Published online Oct 7, 2015. doi: 10.3748/wjg.v21.i37.10487
Peer-review started: January 30, 2015
First decision: April 14, 2015
Revised: April 28, 2015
Accepted: August 31, 2015
Article in press: August 31, 2015
Published online: October 7, 2015
Processing time: 242 Days and 4.4 Hours
The intestinal microbiome is emerging as a crucial mediator between external insults and systemic infections. New research suggests that our intestinal microorganisms contribute to critical illness and the development of non-gastrointestinal infectious diseases. Common pathways include a loss of fecal intestinal bacterial diversity and a disproportionate increase in toxogenic bacterial species. Therapeutic interventions targeting the microbiome - primarily probiotics - have yielded limited results to date. However, knowledge in this area is rapidly expanding and microbiome-based therapy such as short-chain fatty acids may eventually become a standard strategy for preventing systemic infections in the context of critical illness.
Core tip: The role of the intestinal microbiome in the development and treatment of Clostridium difficile (C. difficile) infection is well established. However, the intestinal microbiome is emerging as a crucial mediator in the development of systemic disease and non-gastrointestinal infection. If the pathways linking gut bacteria to systemic infections can be elucidated, it may become possible to intervene upon the microbiome before disease occurs. This understanding would move clinicians beyond fecal microbial transplant for C. difficile infection to paradigm-changing treatments for gut-derived systemic infections.
- Citation: Latorre M, Krishnareddy S, Freedberg DE. Microbiome as mediator: Do systemic infections start in the gut? World J Gastroenterol 2015; 21(37): 10487-10492
- URL: https://www.wjgnet.com/1007-9327/full/v21/i37/10487.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i37.10487
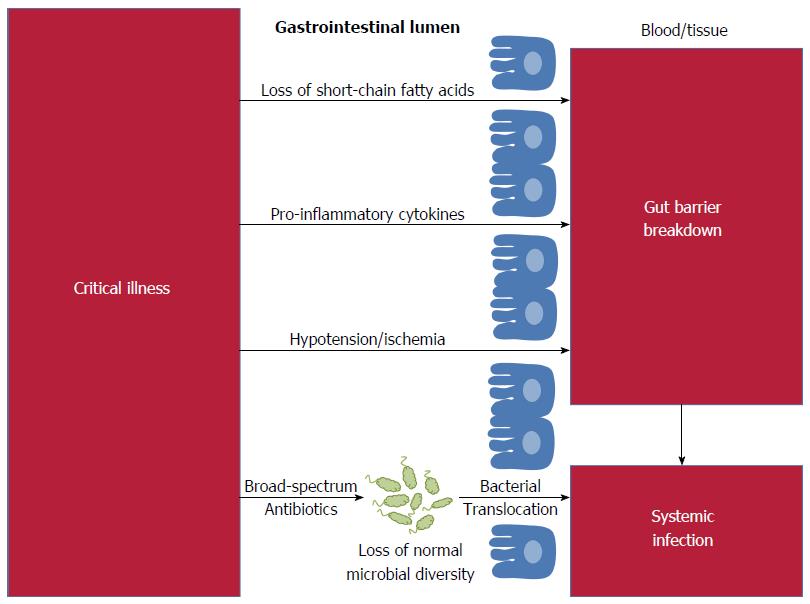
Infections are the leading cause of death in children, the leading cause of death in adults admitted to intensive care units (ICUs) and, in the United States and Europe, the leading cause of iatrogenic death[1]. The relative ease of obtaining fecal specimens and the declining cost of high-throughput sequencing has led to newfound interest in the fecal microbial flora as the source for systemic infections[2]. However, the concept of the gut as a source for infection is not new: over two-thousand years ago, Hippocrates affirmed that “all disease begins in the gut”. Recent studies have demonstrated that gut flora influence a vast array of functions including immunity, energy utilization, and drug metabolism[3]. Disruption to our intestinal microbiome has been linked to systemic infections, including respiratory tract infections, bacteremia and urinary tract infections[4-6]. The gastrointestinal microbiome is emerging not as the cause of disease, but as the crucial mediator between external insults and systemic infections (Figure 1). Data from animal and human studies show that targeted manipulation of the fecal flora has the potential to ameliorate systemic infections.
The quintessential disease of the microbiome remains Clostridium difficile (C. difficile) infection (CDI), which is both caused and cured by altering the intestinal microbiome. Antibiotics are the most important risk factor for the development of CDI, however host microbiome factors have recently been identified that additionally contribute to colonization and infection with C. difficile[7,8]. The associated changes include loss of diversity in the intestinal flora, depletion of Bacteroides, increased Enterococcaceae and Streptococcaceae, reduced non-toxogenic Clostridial species and increased fecal concentrations of C. difficile[3,9-12]. The success of fecal transplant as treatment for recurrent C. Difficile whether by enema, colonoscopic transplant, duodenal infusion, or pills, demonstrates that restoration of a diverse flora is highly effective[3,13,14]. Even more exciting is evidence that targeted restoration of the fecal microbiome with just a handful of specific bacteria can be equally effective[15]. Unfortunately, similar outcomes have yet to be achieved for other systemic or infectious conditions.
Can the knowledge gained in the treatment of CDI be translated into effective treatments in other situations? Perhaps the most promising area is in the relationship between critical illness and the intestinal microbiome. Analysis of stool samples from critically ill patients through Gram staining, culture and 16S rRNA gene amplification has revealed evidence of microbial dysbiosis during critical illness[16,17]. In a study performed by Shimizu et al[16], fecal specimens from patients with severe systemic inflammatory response syndrome (SIRS) were Gram stained and cultured to evaluate bacterial diversity. Patients with fewer bacteria on Gram stain also had fewer obligate and facultative anaerobes upon culture, and patients with stool that was depleted on Gram stain had a significantly increased incidence of enteritis, pneumonia and mortality due to multi-organ dysfunction compared to patients with diverse stool patterns. Zaborin et al[17] achieved similar results using 16S sequencing to evaluate the fecal composition of 14 patients in the ICU, 10 of which had been admitted for a period of over 20 d. In 5 patients, they observed the emergence of ultra-low-diversity communities containing just 1 to 4 bacterial taxa. Decreased intestinal diversity in critically ill patients facilitates colonization by health-care associated pathogens and the development of multi-drug resistant organisms[17,18]. These findings are consistent with the role of intestinal microorganisms in the maintenance of immunity, proliferation of colonic epithelial cells, and energy production[3,18]. During periods of critical illness selective pressure from ischemia, inflammation and altered immunity, decreased intestinal motility and reduced oxygen tension likely contribute to the development of intestinal dysbiosis. Medical therapies, namely antibiotics, also deplete host microbial diversity in critically ill patients through changes in the physiology, structure and gene expression of intestinal bacteria[19,20]. The ability to use fecal microbial characteristics to identify ICU patients at high risk for subsequent systemic infections is a crucial first step towards successful therapeutic interventions through manipulation of the microbiome.
In healthy hosts, the gut epithelium plays a number of important immunological roles including acting as a barrier against invading pathogens, producing hormones and cytokines, producing antimicrobial peptides, and continuously communicating with both the gut-associated lymphoid tissue and with intraluminal bacteria[21]. Critically ill hosts have alterations in gut immune function related to the intestinal microbiome that facilitate systemic infections. On a cellular level, crypt proliferation is markedly decreased and both crypt and villus apoptosis are simultaneously increased following sepsis[22]. Critical illness slows the migration of epithelial cells in a manner dependent on Toll-like receptor-4 (TLR-4) and also induces global alterations in the mucus layer and gut barrier function[23,24]. Patients who are in intensive care invariably receive broad-spectrum antibiotics which affect luminal bacteria even when these antibiotics are given intravenously and secreted into the lumen in exceedingly small amounts[25]. Antibiotics cause loss of bacterial ligands that would normally trigger TLRs and other receptors through microorganism-associated molecular patterns (MAMPs)[26]. Antibiotic-induced loss of specific microbial signals, such as signaling from segmented filamentous bacteria, impairs normal maintenance of T-helper cells and other T-cell subsets[27].
In addition to the direct effects of antibiotics, critical illness itself may alter adaptive immunity including changes in gut secretion of IgA. Intestinal bacterial colonization is a potent inducer of secretory IgA, and IgA is especially important at mucosal surfaces. A functional role for IgA in mucosal infections has been demonstrated in certain infections such as rotavirus, influenza, and cholera toxin[28]. In addition, it has been demonstrated that bacterial specific IgA is induced following bacterial colonization and that the IgA repertoire decreases when there is reduced intestinal microbial diversity, as is the case during critical illness[29]. Another established immune adaptation is the induction of different mucosal CD4+ T cell subsets following intestinal colonization. A variety of functionally distinct CD4+ T cells exists, including regulatory Foxp3+ cells, Th1, Th2, Th17 and T follicular helper cells. Several studies have demonstrated that normal intestinal colonization either with different bacterial communities or individual species such as Bacteroides fragilis also can induce Treg cells[30-32]. The importance of a robust adaptive immune response to colonic bacteria is demonstrated by CDI; compared to patients with a normal serum IgG response after infection, those with a low serum IgG response to C. difficile toxin A are nearly fifty times more likely to experience recurrence[33].
Changes in CD4+ T-cell compartments are likely to impact risk for systemic infections, yet much about the microbiome during critical illness remains uncertain. The most established method of assessing the role of the microbiome in critical illness or other situations is through germ-free mouse models, which completely lack endogenous flora. Germ-free mice have widely varying responses in preclinical models of critical illness. In Pseudomonas aeruginosa pneumonia, for example, germ-free mice have higher mortality and altered intestinal epithelial apoptosis compared to conventional mice; similar results are seen in germ-free mice infected with Klebsiella pneumonia pneumonia, with severity of illness mediated by IL-10[34,35]. In contrast, germ-free mice have 100% survival in a model of ischemia/reperfusion that is lethal in conventional animals, which is associated with abrogation of the systemic inflammatory response[36]. These disparate responses highlight the importance of disease and even pathogen specificity when evaluating host-microbial interactions.
The gastrointestinal microbiome has been implicated in the development of non-gastrointestinal infections in the lungs, blood and urine. Rosen et al[4] evaluated the effect of acid suppression therapy on the abundance and diversity of gastric and lung microflora in children presenting with chronic cough. Acid suppression was associated with gastric bacterial overgrowth of which the predominant species were Streptococcus and Staphylococcus, both known lung pathogens. Although the gastric and pulmonary environments did not correlate with the prevalence and diversity of bacterial species, these results indicate that medications may affect the interaction between the microorganisms of the mouth, upper gastrointestinal tract and the respiratory system.
Taur et al[5] demonstrated an association between the intestinal microbiome and the development of bacteremia in patients undergoing allogeneic hematopoetic stem cell transplant (allo-HSCT). Stool specimens were collected from 94 patients undergoing allo-HSCT from before transplant until 35 d after transplant. After transplant, patients consistently developed reduced bacterial diversity with shifts in the dominating bacterial populations inhabiting the gut to Enterococcus, Streptococcus and various Proteobacteria. Increased Enterococcus domination in gut flora correlated to a nine-fold increase in the risk of Vancomycin-resistant Enterococcus bacteremia. Proteobacterial domination correlated with a five-fold increase in Gram-negative rod bacteremia.
Although the urinary tract was once thought to be sterile, intestinal bacterial composition has now been linked to the development of urinary tract infections and disease. In a study of patients with new urinary catheters, Tambyah et al[6] found that two-thirds of patients developed urinary tract infections from non-urinary sources and, of those, nearly fifty percent had infections that developed after 48 h, suggesting migration of microbes from the perineum. These infections were most commonly caused by Enterococcus, Streptococcus, and Gram-negative rods, which implicates the intestinal microbiome in the development of infection. Small studies investigating the urinary microbiome through the use of 16S rRNA sequencing have demonstrated the presence of bacteria in healthy subjects. Fouts et al[37] showed that in patients at risk of asymptomatic bacteriuria due to spinal cord injury, urinary microbiomes differed from healthy control patients, instead paralleling the fecal microbiome with a predominance of gut microbes including Clostridia and Bacteroidetes[38]. Patients with interstitial cystitis, an inflammatory condition of the bladder, have decreased diversity and a prominence of Lactobacillus compared to healthy controls, again suggesting transmission through the intestinal microbiome[38]. These studies remind us that our intestinal microorganisms have the potential to explain infectious diseases far beyond the gastrointestinal tract.
Despite advances in understanding the role of the intestinal microbiome in the pathogenesis of disease, the use of probiotics as treatment has yielded limited success. Even for treatment and prevention of CDI, probiotics have been disappointing. A 2008 Cochrane review evaluating the use of probiotics for the treatment of CDI in adults found insufficient evidence to support the use of probiotics alone or in conjunction with antibiotics[39]. An updated review evaluating the use of probiotics for the prevention of C. difficile-associated diarrhea (CDAD) in adults and children found moderate quality evidence to suggest that probiotics were both safe and effective for preventing CDAD, but the very modest effect size has deterred use of probiotics for this purpose in clinical practice[40]. To date, there is minimal evidence that probiotics are effective in other gastrointestinal diseases such as ulcerative colitis and Crohn’s disease[41,42].
Probiotics may have the greatest potential for therapeutic benefit in targeted populations such as critically ill patients if key microbial taxa and pathways can be identified. This would allow for possible intervention on the gastrointestinal flora after it has been damaged but before the onset of disease. However, there are barriers to the use of microbiome-based therapeutics in the setting of critical illness given difficulty tolerating oral intake, concerns over the possibility of precipitating infection through the introduction of bacterial species, and incomplete understanding of the correct composition and dosage of probiotic compounds. But several studies now suggest that targeted microbiome-based therapies have the capacity to overcome these hurdles. Among these budding treatments, short-chain fatty acids (SCFAs) may be the best.
SCFAs play a unique role in colonic microbial ecology, especially in the critically ill, where they serve as a direct fuel substrate for colonic epithelial cells and maintain the normal gut barrier. Yamada et al[43] measured fecal SCFAs in critically ill patients with SIRS at the time of ICU admission and for the following 6 wk. Compared to healthy volunteers, patients in the ICU had four-times lower levels of butyrate, the dominant human colonic SCFA, and other SCFAs[43]. Mice models prove that manipulation of the microbiome using SCFAs has the potential to alter bacterial translocation across the gut wall in the setting of critical illness[44,45]. Using a mouse model and an Escherichia coli (E. coli) O157:H7 challenge, Fukuda et al[44] found that Bifidobacterium longum, but not Bifidobacterium adolescentic, was protective against death. This effect was attributed in part to increased production of acetate which resulted in inhibition of translocation of the E. coli O157:H7 from the gut to blood. SCFAs are the main end product of complex carbohydrate metabolism and the authors showed that carbohydrate utilization, as determined by metabolic profiling, was proportional to the degree of protection conferred by the different Bifidobacterium species. Microbiota-derived butyrate was recently shown to stabilize hypoxia-inducible factor, a transcription factor that strengthens the gut barrier in the setting of ischemic colitis or other injuries. This protective mechanism is likely to be of particular importance in critically ill patients[46].
One way to mitigate the potential loss of SCFAs during critical illness is through the administration of fiber-containing feeds. In a study by O’Keefe et al[47] of critically ill patients with necrotizing pancreatitis, administration of fiber-containing enteral feeds increased fecal content of SCFAs. Despite these encouraging findings, few studies have evaluated the use of SCFAs during critical illness. A study performed in rats that had sepsis induced by cecal perforation showed that animals receiving infusion of sodium butyrate had reduced mortality and end organ dysfunction[45]. Similarly, a study of 55 critically ill patients receiving enteral feeding with an experimental supplement containing SCFAs resulted in improved organ function as measured by the Sequential Organ Failure Assessment score over a 10-d period[48]. These studies suggest the potential of SCFAs and fiber as adjunctive therapy in the treatment of critical illness.
The intestinal microbiome is emerging as a crucial mediator between external insults and systemic infection. This new perspective on the microbiome provides a novel platform for describing how pneumonia, bacteremia, urinary tract infections and other systemic conditions are acquired. With this newfound understanding, it may one day be possible to move beyond fecal microbial transplant for C. difficile infection to paradigm-changing treatments for gut-derived systemic infections.
P- Reviewer: Boros M, Gong JP, Kim TI, Sipahi AM, Tasci I S- Editor: Ji FF L- Editor: A E- Editor: Wang CH
| 1. | Vincent JL, Rello J, Marshall J, Silva E, Anzueto A, Martin CD, Moreno R, Lipman J, Gomersall C, Sakr Y. International study of the prevalence and outcomes of infection in intensive care units. JAMA. 2009;302:2323-2329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2205] [Cited by in RCA: 2338] [Article Influence: 146.1] [Reference Citation Analysis (0)] |
| 2. | Tyler AD, Smith MI, Silverberg MS. Analyzing the human microbiome: a “how to” guide for physicians. Am J Gastroenterol. 2014;109:983-993. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 3. | Brandt LJ. American Journal of Gastroenterology Lecture: Intestinal microbiota and the role of fecal microbiota transplant (FMT) in treatment of C. difficile infection. Am J Gastroenterol. 2013;108:177-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 120] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 4. | Rosen R, Amirault J, Liu H, Mitchell P, Hu L, Khatwa U, Onderdonk A. Changes in gastric and lung microflora with acid suppression: acid suppression and bacterial growth. JAMA Pediatr. 2014;168:932-937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 76] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 5. | Taur Y, Xavier JB, Lipuma L, Ubeda C, Goldberg J, Gobourne A, Lee YJ, Dubin KA, Socci ND, Viale A. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis. 2012;55:905-914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 609] [Cited by in RCA: 754] [Article Influence: 58.0] [Reference Citation Analysis (0)] |
| 6. | Tambyah PA, Halvorson KT, Maki DG. A prospective study of pathogenesis of catheter-associated urinary tract infections. Mayo Clin Proc. 1999;74:131-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 137] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 7. | Loo VG, Bourgault AM, Poirier L, Lamothe F, Michaud S, Turgeon N, Toye B, Beaudoin A, Frost EH, Gilca R. Host and pathogen factors for Clostridium difficile infection and colonization. N Engl J Med. 2011;365:1693-1703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 590] [Cited by in RCA: 615] [Article Influence: 43.9] [Reference Citation Analysis (0)] |
| 8. | Surawicz CM, Brandt LJ, Binion DG, Ananthakrishnan AN, Curry SR, Gilligan PH, McFarland LV, Mellow M, Zuckerbraun BS. Guidelines for diagnosis, treatment, and prevention of Clostridium difficile infections. Am J Gastroenterol. 2013;108:478-498; quiz 499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1179] [Cited by in RCA: 1186] [Article Influence: 98.8] [Reference Citation Analysis (0)] |
| 9. | Britton RA, Young VB. Role of the intestinal microbiota in resistance to colonization by Clostridium difficile. Gastroenterology. 2014;146:1547-1553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 328] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 10. | Stein RR, Bucci V, Toussaint NC, Buffie CG, Rätsch G, Pamer EG, Sander C, Xavier JB. Ecological modeling from time-series inference: insight into dynamics and stability of intestinal microbiota. PLoS Comput Biol. 2013;9:e1003388. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 392] [Cited by in RCA: 390] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 11. | Vincent C, Stephens DA, Loo VG, Edens TJ, Behr MA, Dewar K, Manges AR. Reductions in intestinal Clostridiales precede the development of nosocomial Clostridium difficile infection. Microbiome. 2013;1:18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 95] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 12. | Kinnebrew MA, Lee YJ, Jenq RR, Lipuma L, Littmann ER, Gobourne A, No D, van den Brink M, Pamer EG, Taur Y. Early Clostridium difficile infection during allogeneic hematopoietic stem cell transplantation. PLoS One. 2014;9:e90158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 13. | van Nood E, Dijkgraaf MG, Keller JJ. Duodenal infusion of feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:2145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2582] [Cited by in RCA: 2677] [Article Influence: 223.1] [Reference Citation Analysis (0)] |
| 14. | Youngster I, Russell GH, Pindar C, Ziv-Baran T, Sauk J, Hohmann EL. Oral, capsulized, frozen fecal microbiota transplantation for relapsing Clostridium difficile infection. JAMA. 2014;312:1772-1778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 466] [Cited by in RCA: 483] [Article Influence: 43.9] [Reference Citation Analysis (0)] |
| 15. | Petrof EO, Gloor GB, Vanner SJ, Weese SJ, Carter D, Daigneault MC, Brown EM, Schroeter K, Allen-Vercoe E. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: ‘RePOOPulating’ the gut. Microbiome. 2013;1:3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 534] [Cited by in RCA: 523] [Article Influence: 43.6] [Reference Citation Analysis (0)] |
| 16. | Shimizu K, Ogura H, Tomono K, Tasaki O, Asahara T, Nomoto K, Morotomi M, Matsushima A, Nakahori Y, Yamano S. Patterns of Gram-stained fecal flora as a quick diagnostic marker in patients with severe SIRS. Dig Dis Sci. 2011;56:1782-1788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 17. | Zaborin A, Smith D, Garfield K, Quensen J, Shakhsheer B, Kade M, Tirrell M, Tiedje J, Gilbert JA, Zaborina O. Membership and behavior of ultra-low-diversity pathogen communities present in the gut of humans during prolonged critical illness. MBio. 2014;5:e01361-e01314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 247] [Cited by in RCA: 236] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 18. | Shimizu K, Ogura H, Hamasaki T, Goto M, Tasaki O, Asahara T, Nomoto K, Morotomi M, Matsushima A, Kuwagata Y. Altered gut flora are associated with septic complications and death in critically ill patients with systemic inflammatory response syndrome. Dig Dis Sci. 2011;56:1171-1177. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 144] [Cited by in RCA: 157] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 19. | Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 2013;152:39-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 580] [Cited by in RCA: 596] [Article Influence: 49.7] [Reference Citation Analysis (0)] |
| 20. | Leslie JL, Young VB. The rest of the story: the microbiome and gastrointestinal infections. Curr Opin Microbiol. 2015;23:121-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 21. | Clark JA, Coopersmith CM. Intestinal crosstalk: a new paradigm for understanding the gut as the “motor” of critical illness. Shock. 2007;28:384-393. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 366] [Cited by in RCA: 347] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 22. | Coopersmith CM, Stromberg PE, Davis CG, Dunne WM, Amiot DM, Karl IE, Hotchkiss RS, Buchman TG. Sepsis from Pseudomonas aeruginosa pneumonia decreases intestinal proliferation and induces gut epithelial cell cycle arrest. Crit Care Med. 2003;31:1630-1637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 85] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 23. | Neal MD, Sodhi CP, Dyer M, Craig BT, Good M, Jia H, Yazji I, Afrazi A, Richardson WM, Beer-Stolz D. A critical role for TLR4 induction of autophagy in the regulation of enterocyte migration and the pathogenesis of necrotizing enterocolitis. J Immunol. 2013;190:3541-3551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 119] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 24. | Fink MP. Intestinal epithelial hyperpermeability: update on the pathogenesis of gut mucosal barrier dysfunction in critical illness. Curr Opin Crit Care. 2003;9:143-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 169] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 25. | van Ogtrop ML, Guiot HF, Mattie H, van Strijen E, Sekh BR, van Furth R. Modulation of the intestinal flora of mice by treatment with aztreonam and tigemonam. Antimicrob Agents Chemother. 1991;35:983-985. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 26. | Willing BP, Russell SL, Finlay BB. Shifting the balance: antibiotic effects on host-microbiota mutualism. Nat Rev Microbiol. 2011;9:233-243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 473] [Cited by in RCA: 535] [Article Influence: 38.2] [Reference Citation Analysis (0)] |
| 27. | Hill DA, Hoffmann C, Abt MC, Du Y, Kobuley D, Kirn TJ, Bushman FD, Artis D. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2010;3:148-158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 300] [Cited by in RCA: 330] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 28. | Pabst O. New concepts in the generation and functions of IgA. Nat Rev Immunol. 2012;12:821-832. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 416] [Cited by in RCA: 484] [Article Influence: 37.2] [Reference Citation Analysis (0)] |
| 29. | Hapfelmeier S, Lawson MA, Slack E, Kirundi JK, Stoel M, Heikenwalder M, Cahenzli J, Velykoredko Y, Balmer ML, Endt K. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328:1705-1709. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 533] [Cited by in RCA: 608] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 30. | Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2568] [Cited by in RCA: 2877] [Article Influence: 191.8] [Reference Citation Analysis (0)] |
| 31. | Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, McCoy KD, Macpherson AJ. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794-806. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 599] [Cited by in RCA: 656] [Article Influence: 46.9] [Reference Citation Analysis (0)] |
| 32. | Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107:12204-12209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1484] [Cited by in RCA: 1674] [Article Influence: 111.6] [Reference Citation Analysis (0)] |
| 33. | Kyne L, Warny M, Qamar A, Kelly CP. Association between antibody response to toxin A and protection against recurrent Clostridium difficile diarrhoea. Lancet. 2001;357:189-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 626] [Cited by in RCA: 609] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 34. | Fox AC, McConnell KW, Yoseph BP, Breed E, Liang Z, Clark AT, O’Donnell D, Zee-Cheng B, Jung E, Dominguez JA. The endogenous bacteria alter gut epithelial apoptosis and decrease mortality following Pseudomonas aeruginosa pneumonia. Shock. 2012;38:508-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 35. | Fagundes CT, Amaral FA, Vieira AT, Soares AC, Pinho V, Nicoli JR, Vieira LQ, Teixeira MM, Souza DG. Transient TLR activation restores inflammatory response and ability to control pulmonary bacterial infection in germfree mice. J Immunol. 2012;188:1411-1420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 171] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 36. | Souza DG, Vieira AT, Soares AC, Pinho V, Nicoli JR, Vieira LQ, Teixeira MM. The essential role of the intestinal microbiota in facilitating acute inflammatory responses. J Immunol. 2004;173:4137-4146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 188] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 37. | Fouts DE, Pieper R, Szpakowski S, Pohl H, Knoblach S, Suh MJ, Huang ST, Ljungberg I, Sprague BM, Lucas SK. Integrated next-generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury. J Transl Med. 2012;10:174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 279] [Cited by in RCA: 356] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 38. | Siddiqui H, Lagesen K, Nederbragt AJ, Jeansson SL, Jakobsen KS. Alterations of microbiota in urine from women with interstitial cystitis. BMC Microbiol. 2012;12:205. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 154] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 39. | Pillai A, Nelson R. Probiotics for treatment of Clostridium difficile-associated colitis in adults. Cochrane Database Syst Rev. 2008;CD004611. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 96] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 40. | Goldenberg JZ, Ma SS, Saxton JD, Martzen MR, Vandvik PO, Thorlund K, Guyatt GH, Johnston BC. Probiotics for the prevention of Clostridium difficile-associated diarrhea in adults and children. Cochrane Database Syst Rev. 2013;5:CD006095. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 141] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 41. | Mallon P, McKay D, Kirk S, Gardiner K. Probiotics for induction of remission in ulcerative colitis. Cochrane Database Syst Rev. 2007;CD005573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 81] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 42. | Rolfe VE, Fortun PJ, Hawkey CJ, Bath-Hextall F. Probiotics for maintenance of remission in Crohn’s disease. Cochrane Database Syst Rev. 2006;CD004826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 79] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 43. | Yamada T, Shimizu K, Ogura H, Asahara T, Nomoto K, Yamakawa K, Hamasaki T, Nakahori Y, Ohnishi M, Kuwagata Y. Rapid and Sustained Long-Term Decrease of Fecal Short-Chain Fatty Acids in Critically Ill Patients With Systemic Inflammatory Response Syndrome. JPEN J Parenter Enteral Nutr. 2015;39:569-577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 66] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 44. | Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543-547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1444] [Cited by in RCA: 1690] [Article Influence: 120.7] [Reference Citation Analysis (0)] |
| 45. | Zhang LT, Yao YM, Lu JQ, Yan XJ, Yu Y, Sheng ZY. Sodium butyrate prevents lethality of severe sepsis in rats. Shock. 2007;27:672-677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 56] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 46. | Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ, Wilson KE, Glover LE, Kominsky DJ, Magnuson A. Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell Host Microbe. 2015;17:662-671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 807] [Cited by in RCA: 1190] [Article Influence: 119.0] [Reference Citation Analysis (0)] |
| 47. | O’Keefe SJ, Ou J, Delany JP, Curry S, Zoetendal E, Gaskins HR, Gunn S. Effect of fiber supplementation on the microbiota in critically ill patients. World J Gastrointest Pathophysiol. 2011;2:138-145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 41] [Cited by in RCA: 40] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 48. | Beale RJ, Sherry T, Lei K, Campbell-Stephen L, McCook J, Smith J, Venetz W, Alteheld B, Stehle P, Schneider H. Early enteral supplementation with key pharmaconutrients improves Sequential Organ Failure Assessment score in critically ill patients with sepsis: outcome of a randomized, controlled, double-blind trial. Crit Care Med. 2008;36:131-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 115] [Article Influence: 6.8] [Reference Citation Analysis (0)] |