Published online Mar 21, 2016. doi: 10.3748/wjg.v22.i11.3268
Peer-review started: November 8, 2015
First decision: November 27, 2015
Revised: December 14, 2015
Accepted: December 30, 2015
Article in press: December 30, 2015
Published online: March 21, 2016
Processing time: 126 Days and 22.2 Hours
AIM: To investigate the frequency of mutations in pre-core (pre-C) and basic core promoter (BCP) regions of hepatitis B virus (HBV) from Shanxi Province, and the association between mutations and disease related indexes.
METHODS: One hundred chronic hepatitis B patients treated at Shanxi Province Hospital of Traditional Chinese Medicine were included in this study. PCR-reverse dot blot hybridization and mismatch amplification mutation assay (MAMA)-PCR were used to detect the mutations in the HBV pre-C and BCP regions. HBV DNA content and liver function were compared between patients with mutant HBV pre-C and BCP loci and those with wild-type loci. The consistency between PCR-reverse dot blot hybridization and MAMA-PCR for detecting mutations in the HBV pre-C and BCP regions was assessed.
RESULTS: Of the 100 serum samples detected, 9.38% had single mutations in the pre-C region, 29.17% had single mutations in the BCP region, 41.67% had mutations in both BCP and pre-C regions, and 19.79% had wild-type loci. The rates of BCP and pre-C mutations were 65.7% and 34.3%, respectively, in hepatitis B e antigen (HBeAg) positive patients, and 84.6% and 96.2%, respectively, in HBeAg negative patients. The rate of pre-C mutations was significantly higher in HBeAg negative patients than in HBeAg positive patients (χ2 = 26.62, P = 0.00), but there was no significant difference in the distribution of mutations in the BCP region between HBeAg positive and negative patients (χ2 = 2.43, P = 0.12). The presence of mutations in the pre-C (Wilcoxon W = 1802.5, P = 0.00) and BCP regions (Wilcoxon W = 2906.5, P = 0.00) was more common in patients with low HBV DNA content. Both AST and GGT were significantly higher in patients with mutant pre-C and BCP loci than in those with wild-type loci (P < 0.05). PCR-reverse dot blot hybridization and MAMA-PCR for detection of mutations in the BCP and pre-C regions had good consistency, and the Kappa values obtained were 0.91 and 0.58, respectively.
CONCLUSION: HBeAg negative patients tend to have HBV pre-C mutations. However, these mutations do not cause increased DNA copies, but associate with damage of liver function.
Core tip: There is no exact evidence about the frequency of mutations in hepatitis B virus (HBV) pre-C and basic core promoter (BCP) regions of HBV in Shanxi Province and the correlation between mutations and disease-related indicators. Here, we found that the frequency of pre-C mutations was higher in hepatitis B e antigen (HBeAg) negative patients, and liver function parameters AST and GGT in the mutation group were higher than those in the wild-type group; however, DNA replication did not increase in the presence of HBV pre-C and BCP mutations. Taken together, HBeAg-negative patients tend to have HBV pre-C mutation. However, these mutations do not cause increased DNA copies, but associate with liver function damage.
- Citation: Wang XL, Ren JP, Wang XQ, Wang XH, Yang SF, Xiong Y. Mutations in pre-core and basic core promoter regions of hepatitis B virus in chronic hepatitis B patients. World J Gastroenterol 2016; 22(11): 3268-3274
- URL: https://www.wjgnet.com/1007-9327/full/v22/i11/3268.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i11.3268
Hepatitis B virus (HBV) infection is a worldwide epidemic and public health problem. According to data from the World Health Organization, approximately 2 billion people worldwide were ever infected with HBV, including 360 million people with chronic infection[1,2]. Sero-epidemiological data of hepatitis B in 2010 show that there were 93 million hepatitis B surface antigen carriers in China, and the incidence of hepatitis B was 79.46 per 100000 people[3]; in Shanxi Province, the incidence of hepatitis B was 119.34 per 100000 people, and the mortality and case fatality rates were 0.03 per 100000 people and 0.03%, respectively[4,5]. HBV infection not only has an enormous impact on patient health, but also places a huge economic burden to the patient’s family.
At present, hepatitis B e antigen (HBeAg) has been clinically used as one of indicators for active HBV replication in patients with chronic hepatitis B. Main laboratory indicators for monitoring changes in illness status and treatment efficacy include serum immunological indicators, liver function and HBV DNA load; however, it is difficult to explain why patients with serum immunological indicators turning negative have no actual disease remission and why immunological indicators are not consistent with biochemistry indexes (liver function) or viral load. Many scholars have found that the condition of some HBeAg negative hepatitis B patients is not stable and is even aggravated[6-8]. Such HBeAg negative patients are in a state of HBV infection in their whole life, and have active HBV replication and sustained liver function injury. This phenomenon is related with mutations in the pre-core (pre-C; nts1814-1900) and core promoter (CP; nts1613-1849) regions of HBV genome[9-15]. Among these mutations, the pre-C mutation nt1896G/A is common, resulting in the early termination of HBeAg translation[16,17]. The simultaneous presence of nt1762A/T and nt1764G/A mutations in the basic core promoter (BCP) region of HBV can lead to a 50%-70% decrease in the RNA transcription level in the pre-C region, and the strengthening of viral replication[12,18,19]. However, there are currently no reliable epidemiological data about mutations in the pre-C and BCP regions, and the research on the impact of these mutations on the progression of hepatitis B related diseases is lacking. The present study adopted PCR-reverse dot blot hybridization and mismatch amplification mutation assay (MAMA)-PCR to detect the pre-C mutation nt1896G/A and nt1762A/T and nt1764G/A mutations in the BCP region in chronic hepatitis B patients, investigating the frequency of mutations in the pre-C and BCP regions[20,21]. Based on serum immunological indexes, liver function and HBV DNA load, we also explored the correlation of these mutations with disease related indexes.
One hundred serum samples were collected from patients with chronic hepatitis B treated at Shanxi Provincial Hospital of Traditional Chinese Medicine from April 2014 to October 2014. All patients met the 2010 diagnostic criteria for chronic hepatitis B, and patients with other liver diseases such as autoimmune hepatitis, HCV infection, HDV infection, primary biliary cirrhosis, hemochromatosis, alcoholic hepatitis and uncontrolled diabetes were excluded. The mean age of the patients was 40 ± 17 years. There were 54 men and 47 women. Fasting blood samples (5 mL) were drawn in the morning and centrifuged to separate serum. Serum samples were preserved at -80 °C for detection. The laboratory in which serum samples were detected has obtained the laboratory accreditation certificate issued by the China National Accreditation Service for Conformity Assessment and the certificate of conformity for Clinical Gene Amplification Testing Laboratory.
PCR-reverse dot blot hybridization was performed with a commercial kit (detection sensitivity, 5 × 103 IU/mL) provided by DAAN Gene (Guangzhou, China), using an ABI Prism 7500 fluorescence quantitative PCR system, an Hema 3200 PCR thermal cycler, a DA-8000 nucleic acid hybridization apparatus, and an SHA-CA thermostatic water bath oscillator (Feipu, Changzhou, China). MAMA-PCR was performed with a commercial kit (detection sensitivity, 1 × 103 IU/mL) provided by BIOTGENE (Wuhan, China), using an ABI Prism 7500 fluorescence quantitative PCR system. HBV serum immunological markers were detected with a kit (detection sensitivity, 0.006 NCU/mL) provided by Chemclin Biotech (Beijing, China) using a CHEMCLIN1500 chemiluminescence system. Liver function parameters were detected using reagents for ALT, AST, GGT, ALP, ALB and TP obtained from Beckman Coulter AU, and reagents for DBIL and total bilirubin obtained from Wako (Japan), using a Beckman Coulter AU5821 automatic biochemical analyzer.
Specific primers and probes targeting the pre-C and BCP regions of HBV were designed. Nylon membrane were coated with specific probes in advance. After target fragments were amplified by PCR using biotin labelled primers, PCR products were hybridized with specific probes coated on nylon membranes. Unbound PCR products were then removed by washing, followed by color development to detect the pre-C ntA1896 and ntA1899 mutations, as well as ntT1762/A1764 mutations in the BCP region. If a positive signal was yielded by the wild-type probe, the locus was regarded as the wild-type; if a positive signal was yielded by a mutant probe, the locus was considered to have the corresponding mutation; if a positive signal was yielded only in the CC dot and all other dots had negative signals, HBV-DNA negativity was considered or the content was below the lower detection limit of the kit; if negative signals were yielded by different probes for the same locus and positive signals were yielded by probes 7 and 8, the presence of other mutations was considered. All assays were performed in strict accordance with the instructions provided with the kit.
Two pairs of primers targeting the pre-C region of HBV (primers A and B) and a specific probe were used to perform MAMA-PCR in the presence of thermostable DNA polymerase, dNTPs and PCR buffer. The presence of pre-C ntA1896 and ntT1762/A1764 mutations or not was judged based on the difference in Ct values obtained using primers A and B. If the amplification curve for sample PCR reaction tube A was not in “S” shape or Ct (A) value was > 36, the content of HBV DNA in the sample was considered to be below the lower detection limit; |Ct(A)-Ct(B)| ≤ 7.00 suggested the presence of ntA1896 and ntT1762/A1764 mutations; |Ct(A)-Ct(B)| > 7.00 suggested the presence of wild-type ntA1896 and ntT1762/A1764 loci or the level of mutations was below the lower detection limit.
Statistical analyses were performed by normal distribution test, fourfold table χ2 test (corrected), rank sum test for two independent samples, and consistency test.
The HBV content and liver function indexes for the included patients are presented in Table 1.
| Parameter | HBV DNA (IU/mL) | ALT | AST | TBIL | DBIL | ALP | GGT | ALB | TP |
| Median | 6.64 | 45 | 55 | 15.8 | 5.05 | 96.0 | 36 | 43 | 72 |
| Interquartile range | 2.48 | 69 | 57 | 9.8 | 4.35 | 51.3 | 52 | 5.3 | 7 |
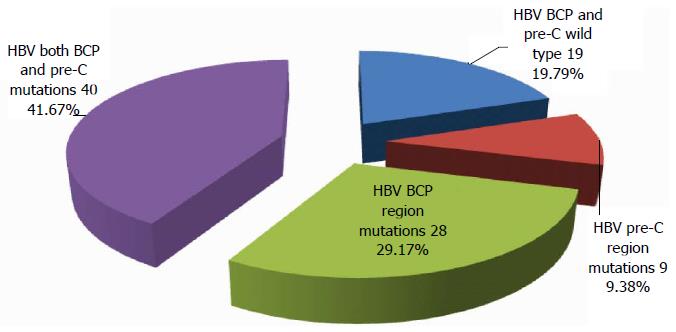
Of the 100 serum samples detected, 100 were positive for HBV DNA, and the HBV DNA content ranged from 3.11 to 8.99 IU/mL; 70 were positive for HBeAg. PCR-reverse dot blot hybridization assay showed that BCP mutations were not detected in 4 cases. Of the remaining 96 cases, 9.38% had single mutations in the pre-C region, 29.17% had single mutations in the BCP region, 41.67% had mutations in both BCP and pre-C regions, and 19.79% had wild-type loci (Figure 1).
The rate of BCP mutations was 65.7% in HBeAg positive patients, and 84.6% in HBeAg negative patients. There was no significant difference in the distribution of mutations in the BCP region of HBV between HBeAg positive and negative patients (continuously corrected χ2 test, P = 0.12). The rate of pre-C mutations was 34.3% in HBeAg positive patients, and 96.2% in HBeAg negative patients. The rate of pre-C mutations was significantly higher in HBeAg negative patients than in HBeAg positive patients (continuously corrected χ2 test, P = 0.00) (Table 2).
| BCP region | Total | χ2(corrected) | P value | Pre-C region | Total | χ2(corrected) | P value | ||||
| Mutations | WT | Mutations | WT | ||||||||
| HBeAg | + | 46 | 24 | 70 | 2.43 | 0.12 | 24 | 46 | 70 | 26.62 | 0.00 |
| - | 22 | 4 | 26 | 25 | 1 | 26 | |||||
| Total | 68 | 28 | 96 | 49 | 47 | 96 | |||||
There were significant differences in HBV DNA contents between patients with mutant BCP or pre-C loci and those with wild-type loci (rank sum test, P = 0.00 for both), and the HBV DNA content was higher in patients without mutations than in those with mutations (Table 3).
| BCP region | No. of cases | Mean rank | Wilcoxon | P value | Pre-C region | No. of cases | Mean rank | Wilcoxon | P value | |
| DNA content | Mutations | 68 | 42.74 | Mutations | 49 | 36.79 | ||||
| WT | 28 | 62.48 | 2906.5 | 0.00 | WT | 47 | 60.71 | 1802.5 | 0.00 | |
| Total | 96 | Total | 96 |
Both AST and GGT were significantly higher in patients with mutant BCP loci than in those with wild-type loci (rank sum test, P = 0.00 for both) (Table 4). Both AST and GGT were also significantly higher in patients with mutant pre-C loci than in those with wild-type loci (rank sum test, P = 0.00 and 0.01, respectively) (Table 5).
| Item | AST | GGT | |||||
| BCP region | No. of cases | Mean rank | Wilcoxon | P value | Mean rank | Wilcoxon | P value |
| WT | 28 | 33.75 | 33.0 | ||||
| Mutant | 68 | 53.96 | 945.0 | 0.00 | 54.8 | 924.0 | 0.00 |
| Total | 96 | ||||||
| Item | AST | GGT | |||||
| No. of cases | Mean rank | Wilcoxon | P value | Mean rank | Wilcoxon | P value | |
| pre-C loci | |||||||
| WT | 47 | 40.55 | 41.33 | ||||
| Mutant | 49 | 55.29 | 1906.0 | 0.00 | 55.38 | 1942.5 | 0.01 |
| Total | 96 | ||||||
The results of PCR-reverse dot blot hybridization (DAAN method) and MAMA-PCR (BIOTGENE method) for detection of mutations in the BCP and pre-C regions of HBV were compared by the consistency test (Table 6). The Kappa values were 0.91 and 0.58 (P = 0.00 for both), respectively. The two methods had good consistency.
| BIOTGENE for BCP region | Total | Kappa | P value | BIOTGENE for pre-C region | Total | Kappa | P value | ||||||
| DAAN for BCP region | WT | Mutant | DAAN for | WT | Mutant | ||||||||
| WT | 23 | 2 | 25 | pre-C region | WT | 29 | 5 | 34 | |||||
| Mutant | 1 | 50 | 51 | 0.91 | 0.00 | Mutant | 11 | 31 | 42 | 0.58 | 0.00 | ||
| Total | 24 | 52 | 76 | Total | 40 | 36 | 76 | ||||||
Patients with HBV infection have diverse clinical manifestations. Some cases of chronic hepatitis may evolve into liver cirrhosis or liver cancer due to virus replication and evasion of the body’s immune system[8,18,22-24]. In the present study, we selected patients with chronic hepatitis B to detect the mutations in the pre-C and BCP regions of HBV. It was found that only 19.79% of chronic hepatitis B patients had wild-type pre-C and BCP loci, and all the others had mutations in the pre-C or BCP region. HBeAg negative patients had a higher rate of mutations in the pre-C region and more severe AST and GGT abnormalities; however, the HBV DNA content was not significantly higher in patients with mutations in the pre-C or/and BCP regions than in those with wild-type pre-C and BCP loci.
The translation of HBeAg precursor begins with the first codon of the pre-C gene. HBeAg precursor consists of 210 amino acids and can form secreted HBeAg, which exists in serum, after the cleavage of a peptide fragment[15]. It is traditionally believed that HBeAg level is consistent with the activity of viral DNA polymerase, and is therefore regarded as one of the markers of HBV replication and infectivity. However, recent studies found that the mutations in the pre-C region can affect the synthesis and secretion of HBeAg by HBV. In this study, HBeAg negative patients had a rate of mutations in the pre-C region as high as 96.2%, as well as liver dysfunction. Thus, the traditional view regarding HBeAg negative patients should be modified, so as not to ignore the continued injury to liver cells caused by HBV in such patients.
HBV BCP region encodes HBcAg, which is expressed on the membrane surface of liver cells, has strong antigenicity and can induce CTL response[25-27]. In this study, the distribution of BCP mutations did not differ significantly between HBeAg positive and negative patients. However, BCP mutations may cause failed CTL activation and thereby aggravate liver injury and liver dysfunction caused by HBV[28].
The finding that the presence of mutations in the pre-C and BCP regions of HBV was more common in patients with low HBV DNA content argues against the traditional view that HBV DNA replication is consistent with the liver injury. Low levels of HBV DNA do not invariably mean mild liver injury. The presence of mutations in the pre-C and BCP regions of HBV in HBeAg negative patients with lower virus replication can also aggravate liver injury, and ignoring this problem may lead to disease progression to liver cancer or cirrhosis[27].
This study also compared PCR-reverse dot blot hybridization and MAMA-PCR for detecting mutations in the pre-C and BCP regions of HBV and found that they had good consistency. However, the consistency between them for detection of mutations in the pre-C region was poorer than that for detection of mutations in the BCP region. PCR-reverse dot blot hybridization can provide more information about mutations in the pre-C and BCP regions, especially the information about heterozygous mutations in the pre-C region. For virus strains with inconsistent results, further DNA sequencing is required.
Hepatitis B virus (HBV) can cause acute and chronic hepatitis, and further develop into cirrhosis, primary liver cancer and end-stage liver failure. Mutations in HBV pre-C and basic core promoter (BCP) regions can cause deletion or reduced synthesis of hepatitis B e antigen (HBeAg), evading of immune surveillance system and persistent infection, increased hepatic cellular immune response, sustained liver function damage and poor treatment. Thus exploring the frequency of mutations in HBV pre-C and BCP regions, as well as the correlation between mutations and disease-related indicators can help improve our understanding of clinical HBV infection status, and provide additional evidence for the diagnosis and treatment of HBV related diseases.
HBV has evolved a unique life cycle that results in the production of enormous viral loads during active replication without actually killing the infected cells directly. New data are emerging that mutant viral genomes emerge frequently because HBV uses reverse transcription to copy its genome. The particular viral mutations or combination of mutations that directly affect the clinical outcome of infection are not known. Further studies are clearly needed to identify the pathogenic basis and clinical sequelae arising from the selection of these particular mutants. Finally, assistance based on the diagnosis and treatment of HBV related diseases could be provided if we know well clinical HBV infection status.
In this study, the method for detection of HBV pre-C and BCP mutations is faster, more accurate, and more specific when comparing real-time fluorescence PCR-reverse dot blot with the traditional method. By discussing HBV 1896, 1899 loci and BCP region mutations of chronic HBV patients from Shanxi Province, the authors will know well clinical HBV infection status and assist in the diagnosis and treatment of HBV related diseases.
The authors explored the relationship between HBV pre-C and BCP region mutations and HBeAg phenotype, DNA content, liver dysfunction and clinical HBV infection status by researching and analyzing mutation rates and distribution in HBV pre-C and BCP regions in Shanxi Province.
BCP gene mutation: A1762/G1764 to T1762/A1764; Pre-C gene mutations: G1896/G1899 to A1896/A1899, A1896/G1899, or G1896/A1899.
This is an interesting and well designed manuscript. In this manuscript, the authors investigated the frequency of mutations in the pre-C and basic core promoter regions of HBV genome in chronic hepatitis B patients from Shanxi Province, and the association between these mutations and disease related indexes. The authors found that in HBeAg negative patients, pre-C mutation rate was 96.2% and there was liver dysfunction, although the distribution of BCP mutations was not significantly associated with HBeAg status.
P- Reviewer: Alric L, Miller C S- Editor: Ma YJ L- Editor: Wang TQ E- Editor: Liu XM
| 1. | World Health Organization. Hepatitis B vaccines. Weekly Epidemiological Record 40: 2009, 405-420. . |
| 2. | Lavanchy D. Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J Viral Hepat. 2004;11:97-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1734] [Cited by in RCA: 1750] [Article Influence: 83.3] [Reference Citation Analysis (0)] |
| 3. | Li HM, Wang JQ, Wang R, Zhao Q, Li L, Zhang JP, Shen T. Hepatitis B virus genotypes and genome characteristics in China. World J Gastroenterol. 2015;21:6684-6697. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 40] [Cited by in RCA: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 4. | National Bureau of Statistics of the People’s Republic of China. China Statistical Yearbook 2011 [EB/OL]. Available from: http://www.stats.gov.cn/tjsj/ndsj/2011/indexeh.htm. |
| 5. | Zhang SHX, Liu BF, Cheng RG, Wang JL, Zhang AG, Hu RW. Direct costs of hepatitis B and related diseases and influencing factors in four cities of Shanxi Province. Zhongguo Xiandai Yisheng. 2014;52:10-14. |
| 6. | Kitab B, Essaid El Feydi A, Afifi R, Trepo C, Benazzouz M, Essamri W, Zoulim F, Chemin I, Alj HS, Ezzikouri S. Variability in the precore and core promoter regions of HBV strains in Morocco: characterization and impact on liver disease progression. PLoS One. 2012;7:e42891. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 7. | Alexopoulou A, Karayiannis P. HBeAg negative variants and their role in the natural history of chronic hepatitis B virus infection. World J Gastroenterol. 2014;20:7644-7652. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 50] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (1)] |
| 8. | Okamoto H, Tsuda F, Akahane Y, Sugai Y, Yoshiba M, Moriyama K, Tanaka T, Miyakawa Y, Mayumi M. Hepatitis B virus with mutations in the core promoter for an e antigen-negative phenotype in carriers with antibody to e antigen. J Virol. 1994;68:8102-8110. [PubMed] |
| 9. | Kitab B, El Feydi AE, Afifi R, Derdabi O, Cherradi Y, Benazzouz M, Rebbani K, Brahim I, Salih Alj H, Zoulim F. Hepatitis B genotypes/subgenotypes and MHR variants among Moroccan chronic carriers. J Infect. 2011;63:66-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 38] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 10. | Funk ML, Rosenberg DM, Lok AS. World-wide epidemiology of HBeAg-negative chronic hepatitis B and associated precore and core promoter variants. J Viral Hepat. 2002;9:52-61. [PubMed] |
| 11. | Fang ZL, Sabin CA, Dong BQ, Wei SC, Chen QY, Fang KX, Yang JY, Wang XY, Harrison TJ. The association of HBV core promoter double mutations (A1762T and G1764A) with viral load differs between HBeAg positive and anti-HBe positive individuals: a longitudinal analysis. J Hepatol. 2009;50:273-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 47] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 12. | Bai X, Zhu Y, Jin Y, Guo X, Qian G, Chen T, Zhang J, Wang J, Groopman JD, Gu J. Temporal acquisition of sequential mutations in the enhancer II and basal core promoter of HBV in individuals at high risk for hepatocellular carcinoma. Carcinogenesis. 2011;32:63-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 13. | Mohamadkhani A, Montazeri G, Poustchi H. The importance of hepatitis B virus genome diversity in Basal core promoter region. Middle East J Dig Dis. 2011;3:13-19. [PubMed] |
| 14. | Yousif M, Mudawi H, Bakhiet S, Glebe D, Kramvis A. Molecular characterization of hepatitis B virus in liver disease patients and asymptomatic carriers of the virus in Sudan. BMC Infect Dis. 2013;13:328. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 15. | Baqai SF, Proudfoot J, Yi DH, Mangahas M, Gish RG. High rate of core promoter and precore mutations in patients with chronic hepatitis B. Hepatol Int. 2015;9:209-217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 16. | Hadziyannis SJ. Natural history of chronic hepatitis B in Euro-Mediterranean and African countries. J Hepatol. 2011;55:183-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 121] [Article Influence: 8.6] [Reference Citation Analysis (1)] |
| 17. | Moradi A, Zhand S, Ghaemi A, Javid N, Bazouri M, Tabarraei A. Mutations in pre-core and basal-core promoter regions of hepatitis B virus in chronic HBV patients from Golestan, Iran. Iran J Basic Med Sci. 2014;17:370-377. [PubMed] |
| 18. | Zhang J, Xu WJ, Wang Q, Zhang Y, Shi M. Prevalence of the precore G1896A mutation in Chinese patients with e antigen negative hepatitis B virus infection and its relationship to pre-S1 antigen. Braz J Microbiol. 2009;40:965-971. [PubMed] |
| 19. | Mumtaz K, Hamid S, Ahmed S, Moatter T, Mushtaq S, Khan A, Mizokami M, Jafri W. A study of genotypes, mutants and nucleotide sequence of hepatitis B virus in Pakistan: HBV genotypes in pakistan. Hepat Mon. 2011;11:14-18. [PubMed] |
| 20. | Ren XD, Lin SY, Wang X, Zhou T, Block TM, Su YH. Rapid and sensitive detection of hepatitis B virus 1762T/1764A double mutation from hepatocellular carcinomas using LNA-mediated PCR clamping and hybridization probes. J Virol Methods. 2009;158:24-29. [PubMed] |
| 21. | Nie H, Evans AA, London WT, Block TM, Ren XD. Quantification of complex precore mutations of hepatitis B virus by SimpleProbe real time PCR and dual melting analysis. J Clin Virol. 2011;51:234-240. [PubMed] |
| 22. | Samal J, Kandpal M, Vivekanandan P. Molecular mechanisms underlying occult hepatitis B virus infection. Clin Microbiol Rev. 2012;25:142-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 111] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 23. | Laeeq SM, Luck NH, Wadhwa RK, Abbas Z, Hasan SM, Younus M, Mubarak M. Left liver lobe diameter albumin ratio as a predictor of esophageal varices in patients with cirrhosis: A preliminary report. J Transl Intern Med. 2014;2:164-167. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 24. | Manzoor M, Wadhwa RK, Abbas Z, Hasan SM, Luck NH, Mubarak M. Unusual presentation of nonalcoholic steatohepatitis-related cirrhosis in a patient with celiac disease and microscopic colitis. J Transl Intern Med. 2014;2:172-174. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 25. | Quarleri J. Core promoter: a critical region where the hepatitis B virus makes decisions. World J Gastroenterol. 2014;20:425-435. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 65] [Cited by in RCA: 65] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 26. | Zhang D, Ma S, Zhang X, Zhao H, Ding H, Zeng C. Prevalent HBV point mutations and mutation combinations at BCP/preC region and their association with liver disease progression. BMC Infect Dis. 2010;10:271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 27. | Sendi H, Mehrab-Mohseni M, Shahraz S, Norder H, Alavian SM, Noorinayer B, Zali MR, Pumpens P, Bonkovsky HL, Magnius LO. CTL escape mutations of core protein are more frequent in strains of HBeAg negative patients with low levels of HBV DNA. J Clin Virol. 2009;46:259-264. [PubMed] |
| 28. | Utama A, Siburian MD, Purwantomo S, Intan MD, Kurniasih TS, Gani RA, Achwan WA, Arnelis SA, Lukito B, Harmono T. Association of core promoter mutations of hepatitis B virus and viral load is different in HBeAg(+) and HBeAg(-) patients. World J Gastroenterol. 2011;17:708-716. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 8] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |