Published online Jul 21, 2016. doi: 10.3748/wjg.v22.i27.6095
Peer-review started: April 18, 2016
First decision: May 12, 2016
Revised: May 23, 2016
Accepted: June 13, 2016
Article in press: June 13, 2016
Published online: July 21, 2016
Processing time: 92 Days and 16.1 Hours
Myanmar is adjacent to India, Bangladesh, Thailand, Laos and China. In Myanmar, the prevalence of hepatitis C virus (HCV) infection is 2%, and HCV infection accounts for 25% of hepatocellular carcinoma. In this study, we reviewed the prevalence of HCV genotypes in Myanmar. HCV genotypes 1, 3 and 6 were observed in volunteer blood donors in and around the Myanmar city of Yangon. Although there are several reports of HCV genotype 6 and its variants in Myanmar, the distribution of the HCV genotypes has not been well documented in areas other than Yangon. Previous studies showed that treatment with peginterferon and a weight-based dose of ribavirin for 24 or 48 wk could lead to an 80%-100% sustained virological response (SVR) rates in Myanmar. Current interferon-free treatments could lead to higher SVR rates (90%-95%) in patients infected with almost all HCV genotypes other than HCV genotype 3. In an era of heavy reliance on direct-acting antivirals against HCV, there is an increasing need to measure HCV genotypes, and this need will also increase specifically in Myanmar. Current available information of HCV genotypes were mostly from Yangon and other countries than Myanmar. The prevalence of HCV genotypes in Myanmar should be determined.
Core tip: We reviewed the prevalence of hepatitis C virus (HCV) genotypes in Myanmar. HCV genotypes 1, 3 and 6 were observed in volunteer blood donors in and around the Myanmar city of Yangon. Although there are several reports of HCV genotype 6 in Myanmar, the distribution of HCV genotypes has not been well documented in areas other than Yangon. Previous studies showed that treatment with peginterferon and a weight-based dose of ribavirin for 24 or 48 wk could lead to an 80%-100% sustained virological response in Myanmar.
- Citation: Win NN, Kanda T, Nakamoto S, Yokosuka O, Shirasawa H. Hepatitis C virus genotypes in Myanmar. World J Gastroenterol 2016; 22(27): 6095-6099
- URL: https://www.wjgnet.com/1007-9327/full/v22/i27/6095.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i27.6095
The hepatitis C virus (HCV) is a single and positive-stranded RNA virus that is approximately 9600 nucleotides in length[1,2]. HCV infection causes acute and chronic hepatitis, cirrhosis and hepatocellular carcinoma (HCC)[3]. HCV is classified into at least 7 confirmed genotypes and 67 subtypes[4].
Myanmar is located in Southeast Asia and has recently emerged as a nation that is pursuing a peaceful transition to democracy[5]. In Myanmar, the prevalence of HCV infection is 2%, and it accounts for 25% of HCC[5]. HCV genotypes influence the outcome and the duration of interferon-based[6,7] and interferon-free treatments[8]. Direct-acting antivirals (DAAs) against HCV could lead to higher sustained virological response (SVR) rates with fewer adverse events[9,10]. HCV genotypes are very important factors in the selection of DAAs and treatment regimens[9,10].
Myanmar is adjacent to India, Bangladesh, Thailand, Laos and China. In India, HCV genotypes 3a, 3b, 1b and 1a have prevalence rates of 50%, 25%, 14% and 10%, respectively, whereas HCV genotype 4 has only a 4% prevalence rate and has been found only in Southern and Western India[11]. The most common HCV genotypes in patients with chronic HCV infection in Bangladesh were types 3 (50%), 3 and 4 (29%), and 1 (14.4%)[12]. In blood donors in Thailand, the most common HCV genotype was 3a (43%), followed by 1b (13%), 6f (13%), 6i (8.7%), 1a (4.4%), 3b (4.4%), 6c (4.4%), 6j (4.4%), and 6n (4.4%)[13]. In Laos, HCV genotypes 1 and 6 have prevalence rates of 5% and 95%, respectively[14]. In Mainland China, 42%, 44% and 14% of patients have HCV genotypes 1, 2 and 3, respectively[15]. In this article, we reviewed the prevalence of HCV genotypes in Myanmar.
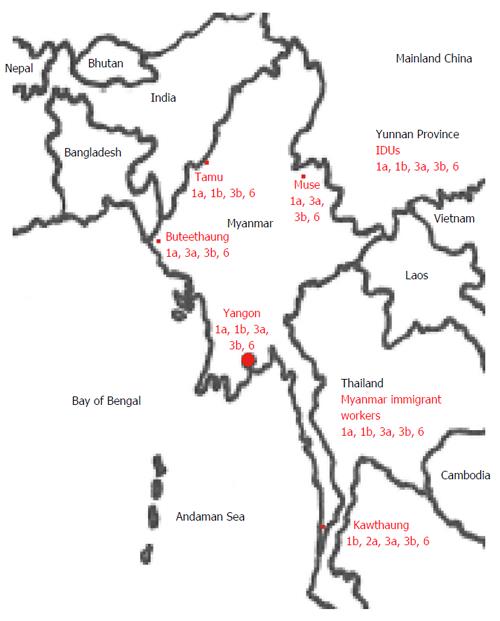
Nakai et al[16] reported that the most common HCV genotype among 24 patients with liver diseases who were examined at Yangon General Hospital, Yangon, Myanmar, was 3b (67%), followed by genotypes 1a (13%) and 3a (8%); the genotypes were determined using PCR with genotype-specific primers. Mellor et al[17] used the PCR-restriction fragment length polymorphism method to show that variants of HCV genotype 6 exist in Myanmar. Shinji et al[18] reported prevalence rates of 31%, 47% and 21% for variants of HCV genotypes 1, 3 and 6 variants, respectively, in volunteer blood donors in and around the Myanmar city of Yangon; these findings were determined using direct sequencing of PCR products. Previous reports[16-19] showed the inconsistent data of the prevalence of HCV genotypes among the different area in Myanmar (Table 1), suggesting that this difference may attribute to the regional difference (Figure 1).
| Ref. | Area/subjects | GT-1a | GT-1b | GT-2a | GT-3a | GT-3b | GT-6 |
| Nakai et al[16], 2001 | Yangon/liver diseases | 13% | 4% | - | 8% | 67% | - |
| Shinji et al[18], 2004 | Yangon/healthy blood donors | 4.5% | 20% | - | 12% | 32% | 21% |
| Lwin et al[19], 2007 | Muse/healthy people | 9% | - | - | 11% | 20% | 60% |
| Lwin et al[19], 2007 | Tamu/healthy people | 2% | 15% | - | - | 25% | 58% |
| Lwin et al[19], 2007 | Kawthaung/healthy people | - | 7% | 7% | 13% | 33% | 40% |
| Lwin et al[19], 2007 | Buteethaung/healthy people | 4% | - | - | 27% | 53% | 16% |
| Akkarathamrongsin et al[23], 2011 | Thailand/immigrant workers from Myanmar | 6.7% | 6.7% | - | 27% | 33% | 27% |
Lwin et al[19] reported that HCV genotype 6 was the most prevalent genotype (49%), followed by HCV genotypes 3 (39%), 1 (11%), and 2 (0.7%). In Myanmar, HCV genotype 6 was most often found in patients in the northern cities and HCV genotype 3 in the southern and western cities, suggesting that there are regional differences in HCV genotype distribution[19].
In the Yunnan province in China, where is located in the far southern part of Mainland China bordering Laos, Vietnam, and Myanmar, HCV genotypes 1a, 1b, 3a, 3b, 6a, 6n, and 6u were found in 1.3%, 20%, 24%, 30%, 5%, 11% and 8.8%, respectively, of patients who were co-infected with HCV and HIV[20,21]. A similar HCV genotype distribution of intravenous drug users was reported in this area[22]. Zhang et al[22] reported that HCV genotype 6 was most common (47%), followed by HCV genotypes 3 (41%) and 1 (12%) in intravenous drug users of the Yunnan province. Lwin et al[19] reported that HCV genotypes 1a, 3a, 3b, and 6 were found in 9%, 11%, 20%, and 60%, respectively, of patients in Muse where is located adjacent to the Yunnan. There seems to be the some association of HCV genotype distribution between Muse in Myanmar and Yunnan province in Mainland China.
In a large number of immigrant workers from Cambodia and Myanmar to Thailand, the predominant HCV genotypes were 1a, 1b, 3a, 3b and 6 (6e, 6f, 6m, 6p and 6r)[23]. The seroprevalence of HCV infection in immigrant workers from Cambodia and Myanmar to Thailand was reported to be 2.3% or 1.7%, respectively. HCV genotypes 1a, 1b, 3a, 3b and 6 were 0%, 24%, 16%, 4% and 56%, respectively, in immigrant workers from Cambodia to Thailand, and those were 6.7%, 6.7%, 26.7%, 33.3% and 26.6%, respectively, in immigrant workers from Myanmar to Thailand. Geographic distribution of HCV genotype 6 covered mainly southern China and the mainland of Southeast Asia, including Vietnam, Laos, Thailand, Cambodia and Myanmar[24-27].
Treatment of peginterferon and a weight-based dose of ribavirin for 24 or 48 wk were given to patients infected with HCV genotypes 2 and 3 or HCV genotypes 1 and 6. SVR rates were 81.2% (39/48), 100% (2/2), 85.5% (94/110), 90.3% (28/31) and 100% (4/4) in patients infected with HCV genotypes 1, 2, 3, 6 and with an indeterminate genotype, respectively[5].
DAAs are currently available and will be available in the near future to treat patients infected with HCV. In regards to the use of DAAs, the importance of measuring HCV genotypes is increasing and will also increase specifically in Myanmar. DAAs against HCV could lead to higher SVR rates (90%-95%) in patients infected with almost all HCV genotypes other than HCV genotype 3[9,10,28]. With current DAAs, HCV genotype 3 is the most difficult-to cure HCV genotype[28]. HCV NS5B polymerase nucleotide inhibitor sofosbuvir plus ribavirin for 12 and 24 wk could lead 61%-68% and 94% SVR rates, respectively, in non-cirrhotic treatment-naïve patients with HCV genotype 3[28]. Those could lead only 21%-34% and 92% SVR rates, respectively, in cirrhotic treatment-naïve patients with HCV genotype 3[28]. In treatment-experienced patients, those treatments could lead to less SVR rates[28]. In patients with HCV genotype 6, HCV NS5A inhibitor ledipasvir plus sofosbuvir could lead to approximately 96% SVR rates[28]. Most of the data of HCV genotypes were from Yangon and countries other than Myanmar. It is important to determine the prevalence of HCV genotypes in Myanmar.
Manuscript source: Invited manuscript
P- Reviewer: Kato T, Yoshioka K S- Editor: Gong ZM L- Editor: A E- Editor: Wang CH
| 1. | Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 973] [Cited by in RCA: 1005] [Article Influence: 55.8] [Reference Citation Analysis (0)] |
| 2. | Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4996] [Cited by in RCA: 4656] [Article Influence: 129.3] [Reference Citation Analysis (0)] |
| 3. | Saito I, Miyamura T, Ohbayashi A, Harada H, Katayama T, Kikuchi S, Watanabe Y, Koi S, Onji M, Ohta Y. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA. 1990;87:6547-6549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 826] [Cited by in RCA: 840] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 4. | Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology. 2014;59:318-327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 965] [Cited by in RCA: 981] [Article Influence: 89.2] [Reference Citation Analysis (1)] |
| 5. | Naing C, Sitt T, Aung AT, Aung K. Sustained Virologic Response to a Dual Peginterferon alfa-2a and Ribavirin in Treating Chronic hepatitis C Infection: A Retrospective Cohort Study. Medicine (Baltimore). 2015;94:e1234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 6. | Kanda T, Imazeki F, Yokosuka O. New antiviral therapies for chronic hepatitis C. Hepatol Int. 2010;4:548-561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 98] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 7. | Omata M, Kanda T, Yu ML, Yokosuka O, Lim SG, Jafri W, Tateishi R, Hamid SS, Chuang WL, Chutaputti A. APASL consensus statements and management algorithms for hepatitis C virus infection. Hepatol Int. 2012;6:409-435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 142] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 8. | Kanda T, Yokosuka O, Omata M. Treatment of hepatitis C virus infection in the future. Clin Transl Med. 2013;2:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 9. | Lok AS, Gardiner DF, Lawitz E, Martorell C, Everson GT, Ghalib R, Reindollar R, Rustgi V, McPhee F, Wind-Rotolo M. Preliminary study of two antiviral agents for hepatitis C genotype 1. N Engl J Med. 2012;366:216-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 478] [Cited by in RCA: 476] [Article Influence: 36.6] [Reference Citation Analysis (0)] |
| 10. | Zeuzem S, Dusheiko GM, Salupere R, Mangia A, Flisiak R, Hyland RH, Illeperuma A, Svarovskaia E, Brainard DM, Symonds WT. Sofosbuvir and ribavirin in HCV genotypes 2 and 3. N Engl J Med. 2014;370:1993-2001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 660] [Cited by in RCA: 638] [Article Influence: 58.0] [Reference Citation Analysis (0)] |
| 11. | Das BR, Kundu B, Khandapkar R, Sahni S. Geographical distribution of hepatitis C virus genotypes in India. Indian J Pathol Microbiol. 2002;45:323-328. [PubMed] |
| 12. | Islam MS, Miah MR, Roy PK, Rahman O, Siddique AB, Chowdhury J, Ahmed F, Rahman S, Khan MR. Genotypes of hepatitis C virus infection in Bangladeshi population. Mymensingh Med J. 2015;24:143-151. [PubMed] |
| 13. | Wasitthankasem R, Posuwan N, Vichaiwattana P, Theamboonlers A, Klinfueng S, Vuthitanachot V, Thanetkongtong N, Saelao S, Foonoi M, Fakthongyoo A. Decreasing Hepatitis C Virus Infection in Thailand in the Past Decade: Evidence from the 2014 National Survey. PLoS One. 2016;11:e0149362. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 14. | Hübschen JM, Jutavijittum P, Thammavong T, Samountry B, Yousukh A, Toriyama K, Sausy A, Muller CP. High genetic diversity including potential new subtypes of hepatitis C virus genotype 6 in Lao People’s Democratic Republic. Clin Microbiol Infect. 2011;17:E30-E34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 15. | Omata M, Kanda T, Yokosuka O, Crawford D, Al-Mahtab M, Wei L, Ibrahim A, Lau GK, Sharma BC, Hamid SS. Features of hepatitis C virus infection, current therapies and ongoing clinical trials in ten Asian Pacific countries. Hepatol Int. 2015;9:486-507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 16. | Nakai K, Win KM, Oo SS, Arakawa Y, Abe K. Molecular characteristic-based epidemiology of hepatitis B, C, and E viruses and GB virus C/hepatitis G virus in Myanmar. J Clin Microbiol. 2001;39:1536-1539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 17. | Mellor J, Walsh EA, Prescott LE, Jarvis LM, Davidson F, Yap PL, Simmonds P. Survey of type 6 group variants of hepatitis C virus in Southeast Asia by using a core-based genotyping assay. J Clin Microbiol. 1996;34:417-423. [PubMed] |
| 18. | Shinji T, Kyaw YY, Gokan K, Tanaka Y, Ochi K, Kusano N, Mizushima T, Fujioka S, Shiraha H, Lwin AA. Analysis of HCV genotypes from blood donors shows three new HCV type 6 subgroups exist in Myanmar. Acta Med Okayama. 2004;58:135-142. [PubMed] |
| 19. | Lwin AA, Shinji T, Khin M, Win N, Obika M, Okada S, Koide N. Hepatitis C virus genotype distribution in Myanmar: Predominance of genotype 6 and existence of new genotype 6 subtype. Hepatol Res. 2007;37:337-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 57] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 20. | Xia X, Lu L, Tee KK, Zhao W, Wu J, Yu J, Li X, Lin Y, Mukhtar MM, Hagedorn CH. The unique HCV genotype distribution and the discovery of a novel subtype 6u among IDUs co-infected with HIV-1 in Yunnan, China. J Med Virol. 2008;80:1142-1152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 72] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 21. | Xia X, Zhao W, Tee KK, Feng Y, Takebe Y, Li Q, Pybus OG, Lu L. Complete genome sequencing and phylogenetic analysis of HCV isolates from China reveals a new subtype, designated 6u. J Med Virol. 2008;80:1740-1746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 22. | Zhang Z, Yao Y, Wu W, Feng R, Wu Z, Cun W, Dong S. Hepatitis C virus genotype diversity among intravenous drug users in Yunnan Province, Southwestern China. PLoS One. 2013;8:e82598. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 23. | Akkarathamrongsin S, Praianantathavorn K, Hacharoen N, Theamboonlers A, Tangkijvanich P, Poovorawan Y. Seroprevalence and genotype of hepatitis C virus among immigrant workers from Cambodia and Myanmar in Thailand. Intervirology. 2011;54:10-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 24. | Pham DA, Leuangwutiwong P, Jittmittraphap A, Luplertlop N, Bach HK, Akkarathamrongsin S, Theamboonlers A, Poovorawan Y. High prevalence of Hepatitis C virus genotype 6 in Vietnam. Asian Pac J Allergy Immunol. 2009;27:153-160. [PubMed] |
| 25. | Antaki N, Craxi A, Kamal S, Moucari R, Van der Merwe S, Haffar S, Gadano A, Zein N, Lai CL, Pawlotsky JM. The neglected hepatitis C virus genotypes 4, 5 and 6: an international consensus report. Liver Int. 2010;30:342-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 120] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 26. | Mixson-Hayden T, Lee D, Ganova-Raeva L, Drobeniuc J, Stauffer WM, Teshale E, Kamili S. Hepatitis B virus and hepatitis C virus infections in United States-bound refugees from Asia and Africa. Am J Trop Med Hyg. 2014;90:1014-1020. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 27. | Wasitthankasem R, Vongpunsawad S, Siripon N, Suya C, Chulothok P, Chaiear K, Rujirojindakul P, Kanjana S, Theamboonlers A, Tangkijvanich P. Genotypic distribution of hepatitis C virus in Thailand and Southeast Asia. PLoS One. 2015;10:e0126764. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 68] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 28. | Omata M, Kanda T, Wei L, Yu ML, Chuang WL, Ibrahim A, Lesmana CR, Sollano J, Kumar M, Jindal A. APASL consensus statements and recommendation on treatment of hepatitis C. Hepatol Int. 2016; Epub ahead of print. [PubMed] |