Published online Feb 21, 2015. doi: 10.3748/wjg.v21.i7.2199
Peer-review started: May 4, 2014
First decision: May 14, 2014
Revised: May 25, 2014
Accepted: June 12, 2014
Article in press: June 13, 2014
Published online: February 21, 2015
Processing time: 283 Days and 6.6 Hours
AIM: To determine the expression and significance of filamin A (FLNa) in colorectal adenocarcinoma tissue.
METHODS: The expression of FLNa in 46 colorectal cancer tissues and normal tissues was detected by immunohistochemistry, reverse transcription polymerase chain reaction (RT-PCR) and Western blotting, and its relationship with clinical parameters and prognosis was analyzed.
RESULTS: The positive expression of FLNa in cancer tissues was lower than that in normal mucosa, and the difference was statistically significant. The expression of FLNa correlated with liver metastasis, lymph node metastasis and rectal invasion depth, regardless of sex, age, tumor location, tumor size, gross shape and histological type of colorectal carcinoma. Multivariate analysis showed that FLNa was an independent risk factor for postoperative survival of patients with colorectal adenocarcinoma. Moreover, survival analysis showed that the expression level of FLNa was closely related with survival of patients with colorectal adenocarcinoma. The results of RT-PCR and Western blotting were consistent with those of immunohistochemistry.
CONCLUSION: FLNa showed low expression in colorectal adenocarcinoma, high correlation with the incidence and development of colorectal cancer, and was considered an indicator of prognosis.
Core tip: The expression of filamin A (FLNa) was correlated with liver metastasis, lymph node metastasis and rectal invasion depth, regardless of sex, age, tumor location, tumor size, gross shape and histological type of colorectal carcinoma. FLNa was an independent risk factor for postoperative survival of patients with colorectal adenocarcinoma. FLNa showed low expression in colorectal adenocarcinoma, high correlation with the incidence and development of colorectal cancer, and was considered an indicator of prognosis.
- Citation: Tian ZQ, Shi JW, Wang XR, Li Z, Wang GY. New cancer suppressor gene for colorectal adenocarcinoma: Filamin A. World J Gastroenterol 2015; 21(7): 2199-2205
- URL: https://www.wjgnet.com/1007-9327/full/v21/i7/2199.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i7.2199
Colorectal cancer is a common malignancy. Tumor invasion and metastasis are the leading causes of death in patients with colorectal cancer[1]. Filamin A (FLNa) is a non-muscle actin-binding protein involved in regulating cell proliferation and migration, has an important role in tumor formation and development and is involved in integrating cell movement and signal transduction[2,3]. Studies have shown that FLNa has low expression in colorectal cancer tissues, high correlation with the incidence and development of colorectal cancer[4-6], and is considered to be a classification indicator of colorectal cancer subtype[7,8]. In this study, the expression of FLNa gene in colorectal adenocarcinoma tissues and normal colorectal mucosa tissues was detected by immunohistochemistry, reverse transcription polymerase chain reaction (RT-PCR) and Western blotting to determine its correlation with clinicopathological parameters and explore its value in the prognosis of patients with colorectal cancer.
From January 2005 to December 2006, 46 colorectal adenocarcinoma tissues and normal colorectal mucosa tissues resected during surgery were randomly selected from patients treated in the Department of the Second General Surgery of the Fourth Hospital of Hebei Medical University. These tumor tissues were taken from the center of the tumor, while the normal tissues were taken at least 10 cm away from the edge of the tumor. All patients enrolled in this study were diagnosed by histopathological examination, had no other primary tumors, no history of preoperative radiotherapy, chemotherapy or immunotherapy, and had complete clinical and follow-up data. The follow-up period ended in July 2011.
The immunohistochemistry kit was purchased from Beijing Zhongshan Golden Bridge Biotechnology Co. Ltd. DAB chromogenic reagent was purchased from TIANGEN Company. Total RNA extraction reagent and radioimmunoprecipitation (radioimmunoprecipitation assay, RIPA) lysis buffer were purchased from Solarbio Company. Primers were synthesized by Shanghai Biological Engineering Company. The reverse transcription kit was purchased from Fermentas Company. The PCR kit was purchased from Promega Company. Rabbit anti-human FLNa monoclonal antibody was purchased from Epitomics Company. Rabbit polyclonal anti-human GAPDH antibody was purchased from Santa Cruz Company. SDS-PAGE standard indicator was purchased from Fermentas Company.
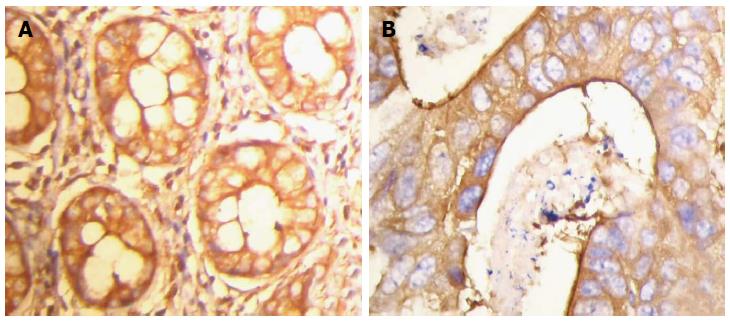
Immunohistochemical staining was performed using the streptomycin affinity biotin-peroxidase immunohistochemistry technique (SP method). Paraffin-embedded tissues were cut into sections 4 μm thick, placed in a container of citrate buffer after dewaxing hydration, then into a microwave oven (95 °C, 15 min) to repair the antigen, and then incubated at room temperature for 20 min with 3% hydrogen peroxide. After washing with PBS, the specimens were incubated with normal goat serum, then rabbit anti-human FLNa monoclonal antibody (1:150 dilution), and left in the refrigerator overnight at 4 °C. After washing with PBS, the specimens were incubated with biotinylated secondary antibody for 30 min at room temperature and washed with PBS. The specimens were then incubated with enzyme-labeled streptavidin horseradish peroxidase for 30 min at room temperature, washed with PBS, colored by DAB, rinsed with water, hematoxylin stained, gradient dehydrated, treated with xylene and mounted with neutral gum. Human uterine tissue was used as a positive control for FLNa, and PBS instead of primary antibody as a negative control for FLNa. The specimens were observed under an optical microscope, where 200 cells in five selected horizons were observed at high magnification, and the average rate of positive cells was calculated. Staining intensity was divided into three levels: light yellow, yellow and brown. Particle status was divided into three categories: no obvious particles, sparse particles and diffuse particles. The immunostaining results were divided into three categories by comprehensive analysis of positive cell rate and staining intensity of each slice: the number of positive cells < 25%, lighter yellow staining and no obvious particles were defined as negative (-); positive cells ≥ 25%- < 50%, yellow staining, and sparse particles were defined as weakly positive (+); positive cells ≥ 50%, brown staining, and diffuse particles were defined as strongly positive (++). Two experienced clinical pathologists determined the results.
Total RNA was extracted using the total RNA extraction kit and its concentration was determined. The primer was set according to the full-length cDNA of human FLNa and GAPDH in GenBank. The upstream primer of FLNa was 5’-AGC CTCCACGAGACATCATC-3’, and the downstream primer was 5’-CCAGTGTGTACTC CCCCTTG-3’; The upstream primer of internal reference GADPH was 5’-GAAGGTG AAGGTCGGAGTC-3’, and the downstream primer was 5’-GAAG ATGGTGATGGGATTTC-3’. One microgram of total RNA was added to 1 μL of random primers, 4 μL of 5 × RT-PCR Buffer, 1 μL of RNA inhibitors, 2 μL of dNTP, 1 μL of reverse transcriptase, followed by diethylpyrocarbonate (DEPC) treated water to a total volume of 20 μL. The reverse transcription steps were as follows: 30 °C for 10 min, 45 °C for 5 min, and 95 °C for 5 min after completion of the reaction. The PCR system included the following: 2 μL of cDNA, 1 μL of each upstream and downstream primer, 2.5 μL of 10 × PCR Buffer, 0.2 μL of Taq DNA polymerase, 0.5 μL of dNTP, plus DEPC treated water to a total volume of 25 μL. The PCR steps were as follows: denaturating for 5 min at 95 °C, 94 °C for 30 s, 64 °C for 30 s, 72 °C for 30 s for a total of 35 cycles, and a final extension at 72 °C for 7 min. The PCR product was loaded on a 2% agarose gel and separated by electrophoresis. The product of FLNa and GAPDH PCR were 310 bp and 220 bp. The PCR results were observed in ultraviolet light.
Tissues were homogenized using RIPA lysis buffer. Total protein was extracted and quantified. 50 μg of protein sample was added to 5 × sample loading buffer (volume ratio 4:1), boiled for 5 min, and the sample was then loaded. After separation by SDS-PAGE, the proteins were transferred to a polyvinylidene fluoride membrane. The membrane was placed in TBST containing 5% skim milk, and left at room temperature for 1 h, the specific antibody (FLNa, 1:2000; GAPDH, 1:200), was then added and incubated at -4 °C overnight. The next day, the membrane was washed three times with TBST solution, for 10 min each time, and the appropriate secondary antibody (1:2000) was then added and incubated at 37 °C for 1 h. The protein bands were detected by chemiluminescence.
Data were analyzed using the statistical software SPSS 13.0. T test, χ2 test or Fisher’s exact test were used to detect correlations, and the Cox regression model was used to detect multivariate analysis. The Kaplan-Meier method was used to determine survival in the FLNa negative and positive expression groups. The test level was α = 0.05. P < 0.05 was considered statistically significant.
Positive expression of FLNa was detected in the cytoplasm (Figure 1). The positive expression rate of FLNa in 46 colorectal adenocarcinoma tissue samples was 47.83% (22/46), with a weakly positive expression rate of 26.09% (12/46) and a strongly positive expression rate of 21.74% (10/46). The positive expression rate of FLNa in normal colorectal tissue samples was 91.30% (42/46), with a weakly positive expression rate of 10.87% (5/46) and a strongly positive expression rate of 80.43% (37/46). Compared with that of cancer tissue samples, the difference was statistically significant (χ2 = 32.679; P = 0.000; Table 1).
| Group | Cases | Expression of filamin A protein | χ2 | P | ||
| - | + | + + | ||||
| Cancer group | 46 | 24 | 12 | 10 | 32.679 | 0.000 |
| Normal group | 46 | 4 | 5 | 37 | ||
Statistical analysis of the immunohistochemistry results showed that the expression of FLNa was correlated with liver metastasis, lymph node metastasis and rectal invasion depth (P < 0.05), regardless of gender, age, tumor location and histological type (P > 0.05; Table 2).
| Parameters | Cases | Positive expression of filamin A | |
| Cases | P | ||
| Sex | |||
| Male | 27 | 12 | 0.584 |
| Female | 19 | 10 | |
| Age | |||
| ≥ 60 yr | 26 | 10 | 0.147 |
| < 60 yr | 20 | 12 | |
| Tumor location | |||
| Colon | 17 | 4 | 0.238 |
| Rectum | 29 | 18 | |
| Liver metastasis | |||
| Yes | 9 | 1 | 0.022 |
| No | 37 | 22 | |
| Lymph node metastasis | |||
| Yes | 33 | 10 | 0.000 |
| No | 13 | 12 | |
| Gross shape | |||
| Uplift | 11 | 5 | 0.977 |
| Ulcerated | 23 | 11 | |
| Infiltrative | 12 | 6 | |
| Depth of invasion | |||
| Serosal invasion | 30 | 7 | 0.000 |
| No serosal invasion | 16 | 15 | |
| Histological type | |||
| Papillary adenocarcinoma | 23 | 12 | 0.935 |
| Tubular adenocarcinoma | 12 | 6 | |
| Mucinous adenocarcinoma | 11 | 5 | |
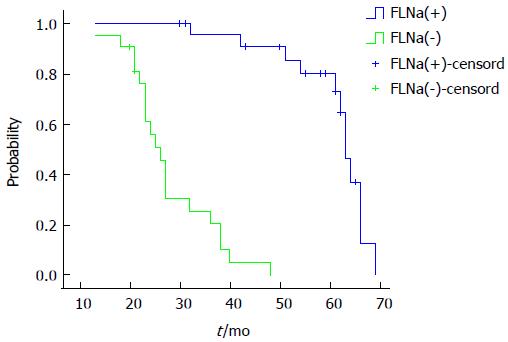
Factors such as age, sex, tumor location, tumor invasion depth, lymph node metastasis and FLNa expression were introduced into the Cox regression model for multivariate analysis. The results showed that lymph node metastasis, tumor invasion depth and FLNa expression were independent factors for prognosis in colorectal cancer patients. The risk of death in the FLNa protein negative expression group was 3.856 times of that of the FLNa protein positive expression group (95%CI: 7.326-19.421; Table 3). The FLNa positive expression group had a median survival time of 63 mo, while the FLNa negative expression group had a median survival time of 26 mo. The log-rank test of survival rate in the two groups showed that the differences were statistically significant (P = 0.000; Figure 2).
| Parameters | B | Wald | df | Sig. | Exp(B) | 95%CI |
| Expression of filamin A protein | -3.274 | 15.114 | 1 | 0.000 | 3.856 | 7.326-19.421 |
| Depth of tumor invasion | 2.685 | 6.163 | 1 | 0.013 | 14.657 | 1.760-122.093 |
| Lymph node metastasis | 2.193 | 10.836 | 1 | 0.001 | 8.958 | 2.428-33.052 |
GAPDH was considered the internal reference, and FLNa/GAPDH was quantified by RT-PCR detection. The results showed that the expression levels of FLNa mRNA in tumor tissues were lower than those in normal tissues [(0.24 ± 0.03) vs (0.95 ± 0.04), P = 0.017], and the difference between the two groups was statistically significant (Figure 3).
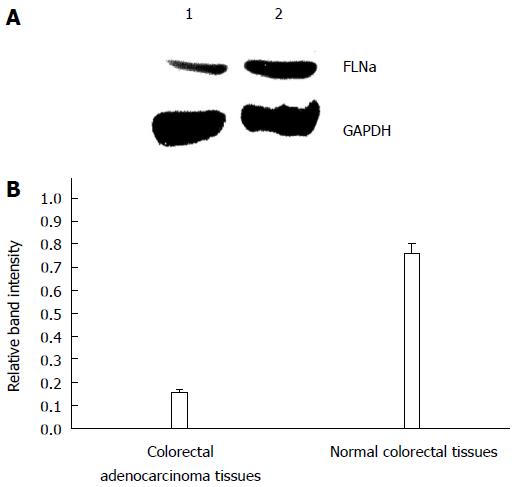
GAPDH was considered the internal reference, and FLNa/GAPDH was quantified by Western blot. The results showed that the expression levels of FLNa mRNA in tumor tissues were lower than those in normal tissues [(0.15 ± 0.02) vs (0.76 ± 0.04), P = 0.013], and the difference between the two groups was statistically significant (Figure 4).
FLNa is an important member of the filament protein family, which has a highly conserved gene structure, a wide range of expression, and has an important role in mammalian growth and development. The molecular weight of the FLNa protein dimer subunit is 280 kDa, with a length of about 80 nm, and includes an actin binding domain at the amino end and a rod-like domain consisting of 24 tandem repeats, which is formed with an interval of two separate hinge structures containing about 30 amino acid residues. Two polypeptide chains are connected to the carboxy terminal end of the 24th repeated sequence and form a “V” type homodimer related to function[9]. FLNa is located mainly in the cytoplasm, the multiple layer structure of the β-sheet in the domain of rod-like molecules provides an interface for protein- protein interactions, which is capable of interacting with a variety of proteins with important functions, and the signal protein which is a cell membrane molecule receptor, is an important signal transduction scaffold protein involved in cell proliferation, adhesion, migration[10,11], initiation and progression of tumor[12], and the regulation of organ development[13].
Currently, there are few studies on the correlation between FLNa and the occurrence and development of cancer. Shi et al[14,15] placed plasmid vectors containing FLNa cDNA into liposomes to transfect human SW480 colon cancer cells over-expressing the FLNa gene, and confirmed that FLNa can inhibit the invasiveness of SW480 cells both in vitro and in vivo. In this study, we used immunohistochemistry to detect the expression of FLNa in colorectal adenocarcinoma tissues and normal colorectal mucosa tissues, and found that the positive expression rate of FLNa protein in cancer tissues was significantly lower than that in normal mucosa. The expression of FLNa correlated with tumor metastasis, lymph node metastasis and depth of rectal invasion, regardless of sex, age, tumor location, size, shape and histological type. Multivariate analysis showed that FLNa was an independent risk factor for postoperative survival of colorectal cancer patients. In addition, survival analysis showed that the survival time of patients with low expression of FLNa protein was significantly shorter than patients with high expression of FLNa. RT-PCR and Western blot results showed that the expression levels of FLNa mRNA and protein in normal colorectal tissues were higher than that in colorectal cancer tissues.
The mechanism by which FLNa suppresses cancer is currently unclear. Kwon et al[16] found that down-regulation of FLNa interacting protein-like 1 was associated with promoter methylation and an invasive phenotype in colon cancer. Vial et al[17] showed that epidermal growth factor regulates the α5β1 integrin activation state through p90RSK-dependent phosphorylation of FLNa. Zhu et al[18] found that FLNa regulates Ras/ERK by the Ras-GRF1 pathway, reduces the levels of intracellular matrix metalloproteinase-9, inhibits degradation of the extracellular matrix, and prevents the migration of tumor cells. Fiori et al[19] found that, compared to M2A7 cells (expressing FLNa), the epidermal growth factor receptor tyrosine ubiquitination level in M2 cells (deletion of FLNa), stimulated by epidermal growth factor, was low. Further study found that the lack of FLNa had an influence on the interactions between epidermal growth factor receptor and the ubiquitin ligase c-Cb1 chain, and degradation of growth factor receptor induced by anti-epidermal growth factor was significantly inhibited in cells, while degradation of growth factor receptor was very active in cells expressing FLNa. Therefore, FLNa inhibits proliferation of tumor cells by reducing the activity of the epidermal growth factor receptor. Sasaki et al[20] found that the interaction between FLNa and the Smads protein family regulates transforming growth factor-β (TGF-β) signaling and inhibits the migration of tumor cells, however, TGF-β signaling is absent in tumor cells not expressing FLNa. In addition, research on the yeast two-hybrid system and signal transduction pathways of tumor cells confirmed that FLNa can specifically combine with G protein-coupled receptors (dopamine receptors and calcium receptors), small GTP-binding proteins (Rho, Rac and CDC42) and its effector molecule, involved in regulating tumor incidence and development[21].
In summary, FLNa is closely related with the incidence and development of colorectal adenocarcinoma, and can be used as an important indicator of prognosis and treatment[22], especially chemotherapy[23], and multiple indicators could provide accurate predictive information, resulting in personalized therapy[24]. FLNa has also been found in feces[25], an elevated level of FLNa mRNA indicates colorectal cancer[26], which can be used as a method of detecting colorectal cancer. However, the tumor suppressing mechanism of FLNa requires further study.
Colorectal cancer is a common malignancy. Tumor invasion and metastasis are the leading causes of death in patients with colorectal cancer. Filamin A (FLNa) is a non-muscle actin-binding protein involved in regulating cell proliferation and migration, and has an important role in tumor formation and development, and is involved in integrating cell movement and signal transduction.
In this study, the expression of FLNa gene in colorectal adenocarcinoma tissues and normal colorectal mucosa tissues was detected by immunohistochemistry, reverse transcription polymerase chain reaction and Western blotting to determine its correlation with clinicopathological parameters and explore its value in the prognosis of patients with colorectal cancer.
This study found that FLNa is an independent risk factor for postoperative survival of patients with colorectal adenocarcinoma, using a series of simple and accurate methods.
This study showed that FLNa is an independent risk factor for postoperative survival of patients with colorectal adenocarcinoma. Thus, the authors could predict the prognosis of colorectal cancer by testing the expression of FLNa.
FLNa is a non-muscle actin-binding protein involved in regulating cell proliferation and migration, and has an important role in tumor formation and development, and is involved in integrating cell movement and signal transduction.
This manuscript is very well written. The authors observed the expression and significance of FLNa in the tissue of colorectal adenocarcinoma. The research design is good, and the results are acceptable.
P- Reviewer: Kim KC, Pizzirusso F S- Editor: Song XX L- Editor: Webster JR E- Editor: Liu XM
| 1. | Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8283] [Cited by in RCA: 8225] [Article Influence: 483.8] [Reference Citation Analysis (0)] |
| 2. | Kolahi KS, Mofrad MR. Molecular mechanics of filamin’s rod domain. Biophys J. 2008;94:1075-1083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 24] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 3. | Stevenson RP, Veltman D, Machesky LM. Actin-bundling proteins in cancer progression at a glance. J Cell Sci. 2012;125:1073-1079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 121] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 4. | Shi JW, Wang GY, Zhang HB, Yu YM, Wang XL, Yang HC, Wang SJ. Specific Expression of FLNa in Colorectal Adenocarcinoma. Zhongguo Zhongliu Linchuang. 2010;37:1342-1343. |
| 5. | Shi JW, Liu HY, Wang GY, Yu YM, Wang XL, Yang HC, Wang SJ. Expression of FLNa in Colorectal Adenocarcinoma its clinical significance. Zhongguo Laonianxue Zazhi. 2010;13:009. |
| 6. | Shi JW, Yu YM, Wang SJ, Wang XL, Zhang J, Wang GY. Expressiom of Filamin A and Matrix Metalloproteinase-9 in Colorectal Adenocarcinoma and Their Significance. Zhongliu. 2011;30:768-773. |
| 7. | Burgess DJ. Gene expression: colorectal cancer classifications. Nat Rev Cancer. 2013;13:380-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 8. | Sadanandam A, Lyssiotis CA, Homicsko K, Collisson EA, Gibb WJ, Wullschleger S, Ostos LC, Lannon WA, Grotzinger C, Del Rio M. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19:619-625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 683] [Cited by in RCA: 763] [Article Influence: 63.6] [Reference Citation Analysis (0)] |
| 9. | Nakamura F, Osborn TM, Hartemink CA, Hartwig JH, Stossel TP. Structural basis of filamin A functions. J Cell Biol. 2007;179:1011-1025. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 203] [Cited by in RCA: 214] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 10. | Robertson SP, Twigg SR, Sutherland-Smith AJ, Biancalana V, Gorlin RJ, Horn D, Kenwrick SJ, Kim CA, Morava E, Newbury-Ecob R. Localized mutations in the gene encoding the cytoskeletal protein filamin A cause diverse malformations in humans. Nat Genet. 2003;33:487-491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 320] [Cited by in RCA: 310] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 11. | Kim H, Sengupta A, Glogauer M, McCulloch CA. Filamin A regulates cell spreading and survival via beta1 integrins. Exp Cell Res. 2008;314:834-846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 61] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 12. | Shi JW, Wang SJ, Wang GY. Research Advance on Role of Filamin A in Tumor Progression. Zhongliu Fangzhiyanjiu. 2010;8:029. |
| 13. | Shi JW, Wang GY. Research Advance on Role of Filamin A in regulating development of organs. Aibian Jibian Tubian. 2012;24:78-80. |
| 14. | Shi JW, Yu YM, Wang SJ, Yang S, Kong FL, Zhang J, Wang GY. Inhibitory effect of filamin A on invasion and metastasis of human colon carcinoma cell line SW480 in vitro. Jichuyixue yu Linchuang. 2011;31:1000-1005. |
| 15. | Shi JW, Yu YM, Wang SJ, Wang XL, Wang GY. Inhibitory effect of filamin A on tumor formation activity of human colon carcinoma SW480 cells in nude mice. Zhongliu. 2011;31:701-706. |
| 16. | Kwon M, Lee SJ, Reddy S, Rybak Y, Adem A, Libutti SK. Down-regulation of Filamin A interacting protein 1-like Is associated with promoter methylation and an invasive phenotype in breast, colon, lung and pancreatic cancers [corrected]. PLoS One. 2013;8:e82620. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 17. | Vial D, McKeown-Longo PJ. Epidermal growth factor (EGF) regulates α5β1 integrin activation state in human cancer cell lines through the p90RSK-dependent phosphorylation of filamin A. J Biol Chem. 2012;287:40371-40380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 36] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 18. | Zhu TN, He HJ, Kole S, D’Souza T, Agarwal R, Morin PJ, Bernier M. Filamin A-mediated down-regulation of the exchange factor Ras-GRF1 correlates with decreased matrix metalloproteinase-9 expression in human melanoma cells. J Biol Chem. 2007;282:14816-14826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 48] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 19. | Fiori JL, Zhu TN, O’Connell MP, Hoek KS, Indig FE, Frank BP, Morris C, Kole S, Hasskamp J, Elias G. Filamin A modulates kinase activation and intracellular trafficking of epidermal growth factor receptors in human melanoma cells. Endocrinology. 2009;150:2551-2560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 20. | Sasaki A, Masuda Y, Ohta Y, Ikeda K, Watanabe K. Filamin associates with Smads and regulates transforming growth factor-beta signaling. J Biol Chem. 2001;276:17871-17877. [PubMed] |
| 21. | Feng Y, Walsh CA. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat Cell Biol. 2004;6:1034-1038. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 379] [Cited by in RCA: 398] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 22. | Sanz-García E, Elez E, Macarulla T, Dienstmann R, Salazar R, & Tabernero J. Prognosis and Therapeutic Implications for Emerging Colorectal Cancer Subtypes. Current Colorectal Cancer Reports. 2014;1-7. |
| 23. | Kim HJ, Kang UB, Lee H, Jung JH, Lee ST, Yu MH, Kim H, Lee C. Profiling of differentially expressed proteins in stage IV colorectal cancers with good and poor outcomes. J Proteomics. 2012;75:2983-2997. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 24. | Mettu NB, Hurwitz H, Hsu DS. Use of molecular biomarkers to inform adjuvant therapy for colon cancer. Oncology (Williston Park). 2013;27:746-754. [PubMed] |
| 25. | Ang CS, Nice EC. Targeted in-gel MRM: a hypothesis driven approach for colorectal cancer biomarker discovery in human feces. J Proteome Res. 2010;9:4346-4355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 45] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 26. | Buckhaults P, Kinzler KW, Vogelstein B. Methods for detection of colorectal cancer: U.S. Patent: 8029764 2011; . |