Published online May 21, 2016. doi: 10.3748/wjg.v22.i19.4716
Peer-review started: February 13, 2016
First decision: March 7, 2016
Revised: March 10, 2016
Accepted: March 30, 2016
Article in press: March 30, 2016
Published online: May 21, 2016
Processing time: 95 Days and 22.2 Hours
AIM: To investigate the expression pattern of plasma long noncoding RNAs (lncRNAs) in Chrohn’s disease (CD) patients.
METHODS: Microarray screening and qRT-PCR verification of lncRNAs and mRNAs were performed in CD and control subjects, followed by hierarchy clustering, GO and KEGG pathway analyses. Significantly dysregulated lncRNAs were categorized into subgroups of antisense lncRNAs, enhancer lncRNAs and lincRNAs. To predict the regulatory effect of lncRNAs on mRNAs, a CNC network analysis was performed and cross linked with significantly changed lncRNAs. The overlapping lncRNAs were randomly selected and verified by qRT-PCR in a larger cohort.
RESULTS: Initially, there were 1211 up-regulated and 777 down-regulated lncRNAs as well as 1020 up-regulated and 953 down-regulated mRNAs after microarray analysis; a heat map based on these results showed good categorization into the CD and control groups. GUSBP2 and AF113016 had the highest fold change of the up- and down-regulated lncRNAs, whereas TBC1D17 and CCL3L3 had the highest fold change of the up- and down-regulated mRNAs. Six (SNX1, CYFIP2, CD6, CMTM8, STAT4 and IGFBP7) of 10 mRNAs and 8 (NR_033913, NR_038218, NR_036512, NR_049759, NR_033951, NR_045408, NR_038377 and NR_039976) of 14 lncRNAs showed the same change trends on the microarray and qRT-PCR results with statistical significance. Based on the qRT-PCR verified mRNAs, 1358 potential lncRNAs with 2697 positive correlations and 2287 negative correlations were predicted by the CNC network.
CONCLUSION: The plasma lncRNAs profiles provide preliminary data for the non-invasive diagnosis of CD and a resource for further specific lncRNA-mRNA pathway exploration.
Core tip: The pathogenesis of Chrohn’s disease (CD) is unclear while increasing evidence supports the involvement of epigenomic regulation. In this study, we jointly used microarray screening and qRT-PCR verification to achieve the plasma specific long noncoding RNAs expression profile of patients with CD and their potential regulation of downstream mRNAs. Our results would provide preliminary data for non-invasive diagnosis of CD and a reservoir for specific lncRNA-mRNA pathway exploration in the future.
- Citation: Chen D, Liu J, Zhao HY, Chen YP, Xiang Z, Jin X. Plasma long noncoding RNA expression profile identified by microarray in patients with Crohn’s disease. World J Gastroenterol 2016; 22(19): 4716-4731
- URL: https://www.wjgnet.com/1007-9327/full/v22/i19/4716.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i19.4716
Crohn’s Disease (CD) is a chronic and relapsing inflammatory disease that could affect any part of the intestine. The prevalence of CD is increasing in developing and developed countries, making it a global health care problem and an interesting research area[1,2]. However, the mechanism of CD remains vague; the involvement of genetic predisposition, immune response and environmental factors has been advocated[3]. Although the development of 5-aminosalicyte acid, prednisone and anti-inflammatory reagents has improved CD therapy, their effects are much more alleviative and sometimes are ineffective for refractory CD[4]. Therefore, the development of novel CD therapeutics is urgently needed, and the exploration of CD mechanisms is of clinical importance.
Recent genome-wide association studies (GWAS) identified novel susceptibility genes for CD[5], highlighting the important role of genomic factors. However, the majority of these studies focused on protein-coding genes and neglected the noncoding RNAs (ncRNAs) that were previously regarded as junk RNA or transcript noises[6]. With the development of high-throughput technologies, huge numbers of ncRNAs were identified, and many ncRNAs have been shown to be involved in physiological processes that maintain cellular and tissue homeostasis[7-9]. Generally, ncRNAs could be categorized into small ncRNAs (< 200 nt), such as microRNAs (miRNAs), and long noncoding RNAs (lncRNAs, > 200 nt). Research on lncRNAs has increased in recent years, showing their potency in regulating protein coding genes at the level of chromatin remodeling, transcriptional control and post-transcriptional processes[10].
A recent study revealed that lncRNAs play a pivotal role in immune function regulation and the progression of autoimmune related diseases, including CD[11]. An in-depth study by Mirza et al[12] reported the transcriptomic landscape of lncRNAs in inflammatory bowel disease (IBD). Furthermore, Qiao et al[13] identified the increased lncRNA DQ786243 level in CD patients and its effect on the function of regulatory T lymphocytes through changing CREB and Foxp3 levels. However, although evidence for plasma lncRNAs as noninvasive diagnostic biomarkers has accumulated[14,15], none has been reported in CD. Therefore, we conducted microarray screening, qRT-PCR verification and bioinformatics analysis of plasma lncRNAs and mRNAs from CD patients, aiming to provide preliminary data for noninvasive CD diagnosis and investigations into the underlying mechanism of CD.
The protocol on human beings was approved by the institutional review board of the First Affiliated Hospital of Zhejiang University and conducted in accordance with the Declaration of Helsinki. The study design and manuscript preparation fully followed the guidelines from the STROBE statement. Written consent was obtained before beginning the study.
CD patients (n = 12) were selected when first diagnosed in the Department of Gastroenterology, The First Affiliated Hospital of Zhejiang University between January 2013 and December 2014. The diagnosis of CD was based on endoscopy manifestations and biopsy, as adopted by the Asia-pacific consensus, with preclusion of intestinal tuberculosis, ulcerative colitis, Bechet’s disease and ischemic colitis[16]. To reduce the bias caused by different severities and extents of disease, we narrowed our selection to severe CD with small intestine involvement and related comorbidities (3 aphthous, 3 perianal abscess, 2 anal fistula and 2 arthralgia). The CD severity degree was assessed based on Harvey-Bradshaw index (HBI) and HBI > 9 was regarded as severe[17]. In CD patients, the average HBI was 11.3. Control subjects (n = 12) were enrolled from healthy volunteers without any health problems during their health checkups at our hospital during the same period. The authors had access to identifying information during or after data collection (Table 1). In this study, 3 of 12 subjects were randomly chosen from each group for microarray analysis, and ensuing qRT-PCR verification of significantly dysregulated lncRNAs and mRNAs was performed for the whole group. All blood samples were collected in a separate vacuum tube and sequentially centrifuged at 3000 rpm for 10 min and at 12000 rpm for 10 min. The cell-free plasma from supernatant was then stored at -80 °C for further analysis.
Total RNA was isolated from each plasma sample by separately mixing the sample with Polyacryl Carrier (MRC, OH, United States), TRIzol reagent (Invitrogen, Carlsbad, CA, United States) and chloroform, according to the manufacturer’s protocol. RNA purification was routinely performed with an RNeasy Mini Kit (Qiagen, Hilden, Germany). RNA quantity was measured with a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA), and RNA quality was tested with an Agilent 2100 Bioanalyzer (Agilent Technologies).
A human 8 × 60 KLncRNA/mRNAV3.0 microarray (Arraystar, Rockville, Maryland, United States) containing 30586 human lncRNAs and 26109 protein-coding transcripts was used in our study. Each transcript was represented using 1-5 probes to improve the statistical confidence. Generally, sample labeling and array hybridization were performed (Supplementary method) according to the One-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technology, Santa Clara, CA, United States), and Agilent Feature Extraction software (version 11.0.1.1) was used to analyze the acquired array images. The quantile normalization and subsequent data processing were performed with the GeneSpring GX v12.1 software package (Agilent Technologies). Differentially expressed lncRNAs and mRNAs with statistical significance between the two groups were identified through a paired t-test (P < 0.05), multiple hypothesis testing (FDR < 0.05) and fold change filtering (≥ 2.0 or ≤ 0.5). Further hierarchical clustering was performed to visualize numerical changes of lncRNAs and samples. The lncRNAs expression data have been deposited into Gene Expression Omnibus (GEO) under accession number GSE75459.
Based on the microarray data, the significantly differentially expressed lncRNAs were further categorized into antisense_LncRNAs, enhancer_LncRNAs and LincRNAs according to their potential effects and associations with downstream mRNAs. For the mRNA analysis, Gene Ontology (GO) that describes genes and gene products in any organism was used, covering the domains of Biological Process (BP), Cellular Component (CC) and Molecular Function (MF). Pathway analysis was also carried out for a functional analysis of mapping genes to KEGG pathways. Fisher’s exact/χ2 test and FDR were jointly used for significance detection. The P-value denotes the significance of GO term and Pathway correlated to the conditions. The lower the P-value, the more significant the Pathway and the GO Term. The FDR indicates the false discovery rate; a smaller FDR indicates smaller error in judging the P-value.
The total RNA isolated from the CD and control groups was reverse transcribed using a PrimeScript RTreagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa, Dalian, China) in accordance with the manufacturer’s instructions. U6 snRNA was amplified as a normalization control, and the relative amount of each lncRNA/mRNA to U6 RNA was calculated using the equation 2-ΔCT, where ΔCT = CTmiRNA - CTu6. Based on combinational consideration of the fold change, raw data, FDR, P-value and clinical manifestation reported by previous studies, 10 mRNAs were selected for quantitative real-time polymerase chain reaction (qRT-PCR) verification (Table S1) with a SYBR Green PCR kit (TaKaRa), with 3 replicated each. The same procedure was performed on 14 lncRNAs for verification (Table S1); these lncRNAs were selected from the overlap between the CNC network predications and the lncRNAs microarray data, with further preclusion of fold change < 2 and raw data density < 200.
The co-expression network of lncRNA-mRNA was constructed based on the correlation between significantly differentially expressed lncRNAs and mRNAs, as previously reported[18]. In the network, a pink node represents a significantly expressed mRNA, and a blue node represents the related lncRNA. Moreover, the red solid line represents a direct connection of a positive correlation between specific lncRNAs and mRNAs, and the green line represents a direct connection of a negative correlation. SPSS (version 16.0, Chicago, IL, United States) was used for statistical analyses. The data are expressed as the mean ± SD. Variables of the microarray and qRT- PCR data between the two groups were compared by Student’s t-test. In the microarray results, a fold change of lncRNAs/mRNAs ≥ 2.0 was chosen for further analysis, and P < 0.05 was considered statistically significant.
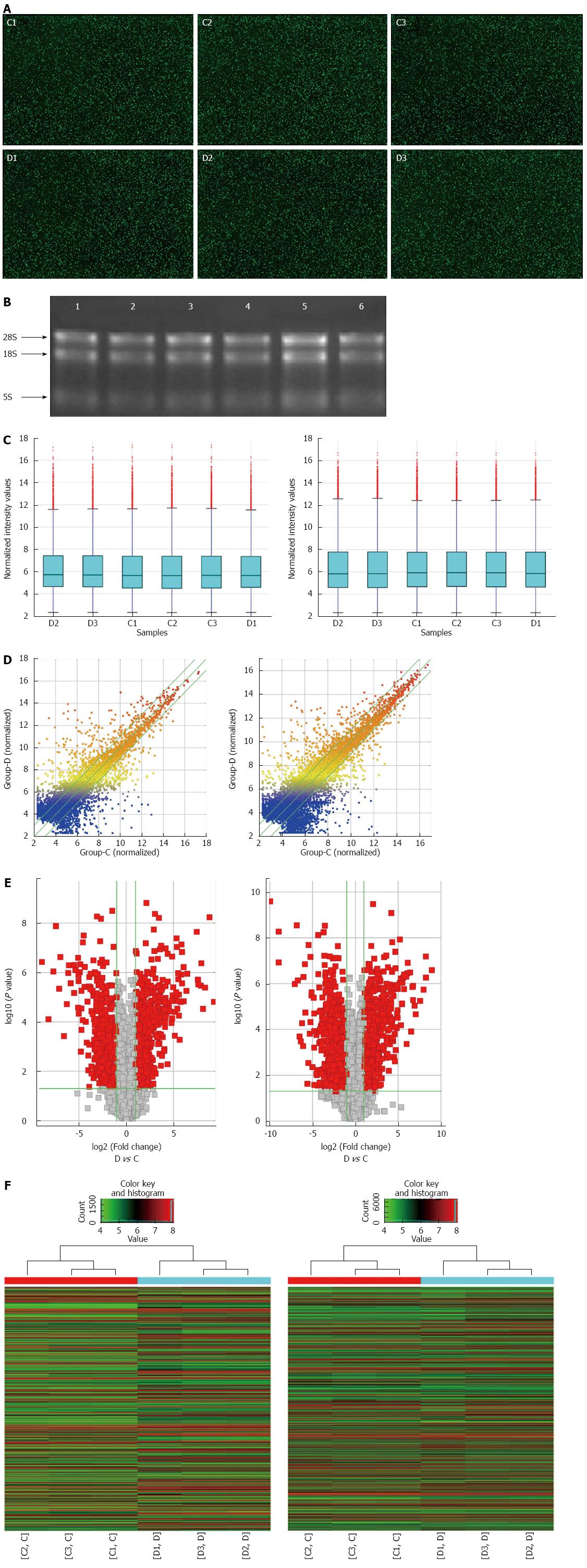
Compared with the control group, the BMI was significantly lower and the WBC and CRP were significantly higher in CD patients (Table 2). A microarray analysis of lncRNAs and mRNAs was carried out in randomly selected subjects. As jointly evaluated by heat map, box plot, scatter plot and volcano plot, the differential expression of lncRNAs and mRNAs was well categorized in the CD and control groups with good RNA quality and microarray image control (Figure 1). Compared with the control group, there were 1211 up-regulated and 777 down-regulated lncRNAs in the CD group (fold change ≥ 2.0, P < 0.05; Table S2). Moreover, there were 1020 up-regulated and 953 down-regulated mRNAs (fold change ≥ 2.0, P < 0.05; Table S3). The top 10 dysregulated lncRNAs and mRNAs are summarized in Table 2.
| Gene name | Transcript | Fold change |
| Up-regulated lncRNAs | ||
| GUSBP2 | ENST00000466668 | 626.49 |
| RP5-968D22.1 | ENST00000422548 | 444.43 |
| RP11-68L1.2 | ENST00000502712 | 324.23 |
| RP11-428F8.2 | ENST00000425364 | 245.98 |
| GAS5-AS1 | NR_037605 | 236.81 |
| RP11-923I11.5 | ENST00000562996 | 196.25 |
| DDX11-AS1 | NR_038927 | 192.07 |
| XLOC_005955 | TCONS_00014043 | 175.81 |
| XLOC_005807 | TCONS_00012771 | 87.76 |
| AC009133.20 | ENST00000569039 | 82.92 |
| Down-regulated lncRNAs | ||
| AF113016 | uc001ody.3 | 481.03 |
| ALOX12P2 | ENST00000575787 | 298.81 |
| AGSK1 | uc010bmo.1 | 208.70 |
| CTC-338M12.3 | ENST00000509252 | 172.74 |
| AC064871.3 | ENST00000413954 | 96.13 |
| RP11-510H23.3 | ENST00000431104 | 77.63 |
| LOC729678 | uc011dhd.2 | 69.95 |
| XLOC_010037 | TCONS_00020749 | 66.82 |
| LOC283761 | NR_027074 | 58.70 |
| XLOC_013142 | TCONS_00027621 | 45.60 |
| Up-regulated mRNAs | ||
| TBC1D17 | NM_024682 | 488.98 |
| GALNT8 | NM_017417 | 303.23 |
| DENND1A | NM_024820 | 301.75 |
| VANGL1 | NM_001172411 | 247.83 |
| VPS29 | NM_057180 | 189.17 |
| EHD1 | NM_006795 | 132.08 |
| FAM84A | NM_145175 | 121.69 |
| SAA4 | NM_006512 | 105.50 |
| ZNF33A | NM_006974 | 101.84 |
| GKN1 | NM_019617 | 91.51 |
| Down-regulated mRNAs | ||
| CCL3L3 | NM_001001437 | 994.63 |
| BGLAP | NM_199173 | 501.53 |
| SLC51B | NM_178859 | 499.44 |
| BAG4 | NM_004874 | 134.54 |
| MAU2 | NM_015329 | 115.54 |
| TSNARE1 | NM_145003 | 106.10 |
| DIXDC1 | NM_033425 | 93.11 |
| ZBTB25 | NM_006977 | 82.42 |
| CMTM8 | NM_178868 | 75.71 |
| ANXA1 | NM_000700 | 69.07 |
To narrow the large number of lncRNAs retrieved from the microarray data, we further carried out lncRNAs subgroup analyses. Based on the association between an lncRNA and its nearby mRNA, lncRNAs were categorized into antisense lncRNAs, enhancer lncRNAs and lincRNAs, providing a more accurate source for further functional study (Table S4). Briefly, among the 15 antisense lncRNAs, up-regulated ENST00000569039 had the highest fold change of 82.92, targeting the nearby gene NM_001042539 (myc-associated zinc finger protein isoform 2); the down-regulated ENST00000555407 had the highest fold change of 4.97, targeting the nearby gene NM_001085471 (forkhead box protein N3 isoform 1). Of the 81 enhancer lncRNAs, up-regulated ENST00000422548 had the highest fold change of 444.43, targeting the nearby gene ENST00000367818 [chemokine (C motif) ligand 1], and the down-regulated ENST00000427085 had the highest fold change of 24.29, targeting the nearby gene ENST00000534062 (retrotransposon-like 1). Finally, in the 161 lincRNAs, the up-regulated TCONS_00027580 had the highest fold change of 54.09, targeting the nearby gene NM_001102599 (carcinoembryonic antigen-related cell adhesion molecule 20 isoform 4L precursor), and the down-regulated TCONS_00027621 had the highest fold change of 45.60, targeting the nearby gene NM_001164309 (zinc finger protein 415 isoform 2).
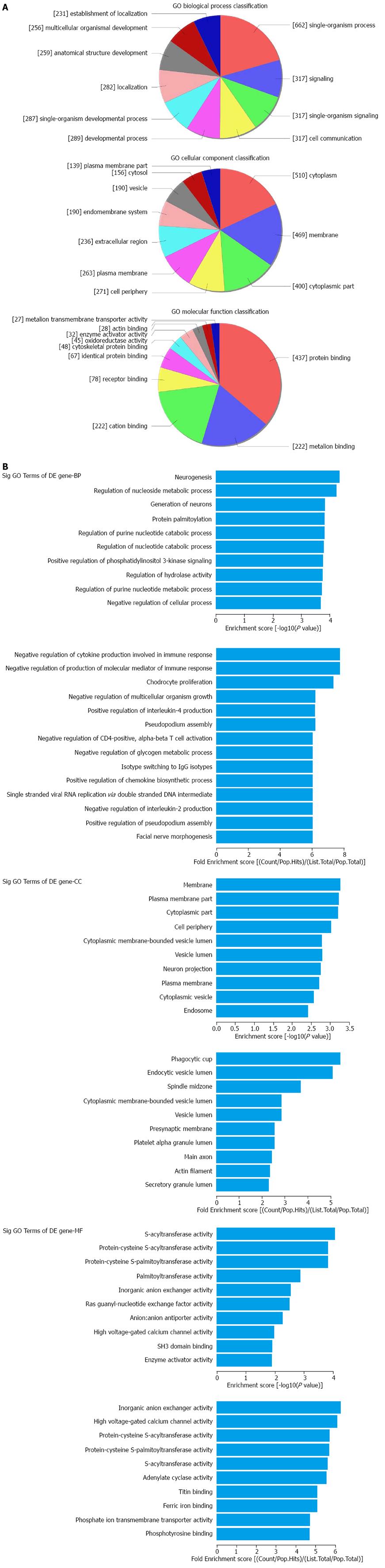
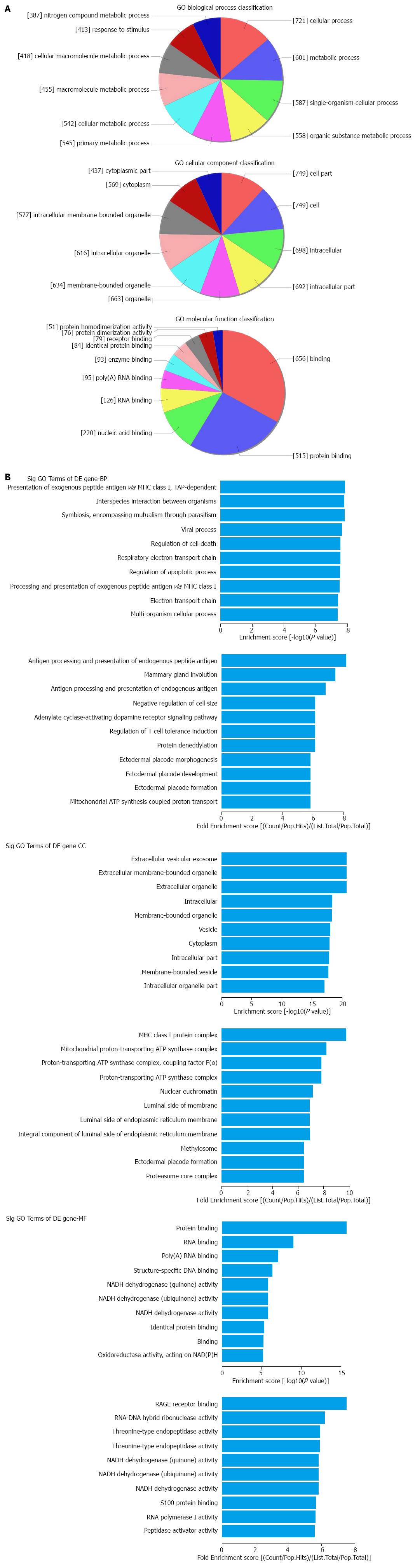
The top 10 dysregulated GO processes of each subgroup (BP, CC and MF) are presented in Figures 2 and 3. Because the GO manifestations in up- and down-regulated mRNAs varied, we analyzed them separately. In the up-regulated mRNAs as shown in Figure 2, the largest GO processes included single-organism process, neurogenesis and negative regulation of cytokine production involved in immune response in BP; cytoplasm, membrane and phagocytic cup in CC; protein binding, S-acyltransferase activity and inorganic anion exchanger activity in MF, according to the different algorithms of routine classification, enrichment score and fold enrichment. Similarly, the largest GO processes of the down-regulated mRNAs were cellular process, presentation of exogenous peptide antigen via MHC class I TAP dependent, antigen processing and presentation of endogenous peptide antigen in BP; cell part, extracellular vesicular exosome and MHC class I protein complex in CC and binding, protein binding and RAGE receptor binding in MF (Figure 3).
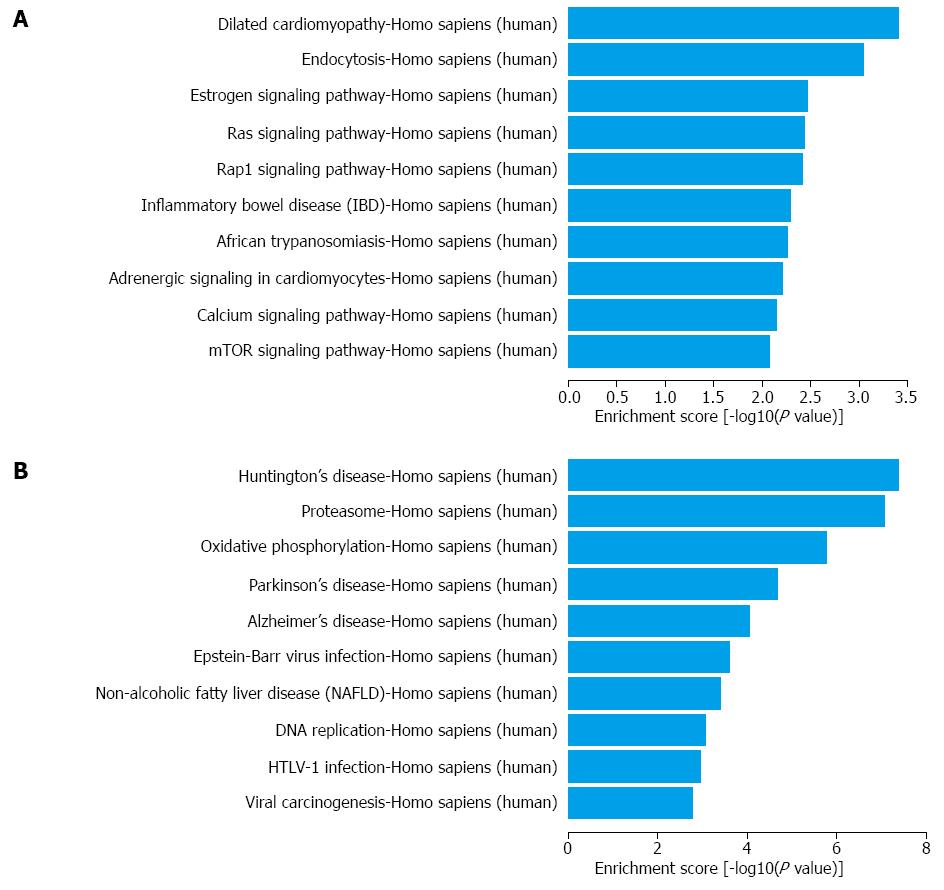
As further shown in Table S5, through the KEGG pathway analysis, 37 gene pathways were found to be targeted in up-regulated mRNAs; the top 3 processes were dilated cardiomyopathy, endocytosis and the estrogen signaling pathway (Figure 4A). More importantly, the IBD process itself was at the sixth of the top 10 pathways according to the enrichment score. Similarly, 32 gene pathways were found in down-regulated mRNAs; the top 3 processes were Huntington’s disease, proteasome and oxidative phosphorylation (Figure 4B).
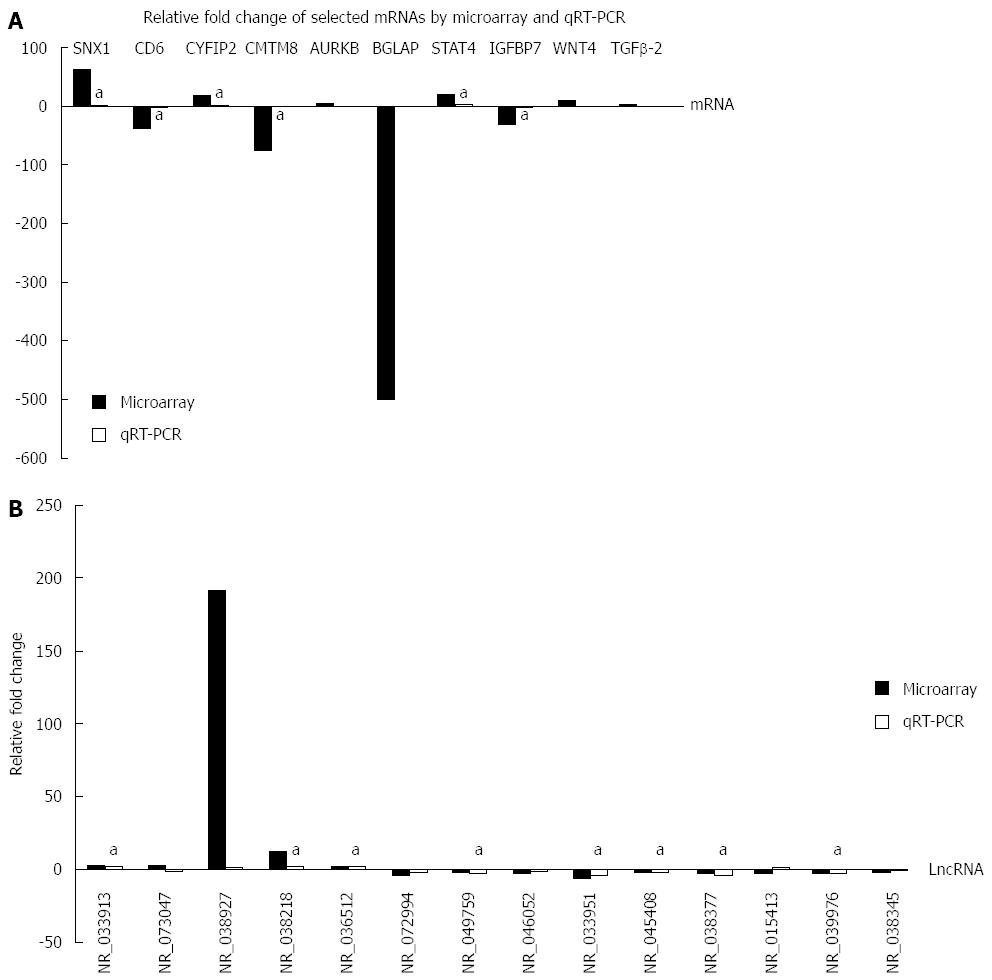
Based on the high normalized intensity in the raw data, high fold change, significant P-value and clinical meanings, we selected 10 mRNAs (SNX1, CYFIP2, CD6, CMTM8, AURKB, BGLAP, STAT4, WNT4, IGFBP7 and TGFβ-2) for qRT-PCR verification. As shown in Figure 5A, six mRNAs (SNX1, CYFIP2, CD6, CMTM8, STAT4 and IGFBP7) showed the same change tendency between the microarray and qRT-PCR results with statistical significance. AURKB and WNT4 showed the opposite change between the microarray and qRT-PCR results without statistical significance.
Because lncRNAs participate in the regulation of gene expression in transcriptional, epigenetic and posttranscriptional stages, it is plausible that certain lncRNAs are involved in CD pathogenesis. Based on the six qRT-PCR verified mRNAs, we predicted 1358 potential lncRNAs with 2697 positive correlations and2287 negative correlations between mRNA and lncRNAs by CNC network construction (Figure S1 and Table S6). After cross-linking between CNC predicted lncRNAs and lncRNAs microarray results, we selected 14 lncRNAs (NR_033913, NR_073047, NR_038927, NR_038218, NR_036512, NR_072994, NR_049759, NR_046052, NR_033951, NR_045408, NR_038377, NR_015413, NR_039976 and NR_038345) for qRT-PCR verification based on the selection criteria used for mRNAs. As shown in Figure 5B, 8 lncRNAs (NR_033913, NR_038218, NR_036512, NR_049759, NR_033951, NR_045408, NR_038377 and NR_039976) showed the same change tendency between the microarray and qRT-PCR results with statistical significance. Two lncRNAs (NR_073047 and NR_015413) showed the opposite change tendency without statistical significance. Four lncRNAs (NR_038927, NR_072994, NR_046052 and NR_038345) showed the same change tendency as in microarray results, but the results did not reach statistical significance (See detailed qRT-PCR and microarray results in Table S7).
CD is an important subtype of IBD with characteristics of intestinal full-thickness lesions and severe complications including perforation, fistula formation, malnutrition and carcinogenesis. Currently, next-generation sequencing and high-density microarrays have provided novel methods for CD study. For example, the NOD2/CARD15 gene mutation was identified to be associated with a CD phenotype[19], whereas a novel CD locus mapping to a gene desert on 5p 13.1 was also reported[20]. Nevertheless, according to the recent meta-analysis of GWAS studies, the number of confirmed genetic loci associated with CD was 150, although an overwhelming majority of these loci are located in noncoding regions[5], suggesting the importance of ncRNAs in CD research. A recent study showed that the loss of endogenous intestinal miRNAs caused impairment of epithelial barrier function, resulting in acute inflammation[21]. Further accumulating data supported an active role of miRNAs in the pathogenesis of IBD[22]. However, research into lncRNAs, another subgroup of ncRNAs with ability in binding protein, RNA and DNA, in CD has been rarely reported.
In this study, we used microarray screening and qRT-PCR verification to obtain the profile of plasma lncRNAs in carefully selected CD patients. We identified a total of 1988 and 2993 dysregulated lncRNAs and mRNAs between the CD and control groups; the numbers in our study were much higher than 450 lncRNAs and 1100 mRNAs from Mirza’s report[12]. This difference might be due to the relatively lower sample size of our study, which makes the data more dispersed. Moreover, there were no overlap of dysregulated lncRNAs between our results and the top 10 dysregulated lncRNAs we extracted from Mirza’s paper, except one lncRNA - DIO3OS that was up-regulated in our but down-regulated in their study. One possible explanation might be the secretion of DIO3OS from intestinal tissue to circulation, which needs further investigation. The intrinsic difference between our and Mirza’s results is the source of the lncRNAs. We reported, for the first time, the plasma lncRNAs in CD patients. If we could enlarge sample size and narrow down the scale of plasma lncRNAs, this method would become a good candidate for the non-invasive diagnosis of CD. Circulating lncRNAs (serum and plasma) have already been used as non-invasive biomarkers for liver cancer[23], breast cancer[24] and lung cancer[25], implying their wide application potential. It is theoretically plausible to use plasma lncRNAs because they are quite stable when included in lipid or lipoprotein vesicles in the circulation. A recent study showed that lncRNAs might be protected by exosomes in blood[26]. Nevertheless, the detailed secretion mechanisms of lncRNAs remain vague.
The application of bioinformatics is pivotal for in-depth analyses of huge data from microarray results. For lncRNAs, we used subgroup analyses based on the category of antisense lncRNAs, enhancer lncRNAs and lincRNAs, according to the effect and gene locus of the lncRNAs. For mRNAs, we combined the GO and KEGG pathway for enrichment analysis. In the GO analysis, we found that the largest portion of mRNAs was located in the cytoplasm and was involved in single-organism process and protein binding activity. In the KEGG pathways, we found IBD and oxidative phosphorylationin the list of the top 10 dysregulated pathways. The former finding supported the effectiveness of our study whereas the latter finding revealed the possibility of oxidative stress and energy metabolism in CD pathogenesis.
Of the top 10 up- and down-regulated lncRNAs (Table 2), only GAS5-AS1 was reported to be associated with various cancers[27], leaving a large blank area for further study. Of the top 10 up-regulated mRNAs, VANGL1 is a planar cell polarity component with a developing role in cancer[28], and FAM84A is associated with the enhanced migration of human colon cancer cells[29]. These mRNAs might contribute to the more frequent occurrence of colon cancer in the background of CD. Plasma SAA4 is present mostly in high density lipoproteins[30]. Therefore, the increased expression of SAA4 in CD might imply its potential role in assisting the stable existence of lncRNAs in the circulation. Of the top 10 down-regulated mRNAs, CMTM8 was revealed to induce caspase related apoptosis through a mitochondrial–mediated pathway[31], whereas BAG4 is involved in mitochondrial apoptosis[32], indicating the importance of apoptosis in the aetiology of CD. Moreover, ZBTB25 was found as a novel NF-AT repressor that participates in T-cell development, differentiation and lineage-specific transcription[33], emphasizing the importance of T cell mediated immune imbalance in CD pathogenesis.
Because we listed the top 10 dysregulated lncRNAs and mRNAs according to the relative fold change, there might be some genes that actually have raw data of low density (< 200). Therefore, we first selected 10 mRNAs for qRT-PCR verification based on the joint consideration of the P-value (< 0.05), fold change (> 2) and raw data (> 200). Then, we used CNC network analysis with combination of the mRNAs selection criteria and selected 14 lncRNAs for further qRT-PCR verification. Of the finally verified 6 mRNAs, a significantly increased STAT4 level was also reported in previous report[34], implying its importance in CD. Moreover, CD6 acts as a cell surface receptor and a target for regulating immune responses[35], whereas CYFIP2 is involved in T cell adhesion[36], further emphasizing the importance of T cell mediated immunity in CD. In 8 of 12 verified lncRNAs, NR_049759 is the transcript variant 2 of the coding gene IFITM3 that was up-regulated in colon mucosa of DSS induced mice[37], an animal model of IBD. NR_045408 is the antisense RNA of the coding gene RCNA3 that acts as a tumor suppressor[38], indicating the possibility of the NR_045408-RCNA3 pathway involvement in the occurrence of cancer in CD.
We identified the expression pattern of plasma lncRNAs based on microarray data. Further bioinformatics analyses successfully categorized the subjects into CD and control groups according to the lncRNAs profile, supporting the possibility of using this method as a non-invasive method for CD diagnosis. However, due to the small sample size, the retrieved profile contains many lncRNAs, which might become an obstacle for further application in clinics. Therefore, enlarging the experimental size to narrow the enrolled lncRNAs is urgently needed. Furthermore, because we only focused on severe CD subjects in this study, investigating lncRNAs expression in mild and moderate CD is suggested. Finally, it is better if we can compare the lncRNAs in plasma and intestinal tissue, which may be helpful for the mechanism exploration of CD. For the mRNA data, the GO and KEGG pathways were used to obtain more information. Approximately 60% of the microarray retrieved lncRNAs and mRNAs were verified by qRT-PCR, supporting the effectiveness of microarray screening. Several qRT-PCR verified lncRNAs and mRNAs were related with cancer or T-cell mediated immunity. Therefore, our data also provide a resource for further study of the lncRNA-mRNA pathway in CD pathogenesis.
Crohn’s disease (CD) has been regarded as a chronic and relapsing inflammatory disease that could affect any part of the intestine. The prevalence of CD is increasing in developing and developed countries, making it a global health care problem and an interesting research area.
The mechanism of CD remains vague; the involvement of genetic predisposition, immune response and environmental factors has been advocated. Currently, with the development of high-throughput technologies, the effect of noncoding RNAs, mainly divided into microRNAs and long noncoding RNAs, has been intensively investigated in CD pathogenesis.
Although evidence for plasma long noncoding RNAs (lncRNAs) as noninvasive diagnostic biomarkers has accumulated, none have been reported in CD. Therefore, we conducted, for the first time, microarray screening, qRT-PCR verification and bioinformatics analysis of plasma lncRNAs and mRNAs from CD patients, aiming to provide preliminary data for noninvasive CD diagnosis and investigations into the underling mechanism of CD.
Although further verification is needed in a larger independent cohort study, the profile of plasma lncRNAs would provide data for noninvasive diagnosis of CD and the potential lncRNA-mRNA pairs may shed light on the pathogenesis of CD.
LncRNAs are a group of RNAs that have the length > 200 nt and do not encodes proteins but exert function of post-transcriptional regulation.
In this work, authors investigated the expression pattern of plasma lncRNAs in CD patients by microarray screening and qRT-PCR verification of lncRNAs and mRNAs, followed by hierarchy clustering, GO and KEGG pathway analysis.
P- Reviewer: Kim SS, Mendez-Sanchez N S- Editor: Qi Y L- Editor: Wang TQ E- Editor: Ma S
| 1. | Baumgart DC, Bernstein CN, Abbas Z, Colombel JF, Day AS, D’Haens G, Dotan I, Goh KL, Hibi T, Kozarek RA. IBD Around the world: comparing the epidemiology, diagnosis, and treatment: proceedings of the World Digestive Health Day 2010--Inflammatory Bowel Disease Task Force meeting. Inflamm Bowel Dis. 2011;17:639-644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 54] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 2. | Ahuja V, Tandon RK. Inflammatory bowel disease in the Asia-Pacific area: a comparison with developed countries and regional differences. J Dig Dis. 2010;11:134-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 152] [Article Influence: 10.1] [Reference Citation Analysis (1)] |
| 3. | Elson CO. Genes, microbes, and T cells--new therapeutic targets in Crohn’s disease. N Engl J Med. 2002;346:614-616. [PubMed] |
| 4. | Bernstein CN. Treatment of IBD: where we are and where we are going. Am J Gastroenterol. 2015;110:114-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 171] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 5. | Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119-124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3979] [Cited by in RCA: 3597] [Article Influence: 276.7] [Reference Citation Analysis (0)] |
| 6. | Wright MW, Bruford EA. Naming ‘junk’: human non-protein coding RNA (ncRNA) gene nomenclature. Hum Genomics. 2011;5:90-98. [PubMed] |
| 7. | Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40:6391-6400. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 463] [Cited by in RCA: 524] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 8. | Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46-58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1483] [Cited by in RCA: 1449] [Article Influence: 96.6] [Reference Citation Analysis (0)] |
| 9. | Kovacic JC. Unlocking the Many Secrets of Noncoding RNA. J Am Coll Cardiol. 2015;65:2538-2541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 10. | Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3924] [Cited by in RCA: 4439] [Article Influence: 277.4] [Reference Citation Analysis (0)] |
| 11. | Wu GC, Pan HF, Leng RX, Wang DG, Li XP, Li XM, Ye DQ. Emerging role of long noncoding RNAs in autoimmune diseases. Autoimmun Rev. 2015;14:798-805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 211] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 12. | Mirza AH, Berthelsen CH, Seemann SE, Pan X, Frederiksen KS, Vilien M, Gorodkin J, Pociot F. Transcriptomic landscape of lncRNAs in inflammatory bowel disease. Genome Med. 2015;7:39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 147] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 13. | Qiao YQ, Huang ML, Xu AT, Zhao D, Ran ZH, Shen J. LncRNA DQ786243 affects Treg related CREB and Foxp3 expression in Crohn’s disease. J Biomed Sci. 2013;20:87. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 14. | Chen Z, Luo Y, Yang W, Ding L, Wang J, Tu J, Geng B, Cui Q, Yang J. Comparison Analysis of Dysregulated LncRNA Profile in Mouse Plasma and Liver after Hepatic Ischemia/Reperfusion Injury. PLoS One. 2015;10:e0133462. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 15. | Hu X, Bao J, Wang Z, Zhang Z, Gu P, Tao F, Cui D, Jiang W. The plasma lncRNA acting as fingerprint in non-small-cell lung cancer. Tumour Biol. 2015;Epub ahead of print. [PubMed] |
| 16. | Ouyang Q, Tandon R, Goh KL, Pan GZ, Fock KM, Fiocchi C, Lam SK, Xiao SD. Management consensus of inflammatory bowel disease for the Asia-Pacific region. J Gastroenterol Hepatol. 2006;21:1772-1782. [PubMed] |
| 17. | Best WR. Predicting the Crohn’s disease activity index from the Harvey-Bradshaw Index. Inflamm Bowel Dis. 2006;12:304-310. [PubMed] |
| 18. | Yu G, Yao W, Wang J, Ma X, Xiao W, Li H, Xia D, Yang Y, Deng K, Xiao H. LncRNAs expression signatures of renal clear cell carcinoma revealed by microarray. PLoS One. 2012;7:e42377. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 97] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 19. | Mendoza JL, Murillo LS, Fernández L, Peña AS, Lana R, Urcelay E, Cruz-Santamaría DM, de la Concha EG, Díaz-Rubio M, García-Paredes J. Prevalence of mutations of the NOD2/CARD15 gene and relation to phenotype in Spanish patients with Crohn disease. Scand J Gastroenterol. 2003;38:1235-1240. [PubMed] |
| 20. | Libioulle C, Louis E, Hansoul S, Sandor C, Farnir F, Franchimont D, Vermeire S, Dewit O, de Vos M, Dixon A. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3:e58. [PubMed] |
| 21. | McKenna LB, Schug J, Vourekas A, McKenna JB, Bramswig NC, Friedman JR, Kaestner KH. MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function. Gastroenterology. 2010;139:1654-164, 1664.e1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 253] [Cited by in RCA: 256] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 22. | Kalla R, Ventham NT, Kennedy NA, Quintana JF, Nimmo ER, Buck AH, Satsangi J. MicroRNAs: new players in IBD. Gut. 2015;64:504-517. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 188] [Cited by in RCA: 207] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 23. | Lu J, Xie F, Geng L, Shen W, Sui C, Yang J. Investigation of serum lncRNA-uc003wbd and lncRNA-AF085935 expression profile in patients with hepatocellular carcinoma and HBV. Tumour Biol. 2015;36:3231-3236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 57] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 24. | Xu N, Chen F, Wang F, Lu X, Wang X, Lv M, Lu C. Clinical significance of high expression of circulating serum lncRNA RP11-445H22.4 in breast cancer patients: a Chinese population-based study. Tumour Biol. 2015;36:7659-7665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 54] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 25. | Wang HM, Lu JH, Chen WY, Gu AQ. Upregulated lncRNA-UCA1 contributes to progression of lung cancer and is closely related to clinical diagnosis as a predictive biomarker in plasma. Int J Clin Exp Med. 2015;8:11824-11830. [PubMed] |
| 26. | Li Q, Shao Y, Zhang X, Zheng T, Miao M, Qin L, Wang B, Ye G, Xiao B, Guo J. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol. 2015;36:2007-2012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 319] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 27. | Ma C, Shi X, Zhu Q, Li Q, Liu Y, Yao Y, Song Y. The growth arrest-specific transcript 5 (GAS5): a pivotal tumor suppressor long noncoding RNA in human cancers. Tumour Biol. 2015;Epub ahead of print. [PubMed] |
| 28. | Hatakeyama J, Wald JH, Printsev I, Ho HY, Carraway KL. Vangl1 and Vangl2: planar cell polarity components with a developing role in cancer. Endocr Relat Cancer. 2014;21:R345-R356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 29. | Kobayashi T, Masaki T, Sugiyama M, Atomi Y, Furukawa Y, Nakamura Y. A gene encoding a family with sequence similarity 84, member A (FAM84A) enhanced migration of human colon cancer cells. Int J Oncol. 2006;29:341-347. [PubMed] |
| 30. | Yamada T, Wada A, Yamaguchi T, Itoh Y, Kawai T. Automated measurement of a constitutive isotype of serum amyloid A/SAA4 and comparison with other apolipoproteins. J Clin Lab Anal. 1997;11:363-368. [PubMed] |
| 31. | Jin C, Wang Y, Han W, Zhang Y, He Q, Li D, Yin C, Tian L, Liu D, Song Q. CMTM8 induces caspase-dependent and -independent apoptosis through a mitochondria-mediated pathway. J Cell Physiol. 2007;211:112-120. [PubMed] |
| 32. | Du Y, Huang Y, Gao Y, Song B, Mao J, Chen L, Bai L, Tang J. Annexin A7 modulates BAG4 and BAG4-binding proteins in mitochondrial apoptosis. Biomed Pharmacother. 2015;74:30-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 33. | Benita Y, Cao Z, Giallourakis C, Li C, Gardet A, Xavier RJ. Gene enrichment profiles reveal T-cell development, differentiation, and lineage-specific transcription factors including ZBTB25 as a novel NF-AT repressor. Blood. 2010;115:5376-5384. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 109] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 34. | Jabeen R, Miller L, Yao W, Gupta S, Steiner S, Kaplan MH. Altered STAT4 Isoform Expression in Patients with Inflammatory Bowel Disease. Inflamm Bowel Dis. 2015;21:2383-2392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 35. | Brown MH. CD6 as a Cell Surface Receptor and As a Target for Regulating Immune Responses. Curr Drug Targets. 2016;17:619-629. [PubMed] |
| 36. | Mayne M, Moffatt T, Kong H, McLaren PJ, Fowke KR, Becker KG, Namaka M, Schenck A, Bardoni B, Bernstein CN. CYFIP2 is highly abundant in CD4+ cells from multiple sclerosis patients and is involved in T cell adhesion. Eur J Immunol. 2004;34:1217-1227. [PubMed] |
| 37. | Koyama N, Hakura A, Toritsuka N, Sonoda J, Seki Y, Tohyama O, Asakura S, Nakano-Ito K, Hosokawa S. Wif1 and Ifitm3 gene expression preferentially altered in the colon mucosa of benzo[a]pyrene pre-treated mice following exposure to dextran sulfate sodium. Chem Biol Interact. 2015;240:164-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 38. | Martínez-Høyer S, Solé-Sánchez S, Aguado F, Martínez-Martínez S, Serrano-Candelas E, Hernández JL, Iglesias M, Redondo JM, Casanovas O, Messeguer R. A novel role for an RCAN3-derived peptide as a tumor suppressor in breast cancer. Carcinogenesis. 2015;36:792-799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |