Published online Sep 14, 2015. doi: 10.3748/wjg.v21.i34.9853
Peer-review started: April 27, 2015
First decision: June 2, 2015
Revised: June 23, 2015
Accepted: July 18, 2015
Article in press: July 18, 2015
Published online: September 14, 2015
Processing time: 141 Days and 3.5 Hours
Hepatocellular carcinoma (HCC) is an aggressive malignancy and the second leading cause of cancer-related deaths worldwide. Conventional biomarkers exhibit poor performance in the surveillance, diagnosis, and prognosis of HCC. MicroRNAs (miRNAs) are a class of evolutionarily conserved small non-coding RNAs that are involved in the regulation of gene expression and protein translation, and they play critical roles in cell growth, differentiation, and the development of various types of cancers, including HCC. Recent evidence revealed the role of miRNAs as potential novel and ideal biomarkers for HCC. miRNAs are released to extracellular spaces, and they are extremely stable in bodily fluids, including serum or plasma, where they are packaged into various microparticles or associated with RNA-binding proteins. Numerous studies have demonstrated that circulating miRNAs have potential applications as minimally invasive biomarkers for HCC diagnosis and prognosis. The present review highlights current understanding of miRNA biogenesis and the origins and types of circulating miRNAs. We summarize recent progress in the use of circulating miRNAs as diagnostic and prognostic biomarkers for HCC. We also discuss the challenges and perspectives of the clinical utility of circulating miRNAs in HCC.
Core tip: MicroRNAs (miRNAs) play critical roles in cell growth, differentiation, and the development of hepatocellular carcinoma (HCC). The study of circulating miRNAs is a rapidly growing field of research, indicating the potential applications of miRNAs as minimally invasive biomarkers for HCC diagnosis, recurrence monitoring and prognosis. This review highlights current understanding of miRNA biogenesis and the origins and types of circulating miRNAs and summarizes recent progress in the study of circulating miRNAs as diagnostic and prognostic biomarkers for HCC. We also discuss the challenges and perspectives regarding the clinical utility of circulating miRNAs in HCC.
- Citation: Zhang YC, Xu Z, Zhang TF, Wang YL. Circulating microRNAs as diagnostic and prognostic tools for hepatocellular carcinoma. World J Gastroenterol 2015; 21(34): 9853-9862
- URL: https://www.wjgnet.com/1007-9327/full/v21/i34/9853.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i34.9853
Hepatocellular carcinoma (HCC) is the seventh most common malignancy and the second leading cause of cancer-related deaths worldwide. Globally, there are approximately 750000 new cases and 700000 deaths of HCC reported per annum[1-3]. Two well-known risk factors for HCC are chronic viral hepatitis B (HBV) and C (HCV), which account for 80%-90% of all HCC cases worldwide. Other risks for HCC include obesity, diabetes, vitamin D deficiency, aflatoxin B1 exposure, alcoholic and non-alcoholic liver cirrhosis[4,5]. However, the underlying mechanism of HCC has not been entirely elucidated. Surgical resection and orthotopic liver transplantation are the best curative tools for the long-term survival of HCC patients. However, surgical resection is not feasible in more than 80% of HCC patients because of tumor location, tumor size or severity of the underlying liver disease. Only 5%-15% of HCC patients are potentially resectable[6,7]. The overall five-year survival rate in patients with HCC is very low, ranging from 5% to 9%. The cumulative five-year recurrence rate is approximately 70% to 80% even after curative surgical resection. Recurrence after resection generally results in a high rate of mortality[8]. Therefore, the most urgent needs are the identification of sensitive and specific markers for early diagnosis, the monitoring of recurrence and the prediction of prognosis for HCC.
Current methods for HCC diagnosis are classified into the following main categories: imaging [abdominal ultrasonography, magnetic resonance imaging (MRI), and contrast-enhanced computed tomography (CT)] and laboratory biomarker analysis [serum alpha-fetoprotein (AFP) levels][9,10]. However, the diagnostic performance of imaging technologies is unsatisfactory, particularly for the diagnosis of small lesions and early-stage HCC[11,12]. AFP is the most commonly used tumor marker for HCC diagnosis and prognosis prediction, but the false negative rate using AFP level alone is as high as 40% for patients with early-stage HCC. AFP levels remain normal in 15%-30% of all the patients, even patients with advanced HCC[6,13]. Therefore, the American Association for the Study of Liver Disease Practice Guidelines discarded AFP for surveillance and diagnosis because of its low sensitivity (39%-64%) and specificity (76%-94%)[14]. Therefore, the identification of novel and ideal biomarkers with high specificity and sensitivity for HCC diagnosis is desperately needed. Circulating microRNAs (miRNAs) have received remarkable attention because they offer great advantages and good performance as novel biomarkers for HCC diagnosis and prognosis prediction[15].
Emerging evidence demonstrated that miRNAs are an important class of non-coding RNAs as tumor oncogenes or suppressors that are involved in the HCC development[16]. miRNAs are endogenous nucleotides that are found in intra- and extracellular spaces, such as blood, urine and saliva[15,17]. Cellular miRNAs are also released, detected and quantified in the blood[18]. Circulating miRNAs exhibit several obvious advantages as potentially novel and ideal diagnostic biomarkers, including significant stability in various types of body fluids, resistance to endogenous RNase digestion, low cost, ease of analysis, and sensitive detection methods, such as real-time quantitative polymerase chain reaction (qRT-PCR)[19-21]. The above characteristics of circulating miRNAs and the limitations of currently available non-invasive methods to monitor HCC indicate that circulating miRNAs are a rapidly growing field of research that hold great promise for the development of reliable biomarkers for the diagnosis and monitoring of HCC[6,20,22].
This review highlights miRNA biogenesis and the types and origins of circulating miRNAs, summarizes recent findings of circulating miRNAs as prognostic and predictive biomarkers for HCC, and addresses challenges and perspectives for the clinical utility of circulating miRNAs in HCC.
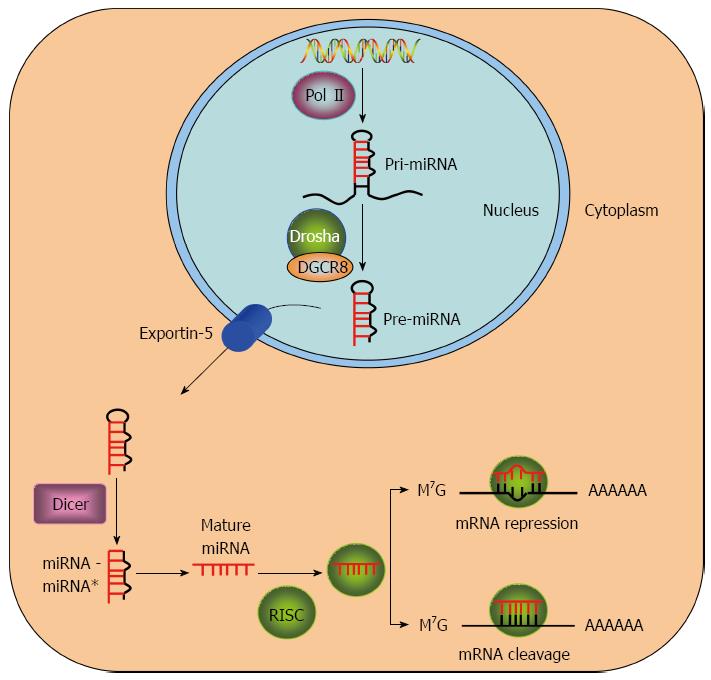
miRNAs are single-stranded, non-coding RNA strands of 19-25 nucleotides that regulate more than 50% of all protein-coding genes in mammals[23,24]. There are 1881 human miRNAs sequences registered in the miRBase database. The synthesis of miRNAs primarily consists of two steps, including nuclear synthesis within and outside of the nucleus[25] (Figure 1). First, RNA polymerase II transcribes miRNAs in the nucleus into primary miRNAs (pri-miRNAs) that are several hundred nucleotides in length[26,27]. Subsequently, the microprocessor complex, which consists of DiGeorge syndrome critical region gene (DGCR8) and the nuclease Drosha, cleaves the pri-miRNAs into miRNA precursors (pre-miRNAs) of approximately 60 to 70 nucleotides[3,28,29]. Exportin-5 transports the pre-miRNAs from the nucleus to the cytoplasm[25,30,31]. The pre-miRNAs are further processed in the cytoplasm by RNase Dicer to final miRNA duplexes (miRNA-miRNA*) of 19-25 nucleotides[32,33]. Finally, the mature single-stranded miRNAs are generated and retained to form RISC (RNA-induced silencing complex), which is destined for mRNA repression/cleavage, but its passenger strand (miRNA*) undergoes degradation[33-35]. Mature miRNAs recognize and bind to the 3′ untranslated region (UTR) of multiple mRNA targets with imperfect or perfect complementarity, which enables one specific miRNA to inhibit the translation of multiple genes[36]. Multiple miRNAs may also regulate one mRNA[17]. miRNAs participate in many key cellular processes, including proliferation, cell cycle, apoptosis, and metastasis[7,37]. The involvement of miRNAs in cancer is well established because miRNAs behave as tumor-suppressor genes or oncogenes, depending on the cellular function of their targets[38,39]. Growing evidence indicates that miRNAs may become a new tool for HCC diagnosis, prognosis, and therapy[38,40,41].
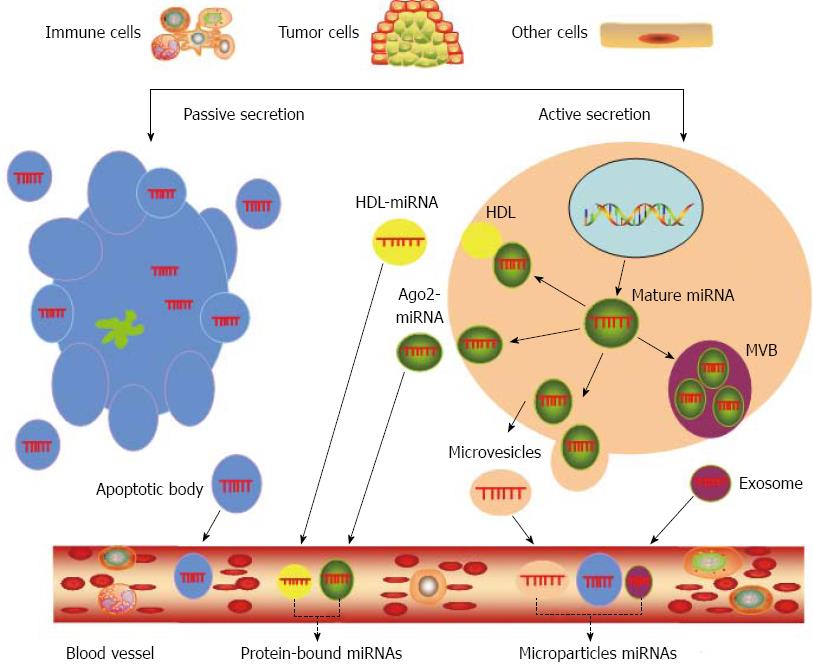
The mechanisms through which circulating miRNAs originate have been established recently. Figure 2 shows that circulating miRNAs originate from the following mechanisms: (1) active secretion via membrane vesicles; (2) active secretion in protein-miRNA complexes and lipoprotein complexes (such as high-density lipoproteins); (3) active secretion via exosomes when a multivesicular body fuses with the cell membrane; and (4) passive secretion originating from cell necrosis and apoptosis[42-46]. Some research has demonstrated that circulating miRNAs exist in two different types, circulating protein-bound miRNAs or circulating microparticle miRNAs[18,43,47,48]. Circulating microparticle miRNAs are circulating miRNAs that are packaged into microparticles. Three different types of microparticles were defined by their size, mechanisms of production, and protein and lipid content[49]: (1) exosomes (40 to 100 nm in diameter; released by exocytosis from multivesicular bodies); (2) microvesicles (100 to 1000 nm in diameter; released via plasma budding); and (3) apoptotic bodies (1000 to 4000 nm in diameter; released by cells executing apoptotic process)[20,43,50,51]. Circulating protein-bound miRNAs are circulating miRNAs that are associated with specific RNA-binding proteins and lipoproteins, such as Ago2[52], which is a component of the RNA-induced silencing complex, and high-density lipoproteins (HDL)[53]. One study demonstrated that the majority of circulating miRNAs were associated with protein complexes rather than vesicles[52], but other reports indicated that circulating miRNAs were selectively sorted to microparticles or protein complexes[54-57]. Notably, both types of circulating miRNAs vary in different types of liver disease. Bala and co-workers reported that circulating miR-122 and miR-155 are predominantly associated with the exosome-rich fraction in alcoholic liver disease and inflammatory liver injury, but these miRNAs are present in the protein-rich fraction in drug-induced liver injury. Therefore, these results suggest that the shift in different types of circulating miRNAs may provide further specificity of the mechanisms of liver pathology[22].
A recent study identified miRNAs in many body fluids, including serum and plasma[58]. Circulating miRNAs in body fluids are packaged into microparticles or bound to proteins, which provides stability and resistance to plasma RNase digestion and enables miRNA transfer from one cell to another during diverse biological processes[59]. Circulating miRNAs exhibit paracrine effects on tumor growth[60,61]. If tumors are defined as relatively homogeneous cancer cells that are formed in an independent microenvironment, circulating miRNAs may play a novel role as regulators of intercellular communications during tumor formation[62,63]. Aside from their functional roles in tumorigenesis, circulating miRNAs represent a less invasive alternative for diagnostic testing. Easily assessable serum-based miRNAs may provide novel biomarkers of diagnostic, prognostic and predictive values for HCC, specifically in the field of hepatology.
The first description of the presence of circulating miRNAs in serum and their potential as cancer markers was reported by Lawrie et al[64] in 2008. This study demonstrated the overexpression of miR-155, miR-21 and miR-210 in patients with diffuse large B-cell lymphoma compared to healthy controls. Notably, high miR-21 levels were associated with the relapse-free survival of these patients[60]. Multiple studies identified circulating miRNAs as diagnostic and prognostic markers for a wide range of malignancies, which primarily used blood plasma or serum for their analyses[48].
Evidence also suggests that miRNAs are widely involved in HCC carcinogenesis, differentiation and metastasis. Aberrant miRNA expression was reported in HCC patients and cell lines. More information on aberrant miRNA expression in HCC is available in comprehensive reviews[3,40]. Therefore, circulating miRNAs should be affected during HCC development and progression. Various reports demonstrated the potential clinical application of circulating miRNAs in HCC diagnosis and prognosis.
The identification of a single or several individual circulating miRNAs as biomarkers for HCC diagnosis is expected. This approach has a unique advantage because it is much simpler and more straightforward compared to the detection of all circulating miRNAs. For example, miR-122 in the liver was the most abundant liver-specific miRNA, and it accounted for 70% of the total hepatic miRNAs[65]. MiR-122 plays a critical role in liver homeostasis and hepatocarcinogenesis[66]. Xu et al[67] and Qi et al[68] found elevated levels of circulating miR-122 in patients with HCC compared to healthy individuals. Qi et al[68] revealed that serum miR-122 was a potential marker for the discrimination of HCC patients from healthy controls with an AUC (the area under the receiver operating characteristic curve) of 0.869, and a cut-off value of 0.475. The sensitivity and specificity for serum miR-122 were 81.6% and 83.3%, respectively. Xu et al[67] found similar results. These results suggest that circulating miR-122, which is a liver-specific miRNA, may serve as a potential marker for HCC diagnosis. MiR-21 is also a promising biochemical biomarker for HCC diagnosis because plasma levels of miR-21 are notably higher in HCC patients than in healthy volunteers and patients with chronic hepatitis[69]. Zhang et al[6] demonstrated that serum miR-143 and miR-215 were valuable biomarkers to distinguish HCC from healthy controls with an AUC of 0.795 (sensitivity and specificity were 71% and 83%, respectively) and 0.816 (sensitivity and specificity were 80% and 91%, respectively), respectively.
However, the specificity of the detection method for a single or several individual HCC-related circulating miRNAs is relatively poor. For example, increased levels of circulating miRNA-122 are found in HCC, HBV infection[70], HCV infection[71], non-alcoholic fatty-liver disease[72], liver cirrhosis[67], and alcohol-related liver disease[73]. Xu et al[67] and Qi et al[68] suggest that serum miR-122 is a potential marker for the discrimination of HCC patients from healthy controls, but not HCC patients from patients with chronic hepatitis. Therefore, these authors proposed serum miR-122 as a novel biomarker for liver injury but not specifically for HCC. HCC is a highly complex, multifactorial and heterogeneous disease, and numerous miRNAs are dysregulated during HCC onset and progression. Therefore, a combination of multiple circulating miRNAs or a plasma/serum miRNA panel instead of a single circulating miRNA may offer more specificity and sensitivity as biomarkers for HCC diagnosis and prognosis prediction.
Jiang et al[74] demonstrated that three serum miRNAs (miR-10b, miR-106b, and miR-181a) discriminated HCC patients from normal controls (AUC of 0.85, 0.82, and 0.89, respectively). However, the ability of single serum miRNA to differentiate CLD patients from normal controls was not satisfactory. This report indicated that the panel of three serum miRNAs displayed better performance compared to a single serum miRNA assay, with an AUC of 0.94 in discriminating HCC patients from normal controls and 0.91 in discriminating HCC patients from CLD.
Tan et al[12] preformed a study of 667 subjects (261 HCC patients, 233 cirrhosis patients, and 173 healthy controls) to identify a serum miRNA panel for use as biomarkers in the diagnosis of HBV-related HCC. Tan et al[12] first used sequencing to analyze miRNAs that were differently expressed in serum samples obtained from three patients with HBV-related HCC compared to sera from three cirrhosis patients, and three healthy controls. Secondly, differently expressed miRNAs were validated using qRT-PCR in 20 HBV-related HCC patients, 20 cirrhosis patients and 20 healthy controls in the biomarker selection stage. Subsequently, 135 HBV-related HCC patients, 132 cirrhosis patients and 90 healthy controls formed a training set. Finally, the established serum miRNA panel was tested in an independent cohort in the validation phase (103 HBV-related HCC patients, 78 cirrhosis patients, and 60 healthy controls). These data identified eight miRNAs (miR-206, miR-141-3p, miR-433-3p, miR-1228-5p, miR-199a-5p, miR-122-5p, miR-192-5p, and miR-26a-5p) and established a serum miRNA panel that provided high diagnostic accuracy for HCC (AUC = 0.887 and 0.879 for training and validation sets, respectively). These results demonstrated that a serum miRNA panel could differentiate HBV-related HCC from healthy controls (AUC = 0.893) and cirrhosis (AUC = 0.892) with a high degree of accuracy.
Similarly, Zhou et al[75] also attempted to identify a plasma miRNA panel for the diagnosis of HBV-related HCC. This study investigated 934 participants (healthy, chronic hepatitis B, cirrhosis, and HBV-related HCC). First, Zhou et al[75] used microarrays to screen 723 miRNAs in 137 plasma samples for the diagnosis of HCC in the discovery stage. Fifteen candidate miRNAs that were discovered via microarray were selected for further testing using qRT-PCR from 102 participants in the training stage. Seven miRNAs that were differentially expressed between the HCC and control groups (healthy, chronic hepatitis B, and cirrhosis) were further tested in an additional 305 participants. Finally, seven miRNAs were used to predict the probability of HCC diagnosis in an independent cohort of 390 patients in the validation phase. The results demonstrated that the plasma miRNA panel (miR-122, miR-192, miR-21, miR-223, miR-26a, miR-27a, and miR-801) provided a high diagnostic accuracy of HCC (AUC = 0.864 and 0.888 for training and validation data set, respectively), and it differentiated HCC from healthy controls (AUC = 0.941), chronic hepatitis B (AUC = 0.842), and cirrhosis (AUC = 0.884), respectively.
AFP is the most widely used and broadly known biomarker for HCC diagnosis, and the combination of AFP measurement and ultrasound at 6-mo intervals are the standard tools for the diagnosis and monitoring of HCC in China. However, the sensitivity and specificity of serum AFP levels are relatively unsatisfactory, especially for AFP-negative HCC and small-size HCC (< 3 cm). The testing of AFP levels was dropped from current surveillance guidelines in Europe and the United States because of its low sensitivity and specificity[12,76,77]. AFP is not an ideal biomarker for HCC, but some reports demonstrated that a combination of serum/plasma miRNAs with alkaline phosphatase (ALP) may be beneficial in HCC patients and improve the sensitivity and specificity of liver cancer diagnosis.
For example, Tomimaru et al[69] measured plasma miR-21 level using qRT-PCR in 126 HCC patients, 30 chronic hepatitis patients, and 50 healthy volunteers. Their results suggested that miR-21 levels were higher in HCC patients than in chronic hepatitis patients (AUC of 77.3; sensitivity of 61.1% and specificity of 83.3%) and healthy volunteers (AUC of 95.3; sensitivity of 87.3% and specificity of 96%). Notably, the combination of plasma miR-21 with AFP improved the differentiation power between HCC patients and patients with chronic hepatitis (AUC of 0.823; sensitivity of 81.0% and specificity of 80.0%) and healthy volunteers (AUC to 0.971; sensitivity of 92.9% and specificity of 94.0%). Qu et al[78] analyzed serum levels of three miRNAs, namely miR-16, miR-195, and miR-199a, either alone or in combination with conventional serum markers (AFP, ALP-L3 and DCP) to differentiate HCC from CLDs. These authors suggested that miR-16 was a more sensitive biomarker for HCC, and the combination of miR-16, AFP, AFP-L3, and DCP identified 92.4% of HCC cases with a high specificity (78.5%). These results demonstrated that the combination of serum/plasma miRNAs with already established markers (such as AFP) may improve the performance of HCC diagnosis.
Circulating miRNAs may also aid the prediction of HCC prognosis. For example, Xu et al[4] analyzed the prognostic role of serum miR-122 levels in 122 HCC patients, and their results revealed a higher overall survival rate in HCC patients with high serum miR-122 levels compared to low miR-122 levels. They also found that high serum miR-122 levels were independently associated with higher overall survival rates in HCC patients. These outcomes suggest that a high serum miR-122 level is a good biomarker of prognosis in HCC patients. Köberle et al[79] also found that HCC patients with higher miR-122 serum levels exhibited longer overall survival than individuals with lower miR-122 serum concentrations, but the serum miR-122 level was not independently associated with overall survival. However, Köberle et al[79] suggested that miR-1 serum levels were independently associated with overall survival, but miR-1 serum levels exhibited no relevant correlation with clinical chemistry liver parameters. Their data indicated that serum miR-1 might improve the predictive value of classical HCC staging scores. Li et al[80] found that high levels of miR-221 expression correlated with tumor size, cirrhosis and tumor stage in HCC patients. Their results also suggested that the overall survival rate of the high miR-221 expression group was significantly lower than that of the low miR-221 expression group. Therefore, serum miR-221 also might provide predictive significance for the prognosis of HCC patients.
Collectively, these data illustrate that circulating miRNAs may be used as non-invasive biomarkers for the diagnosis and prognosis of HCC, and these nucleotides may become promising next-generation biomarkers for HCC detection. The data described above are summarized in Table 1.
| MicroRNAs | Diagnosis/prognosis | Up/downregulated in plasma/serum | Plasma/serum | Relevance | Ref. |
| miR-12 | Yes/No | Upregulated | Serum | Compared with healthy control | [67] |
| miR-122 | Yes/No | Upregulated | Serum | [67] | |
| miR-223 | Yes/No | Upregulated | Serum | [67] | |
| miR-122 | Yes/No | Upregulated | Serum | Compared with healthy control | [68] |
| miR-21 | Yes/No | Upregulated | Plasma | Compared with chronic hepatitis or healthy control | [69] |
| miR-143 | Yes/No | Upregulated | Serum | Compared with healthy control | [6] |
| miR-215 | Yes/No | Upregulated | Serum | [6] | |
| miR-10b | Yes/No | Upregulated | Serum | Compared with CLD or healthy control | [74] |
| miR-106b | Yes/No | Upregulated | Serum | [74] | |
| miR-181a | Yes/No | Downregulated | Serum | [74] | |
| miR-206 | Yes/No | Upregulated | Serum | HBV-related HCC compared with cirrhosis or healthy control | [12] |
| miR-141-3p | Yes/No | Upregulated | Serum | [12] | |
| miR-433-3p | Yes/No | Upregulated | Serum | [12] | |
| miR-1228-5p | Yes/No | Upregulated | Serum | [12] | |
| miR-199a-5p | Yes/No | Downregulated | Serum | [12] | |
| miR-122-5p | Yes/No | Downregulated | Serum | [12] | |
| miR-192-5p | Yes/No | Downregulated | Serum | [12] | |
| miR-26a-5p | Yes/No | Downregulated | Serum | [12] | |
| miR-122 | Yes/No | Downregulated | Plasma | HBV-related HCC compared with chronic hepatitis B or healthy control | [75] |
| miR-192 | Yes/No | Upregulated | Plasma | [75] | |
| miR-21 | Yes/No | Upregulated | Plasma | [75] | |
| miR-223 | Yes/No | Downregulated | Plasma | [75] | |
| miR-26a | Yes/No | Downregulated | Plasma | [75] | |
| miR-27a | Yes/No | Downregulated | Plasma | [75] | |
| miR-801 | Yes/No | Upregulated | Plasma | [75] | |
| miR-16 | Yes/No | Downregulated | Serum | Compared with CLD or healthy control | [78] |
| miR-21 | Yes/No | Upregulated | Plasma | Compared with chronic hepatitis or healthy volunteers | [69] |
| miR-375d | Yes/No | Downregulated | Serum | Compared with healthy control | [87] |
| miR-199a-3p | Yes/No | Downregulated | [87] | ||
| miR-30c-5p | Yes/No | Upregulated | Serum | HCV-related HCC compared with healthy control | [15] |
| miR-223-3p | Yes/No | Downregulated | Serum | [15] | |
| miR-302c-3p | Yes/No | Upregulated | Serum | [15] | |
| miR-17-5p | Yes/No | Upregulated | Serum | [15] | |
| miR-122 | No/Yes | Upregulated | Serum | Improved overall survival | [4] |
| miR-1 | No/Yes | Upregulated | Serum | Improved overall survival | [79] |
| miR-221 | No/Yes | Upregulated | Serum | High serum miR-221 levels decreased survival rate | [81] |
The use of circulating miRNAs as biomarkers for HCC has received attention since the first circulating miRNAs signatures were reported as potential diagnostic tools in oncology in 2008. This review highlights the process of miRNAs biogenesis and elaborates the origins and types of circulating miRNAs. Notably, we have summarized recent advances in circulating miRNAs signatures as non-invasive biomarkers in the diagnosis and prognosis of HCC. Overall, our review illustrates the potential application of circulating miRNAs, either as a single marker or as a panel of circulating miRNAs or in combination with conventional markers, as biomarkers for HCC diagnosis and prognosis predictions.
Encouraging progress in the use of circulating miRNAs in HCC diagnosis and prognosis has been made, and circulating miRNAs are extremely promising biomarkers. However, there are no clinically used miRNAs biomarkers for HCC, and further investigation is required to overcome some of the challenges in their application. First, the origin of circulating miRNAs has not been fully elucidated. Circulating miRNAs in serum/plasma have a heterogeneous origin, including from blood cells, endothelia cells, and other high blood-flow organs, which demonstrates that the expression of tumor-specific miRNA signatures may be masked by circulating miRNAs from other origins[19,81-83]. Another issue is that well-standardized protocols are not implemented for pre-analytical decisions of circulating miRNAs (e.g., sample storage, sample processing, the profiling method) to post-analytical processing (e.g., data normalization), which makes definitive comparisons between studies difficult[43,84]. Unfortunately, there is no widely accepted endogenous normalization control for circulating miRNAs, despite the use of qRT-PCR, which is the most commonly used technology for the detection of miRNAs in circulation. Furthermore, most miRNAs studies use small sample sizes, limited number of screened miRNAs, fail to differentiate HCC from hepatitis B and C, and lack large-scale prospective studies[9,20,74]. Moreover, much work is also required to compare values of circulating protein-bound miRNAs and microparticle miRNAs in different HCC patients. Finally, it is very important to use appropriate controls that are well matched in age, race, gender, etiology and severity of underlying liver disease when evaluating the sensitivity and specificity of circulating miRNAs for the diagnosis and monitoring of HCC[20,85,86].
Research on circulating miRNAs is in its infancy, and there are some challenges in the clinical application of circulating miRNAs. However, an improved understanding of the origins, stability, detection methods, and roles of circulating miRNAs in HCC will support the great potential of circulating miRNAs to become diagnostic and prognostic tools for HCC in the future.
P- Reviewer: Grassi G, Tomizawa M S- Editor: Ma YJ L- Editor: Wang TQ E- Editor: Wang CH
| 1. | Maluccio M, Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J Clin. 2012;62:394-399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 577] [Cited by in RCA: 698] [Article Influence: 53.7] [Reference Citation Analysis (0)] |
| 2. | Zhang Y, Wang Y, Du Z, Wang Q, Wu M, Wang X, Wang L, Cao L, Hamid AS, Zhang G. Recombinant human decorin suppresses liver HepG2 carcinoma cells by p21 upregulation. Onco Targets Ther. 2012;5:143-152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 3. | Borel F, Konstantinova P, Jansen PL. Diagnostic and therapeutic potential of miRNA signatures in patients with hepatocellular carcinoma. J Hepatol. 2012;56:1371-1383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 191] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 4. | Xu Y, Bu X, Dai C, Shang C. High serum microRNA-122 level is independently associated with higher overall survival rate in hepatocellular carcinoma patients. Tumour Biol. 2015;36:4773-4776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 5. | Hamid AS, Tesfamariam IG, Zhang Y, Zhang ZG. Aflatoxin B1-induced hepatocellular carcinoma in developing countries: Geographical distribution, mechanism of action and prevention. Oncol Lett. 2013;5:1087-1092. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 169] [Cited by in RCA: 207] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 6. | Zhang ZQ, Meng H, Wang N, Liang LN, Liu LN, Lu SM, Luan Y. Serum microRNA 143 and microRNA 215 as potential biomarkers for the diagnosis of chronic hepatitis and hepatocellular carcinoma. Diagn Pathol. 2014;9:135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 86] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 7. | Yang Z, Miao R, Li G, Wu Y, Robson SC, Yang X, Zhao Y, Zhao H, Zhong Y. Identification of Recurrence Related microRNAs in Hepatocellular Carcinoma after Surgical Resection. Int J Mol Sci. 2013;14:1105-1118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 8. | Kumamoto T, Tanaka K, Matsuo K, Takeda K, Nojiri K, Mori R, Taniguchi K, Matsuyama R, Ueda M, Akiyama H. Adjuvant hepatic arterial infusion chemotherapy with 5-Fluorouracil and interferon after curative resection of hepatocellular carcinoma: a preliminary report. Anticancer Res. 2013;33:5585-5590. [PubMed] |
| 9. | Yin W, Zhao Y, Ji YJ, Tong LP, Liu Y, He SX, Wang AQ. Serum/plasma microRNAs as biomarkers for HBV-related hepatocellular carcinoma in China. Biomed Res Int. 2015;2015:965185. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 10. | Scaggiante B, Kazemi M, Pozzato G, Dapas B, Farra R, Grassi M, Zanconati F, Grassi G. Novel hepatocellular carcinoma molecules with prognostic and therapeutic potentials. World J Gastroenterol. 2014;20:1268-1288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 58] [Cited by in RCA: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 11. | Murakami T, Kim T, Oi H, Nakamura H, Igarashi H, Matsushita M, Okamura J, Kozuka T. Detectability of hypervascular hepatocellular carcinoma by arterial phase images of MR and spiral CT. Acta Radiol. 1995;36:372-376. [PubMed] |
| 12. | Tan Y, Ge G, Pan T, Wen D, Chen L, Yu X, Zhou X, Gan J. A serum microRNA panel as potential biomarkers for hepatocellular carcinoma related with hepatitis B virus. PLoS One. 2014;9:e107986. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 134] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 13. | Volk ML, Hernandez JC, Su GL, Lok AS, Marrero JA. Risk factors for hepatocellular carcinoma may impair the performance of biomarkers: a comparison of AFP, DCP, and AFP-L3. Cancer Biomark. 2007;3:79-87. [PubMed] |
| 14. | Zinkin NT, Grall F, Bhaskar K, Otu HH, Spentzos D, Kalmowitz B, Wells M, Guerrero M, Asara JM, Libermann TA. Serum proteomics and biomarkers in hepatocellular carcinoma and chronic liver disease. Clin Cancer Res. 2008;14:470-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 152] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 15. | Oksuz Z, Serin MS, Kaplan E, Dogen A, Tezcan S, Aslan G, Emekdas G, Sezgin O, Altintas E, Tiftik EN. Serum microRNAs; miR-30c-5p, miR-223-3p, miR-302c-3p and miR-17-5p could be used as novel non-invasive biomarkers for HCV-positive cirrhosis and hepatocellular carcinoma. Mol Biol Rep. 2015;42:713-720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 74] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 16. | Zhu Z, Zhang X, Wang G, Zheng H. Role of MicroRNAs in Hepatocellular Carcinoma. Hepat Mon. 2014;14:e18672. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 17. | Fujita Y, Kuwano K, Ochiya T, Takeshita F. The impact of extracellular vesicle-encapsulated circulating microRNAs in lung cancer research. Biomed Res Int. 2014;2014:486413. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 18. | Roderburg C, Luedde T. Circulating microRNAs as markers of liver inflammation, fibrosis and cancer. J Hepatol. 2014;61:1434-1437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 92] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 19. | Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014;11:145-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 704] [Cited by in RCA: 829] [Article Influence: 75.4] [Reference Citation Analysis (0)] |
| 20. | Arrese M, Eguchi A, Feldstein AE. Circulating microRNAs: emerging biomarkers of liver disease. Semin Liver Dis. 2015;35:43-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 71] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 21. | Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513-10518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5636] [Cited by in RCA: 6307] [Article Influence: 371.0] [Reference Citation Analysis (0)] |
| 22. | Bala S, Petrasek J, Mundkur S, Catalano D, Levin I, Ward J, Alao H, Kodys K, Szabo G. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology. 2012;56:1946-1957. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 480] [Cited by in RCA: 525] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 23. | Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597-610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3165] [Cited by in RCA: 3671] [Article Influence: 244.7] [Reference Citation Analysis (0)] |
| 24. | Tan YL, Chen WN. MicroRNAs as therapeutic strategy for hepatitis B virus-associated hepatocellular carcinoma: current status and future prospects. World J Gastroenterol. 2014;20:5973-5986. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 11] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 25. | Chu R, Mo G, Duan Z, Huang M, Chang J, Li X, Liu P. miRNAs affect the development of hepatocellular carcinoma via dysregulation of their biogenesis and expression. Cell Commun Signal. 2014;12:45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 26. | Piontek K, Selaru FM. MicroRNAs in the biology and diagnosis of cholangiocarcinoma. Semin Liver Dis. 2015;35:55-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 27. | Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci USA. 2007;104:17719-17724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 377] [Cited by in RCA: 397] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 28. | Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1843] [Cited by in RCA: 1966] [Article Influence: 93.6] [Reference Citation Analysis (0)] |
| 29. | Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3513] [Cited by in RCA: 3629] [Article Influence: 165.0] [Reference Citation Analysis (0)] |
| 30. | Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1918] [Cited by in RCA: 1887] [Article Influence: 89.9] [Reference Citation Analysis (0)] |
| 31. | Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, Yoneda Y, Tsukihara T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 2009;326:1275-1279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 334] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 32. | Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2080] [Cited by in RCA: 2013] [Article Influence: 80.5] [Reference Citation Analysis (0)] |
| 33. | Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3476] [Cited by in RCA: 3274] [Article Influence: 136.4] [Reference Citation Analysis (0)] |
| 34. | Kalla R, Ventham NT, Kennedy NA, Quintana JF, Nimmo ER, Buck AH, Satsangi J. MicroRNAs: new players in IBD. Gut. 2015;64:504-517. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 188] [Cited by in RCA: 207] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 35. | Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631-640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1109] [Cited by in RCA: 1107] [Article Influence: 55.4] [Reference Citation Analysis (0)] |
| 36. | Andersen HH, Duroux M, Gazerani P. MicroRNAs as modulators and biomarkers of inflammatory and neuropathic pain conditions. Neurobiol Dis. 2014;71:159-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 136] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 37. | Ambros V. The functions of animal microRNAs. Nature. 2004;431:350-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7919] [Cited by in RCA: 8596] [Article Influence: 409.3] [Reference Citation Analysis (0)] |
| 38. | Giordano S, Columbano A. MicroRNAs: new tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology. 2013;57:840-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 263] [Cited by in RCA: 297] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 39. | Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 802] [Cited by in RCA: 911] [Article Influence: 70.1] [Reference Citation Analysis (0)] |
| 40. | D’Anzeo M, Faloppi L, Scartozzi M, Giampieri R, Bianconi M, Del Prete M, Silvestris N, Cascinu S. The role of micro-RNAs in hepatocellular carcinoma: from molecular biology to treatment. Molecules. 2014;19:6393-6406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 41. | Scaggiante B, Dapas B, Farra R, Grassi M, Pozzato G, Giansante C, Fiotti N, Grassi G. Improving siRNA bio-distribution and minimizing side effects. Curr Drug Metab. 2011;12:11-23. [PubMed] |
| 42. | Zen K, Zhang CY. Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev. 2012;32:326-348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 337] [Cited by in RCA: 367] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 43. | Lindner K, Haier J, Wang Z, Watson DI, Hussey DJ, Hummel R. Circulating microRNAs: emerging biomarkers for diagnosis and prognosis in patients with gastrointestinal cancers. Clin Sci (Lond). 2015;128:1-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 44. | Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442-17452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1382] [Cited by in RCA: 1588] [Article Influence: 105.9] [Reference Citation Analysis (0)] |
| 45. | Rayner KJ, Hennessy EJ. Extracellular communication via microRNA: lipid particles have a new message. J Lipid Res. 2013;54:1174-1181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 127] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 46. | Ma R, Jiang T, Kang X. Circulating microRNAs in cancer: origin, function and application. J Exp Clin Cancer Res. 2012;31:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 143] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 47. | Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110:483-495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 722] [Cited by in RCA: 789] [Article Influence: 60.7] [Reference Citation Analysis (0)] |
| 48. | Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1009] [Cited by in RCA: 1128] [Article Influence: 80.6] [Reference Citation Analysis (0)] |
| 49. | Lemoinne S, Thabut D, Housset C, Moreau R, Valla D, Boulanger CM, Rautou PE. The emerging roles of microvesicles in liver diseases. Nat Rev Gastroenterol Hepatol. 2014;11:350-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 138] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 50. | Dragovic RA, Gardiner C, Brooks AS, Tannetta DS, Ferguson DJ, Hole P, Carr B, Redman CW, Harris AL, Dobson PJ. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine. 2011;7:780-788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 929] [Cited by in RCA: 973] [Article Influence: 69.5] [Reference Citation Analysis (0)] |
| 51. | Rani S, O’Brien K, Kelleher FC, Corcoran C, Germano S, Radomski MW, Crown J, O’Driscoll L. Isolation of exosomes for subsequent mRNA, MicroRNA, and protein profiling. Methods Mol Biol. 2011;784:181-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 83] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 52. | Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003-5008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2345] [Cited by in RCA: 2635] [Article Influence: 188.2] [Reference Citation Analysis (0)] |
| 53. | Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423-433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2292] [Cited by in RCA: 2185] [Article Influence: 156.1] [Reference Citation Analysis (0)] |
| 54. | Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, Sun F, Lu J, Yin Y, Cai X. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. 2010;39:133-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 891] [Cited by in RCA: 964] [Article Influence: 64.3] [Reference Citation Analysis (0)] |
| 55. | Nolte-’t Hoen EN, Buermans HP, Waasdorp M, Stoorvogel W, Wauben MH, ‘t Hoen PA. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012;40:9272-9285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 481] [Cited by in RCA: 583] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 56. | Jaiswal R, Luk F, Gong J, Mathys JM, Grau GE, Bebawy M. Microparticle conferred microRNA profiles--implications in the transfer and dominance of cancer traits. Mol Cancer. 2012;11:37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 86] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 57. | Li H, Huang S, Guo C, Guan H, Xiong C. Cell-free seminal mRNA and microRNA exist in different forms. PLoS One. 2012;7:e34566. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 65] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 58. | Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733-1741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1810] [Cited by in RCA: 2083] [Article Influence: 138.9] [Reference Citation Analysis (0)] |
| 59. | Wang WT, Chen YQ. Circulating miRNAs in cancer: from detection to therapy. J Hematol Oncol. 2014;7:86. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 104] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 60. | Khoury S, Tran N. Circulating microRNAs: potential biomarkers for common malignancies. Biomark Med. 2015;9:131-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 60] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 61. | Fabbri M, Paone A, Calore F, Galli R, Croce CM. A new role for microRNAs, as ligands of Toll-like receptors. RNA Biol. 2013;10:169-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 115] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 62. | Hergenreider E, Heydt S, Tréguer K, Boettger T, Horrevoets AJ, Zeiher AM, Scheffer MP, Frangakis AS, Yin X, Mayr M. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat Cell Biol. 2012;14:249-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 960] [Cited by in RCA: 1067] [Article Influence: 82.1] [Reference Citation Analysis (0)] |
| 63. | Chen WX, Cai YQ, Lv MM, Chen L, Zhong SL, Ma TF, Zhao JH, Tang JH. Exosomes from docetaxel-resistant breast cancer cells alter chemosensitivity by delivering microRNAs. Tumour Biol. 2014;35:9649-9659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 118] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 64. | Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672-675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1258] [Cited by in RCA: 1332] [Article Influence: 78.4] [Reference Citation Analysis (0)] |
| 65. | Chang J, Guo JT, Jiang D, Guo H, Taylor JM, Block TM. Liver-specific microRNA miR-122 enhances the replication of hepatitis C virus in nonhepatic cells. J Virol. 2008;82:8215-8223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 168] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 66. | Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, Huang Y, Chen HC, Lee CH, Tsai TF. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884-2897. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 612] [Cited by in RCA: 669] [Article Influence: 51.5] [Reference Citation Analysis (0)] |
| 67. | Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, Huang L, Li H, Tan W, Wang C. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011;50:136-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 410] [Cited by in RCA: 450] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 68. | Qi P, Cheng SQ, Wang H, Li N, Chen YF, Gao CF. Serum microRNAs as biomarkers for hepatocellular carcinoma in Chinese patients with chronic hepatitis B virus infection. PLoS One. 2011;6:e28486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 212] [Cited by in RCA: 241] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 69. | Tomimaru Y, Eguchi H, Nagano H, Wada H, Kobayashi S, Marubashi S, Tanemura M, Tomokuni A, Takemasa I, Umeshita K. Circulating microRNA-21 as a novel biomarker for hepatocellular carcinoma. J Hepatol. 2012;56:167-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 285] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 70. | Ji F, Yang B, Peng X, Ding H, You H, Tien P. Circulating microRNAs in hepatitis B virus-infected patients. J Viral Hepat. 2011;18:e242-e251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 122] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 71. | Bihrer V, Friedrich-Rust M, Kronenberger B, Forestier N, Haupenthal J, Shi Y, Peveling-Oberhag J, Radeke HH, Sarrazin C, Herrmann E. Serum miR-122 as a biomarker of necroinflammation in patients with chronic hepatitis C virus infection. Am J Gastroenterol. 2011;106:1663-1669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 160] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 72. | Cermelli S, Ruggieri A, Marrero JA, Ioannou GN, Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One. 2011;6:e23937. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 423] [Cited by in RCA: 466] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 73. | Zhang Y, Jia Y, Zheng R, Guo Y, Wang Y, Guo H, Fei M, Sun S. Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clin Chem. 2010;56:1830-1838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 307] [Cited by in RCA: 323] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 74. | Jiang L, Cheng Q, Zhang BH, Zhang MZ. Circulating microRNAs as biomarkers in hepatocellular carcinoma screening: a validation set from China. Medicine (Baltimore). 2015;94:e603. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 75. | Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, Wang JF, Zhang Z, Lu S, Huang X. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol. 2011;29:4781-4788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 430] [Cited by in RCA: 497] [Article Influence: 35.5] [Reference Citation Analysis (0)] |
| 76. | Wang L, Yao M, Dong Z, Zhang Y, Yao D. Circulating specific biomarkers in diagnosis of hepatocellular carcinoma and its metastasis monitoring. Tumour Biol. 2014;35:9-20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 77. | Schütte K, Schulz C, Link A, Malfertheiner P. Current biomarkers for hepatocellular carcinoma: Surveillance, diagnosis and prediction of prognosis. World J Hepatol. 2015;7:139-149. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 64] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 78. | Qu KZ, Zhang K, Li H, Afdhal NH, Albitar M. Circulating microRNAs as biomarkers for hepatocellular carcinoma. J Clin Gastroenterol. 2011;45:355-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 190] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 79. | Köberle V, Kronenberger B, Pleli T, Trojan J, Imelmann E, Peveling-Oberhag J, Welker MW, Elhendawy M, Zeuzem S, Piiper A. Serum microRNA-1 and microRNA-122 are prognostic markers in patients with hepatocellular carcinoma. Eur J Cancer. 2013;49:3442-3449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 148] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 80. | Li J, Wang Y, Yu W, Chen J, Luo J. Expression of serum miR-221 in human hepatocellular carcinoma and its prognostic significance. Biochem Biophys Res Commun. 2011;406:70-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 168] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 81. | Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223-7233. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1356] [Cited by in RCA: 1520] [Article Influence: 108.6] [Reference Citation Analysis (0)] |
| 82. | Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, Tait JF, Tewari M. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (Phila). 2012;5:492-497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 691] [Cited by in RCA: 722] [Article Influence: 55.5] [Reference Citation Analysis (0)] |
| 83. | Williams Z, Ben-Dov IZ, Elias R, Mihailovic A, Brown M, Rosenwaks Z, Tuschl T. Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proc Natl Acad Sci USA. 2013;110:4255-4260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 284] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 84. | Grasedieck S, Sorrentino A, Langer C, Buske C, Döhner H, Mertens D, Kuchenbauer F. Circulating microRNAs in hematological diseases: principles, challenges, and perspectives. Blood. 2013;121:4977-4984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 110] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 85. | Berger F, Reiser MF. Micro-RNAs as potential new molecular biomarkers in oncology: have they reached relevance for the clinical imaging sciences? Theranostics. 2013;3:943-952. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 86. | Qi J, Wang J, Katayama H, Sen S, Liu SM. Circulating microRNAs (cmiRNAs) as novel potential biomarkers for hepatocellular carcinoma. Neoplasma. 2013;60:135-142. [PubMed] |
| 87. | Yin J, Hou P, Wu Z, Wang T, Nie Y. Circulating miR-375 and miR-199a-3p as potential biomarkers for the diagnosis of hepatocellular carcinoma. Tumour Biol. 2015;36:4501-4507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 57] [Article Influence: 5.7] [Reference Citation Analysis (0)] |