H, Chuah MK, VandenDriessche T, Fukuda H, Kondo A, Ueda M, Seno M. Nanoparticles for the delivery of genes and drugs to human hepatocytes. Nat Biotechnol. 2003;21:885-890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 180] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 33. | Matsuura Y, Yagi H, Matsuda S, Itano O, Aiura K, Kuroda S, Ueda M, Kitagawa Y. Human liver-specific nanocarrier in a novel mouse xenograft model bearing noncancerous human liver tissue. Eur Surg Res. 2011;46:65-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 34. | Liu Q, Jung J, Somiya M, Iijima M, Yoshimoto N, Niimi T, Maturana AD, Shin SH, Jeong SY, Choi EK. Virosomes of hepatitis B virus envelope L proteins containing doxorubicin: synergistic enhancement of human liver-specific antitumor growth activity by radiotherapy. Int J Nanomedicine. 2015;10:4159-4172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 35. | Krone B, Lenz A, Heermann KH, Seifer M, Lu XY, Gerlich WH. Interaction between hepatitis B surface proteins and monomeric human serum albumin. Hepatology. 1990;11:1050-1056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 36. | Ishihara K, Waters JA, Pignatelli M, Thomas HC. Characterisation of the polymerised and monomeric human serum albumin binding sites on hepatitis B surface antigen. J Med Virol. 1987;21:89-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 37. | Itoh Y, Kuroda S, Miyazaki T, Otaka S, Fujisawa Y. Identification of polymerized-albumin receptor domain in the pre-S2 region of hepatitis B virus surface antigen M protein. J Biotechnol. 1992;23:71-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 38. | Kasuya T, Nomura S, Matsuzaki T, Jung J, Yamada T, Tatematsu K, Okajima T, Tanizawa K, Kuroda S. Expression of squamous cell carcinoma antigen-1 in liver enhances the uptake of hepatitis B virus envelope-derived bio-nanocapsules in transgenic rats. FEBS J. 2008;275:5714-5724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 39. | Yamada M, Oeda A, Jung J, Iijima M, Yoshimoto N, Niimi T, Jeong SY, Choi EK, Tanizawa K, Kuroda S. Hepatitis B virus envelope L protein-derived bio-nanocapsules: mechanisms of cellular attachment and entry into human hepatic cells. J Control Release. 2012;160:322-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 40. | Meier A, Mehrle S, Weiss TS, Mier W, Urban S. Myristoylated PreS1-domain of the hepatitis B virus L-protein mediates specific binding to differentiated hepatocytes. Hepatology. 2013;58:31-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 117] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 41. | Somiya M, Liu Q, Yoshimoto N, Iijima M, Tatematsu K, Nakai T, Okajima T, Kuroki K, Ueda K, Kuroda S. Cellular uptake of hepatitis B virus envelope L particles is independent of sodium taurocholate cotransporting polypeptide, but dependent on heparan sulfate proteoglycan. Virology. 2016;497:23-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 42. | Smith AE, Helenius A. How viruses enter animal cells. Science. 2004;304:237-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 561] [Cited by in RCA: 557] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 43. | Baumert TF, Meredith L, Ni Y, Felmlee DJ, McKeating JA, Urban S. Entry of hepatitis B and C viruses - recent progress and future impact. Curr Opin Virol. 2014;4:58-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 44. | Oess S, Hildt E. Novel cell permeable motif derived from the PreS2-domain of hepatitis-B virus surface antigens. Gene Ther. 2000;7:750-758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 87] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 45. | Rodríguez-Crespo I, Gómez-Gutiérrez J, Nieto M, Peterson DL, Gavilanes F. Prediction of a putative fusion peptide in the S protein of hepatitis B virus. J Gen Virol. 1994;75:637-639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 46. | Delgado CL, Núñez E, Yélamos B, Gómez-Gutiérrez J, Peterson DL, Gavilanes F. Study of the putative fusion regions of the preS domain of hepatitis B virus. Biochim Biophys Acta. 2015;1848:895-906. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 47. | Somiya M, Sasaki Y, Matsuzaki T, Liu Q, Iijima M, Yoshimoto N, Niimi T, Maturana AD, Kuroda S. Intracellular trafficking of bio-nanocapsule-liposome complex: Identification of fusogenic activity in the pre-S1 region of hepatitis B virus surface antigen L protein. J Control Release. 2015;212:10-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 48. | Watashi K, Wakita T. Hepatitis B Virus and Hepatitis D Virus Entry, Species Specificity, and Tissue Tropism. Cold Spring Harb Perspect Med. 2015;5:a021378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 42] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 49. | Liu Q, Somiya M, Shimada N, Sakamoto W, Yoshimoto N, Iijima M, Tatematsu K, Nakai T, Okajima T, Maruyama A. Mutational analysis of hepatitis B virus pre-S1 (9-24) fusogenic peptide. Biochem Biophys Res Commun. 2016;474:406-412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |