Published online Jul 21, 2015. doi: 10.3748/wjg.v21.i27.8398
Peer-review started: November 26, 2014
First decision: January 8, 2015
Revised: February 13, 2015
Accepted: April 28, 2015
Article in press: April 28, 2015
Published online: July 21, 2015
Processing time: 238 Days and 23.9 Hours
AIM: To assess the influence of SLIT and NTRK-like family member 3 (SLITRK3) on the prognosis of gastrointestinal stromal tumor (GIST) and determine whether SLITRK3 can help improve current risk stratification systems.
METHODS: We hypothesized that SLITRK3 could be used as a prognostic molecular biomarker for GIST. 35 fresh tumor samples and 417 paraffin-embedded specimens from GIST patients were utilized. SLITRK3 mRNA expression in GIST tumor tissue was detected by real-time polymerase chain reaction, and SLITRK3 protein levels were estimated by immunohistochemistry. The correlation of SLITRK3 expression with various tumor clinicopathological characteristics and follow-up data were analyzed.
RESULTS: GIST tumors had high expression of SLITRK3 compared with adjacent normal tissues and the expression level gradually increased with risk grade. SLITRK3 protein expression was closely associated with gastrointestinal bleeding, tumor site, tumor size, mitotic index, and National Institutes of Health (NIH) classification. Survival analysis showed that SLITRK3 expression was closely correlated with overall survival and disease-free survival of GIST patients. Multivariate analysis also identified SLITRK3 expression, mitotic index, and NIH stage as significant risk factors of GIST recurrence.
CONCLUSION: SLITRK3 expression is a highly significant predictor of GIST recurrence and metastasis. Combinations of SLITRK3 and NIH stage have strong predictive and prognostic value, and are feasible markers for clinical practice in gastrointestinal stromal tumor.
Core tip: Prognostic biomarkers are required to refine risk stratification treatment strategies for gastrointestinal stromal tumor (GIST). In this study, we hypothesized that SLIT and NTRK-like family member 3 (SLITRK3) could be used as a prognostic molecular biomarker for GIST. The results indicated that SLITRK3 expression is a highly significant predictor of GIST recurrence and metastasis. Combinations of SLITRK3 and NIH stage have strong predictive and prognostic value, and are feasible markers for clinical practice in gastrointestinal stromal tumor.
- Citation: Wang CJ, Zhang ZZ, Xu J, Wang M, Zhao WY, Tu L, Zhuang C, Liu Q, Shen YY, Cao H, Zhang ZG. SLITRK3 expression correlation to gastrointestinal stromal tumor risk rating and prognosis. World J Gastroenterol 2015; 21(27): 8398-8407
- URL: https://www.wjgnet.com/1007-9327/full/v21/i27/8398.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i27.8398
Gastrointestinal stromal tumors (GISTs) are the most common non-epithelial tumors[1] and the most common type of gastrointestinal cancer following gastric and colorectal cancer[1-4]. Surgery is the primary treatment option, but patients suffer from high-rates of tumor recurrence or metastasis, resulting in death. The majority of GISTs result from activating mutations in c-KIT and alpha-type platelet-derived growth factor receptor (PDGFRA)[5]. Recent studies have shown that adjuvant therapy with imatinib, a small molecule tyrosine kinase inhibitor, can prolong both survival and time to metastasis following surgery[6]. However, most micro-GISTs (less than 1 cm in diameter) have little malignancy potential despite the presence of KIT or PDGFRA mutations[7]. Furthermore, a 2002 risk assessment for aggressive GISTs showed that tumor growth rates can be affected by numerous factors[8]. Together, this demonstrates the need for additional prognostic molecular biomarkers to better characterize tumor prognosis and guide treatment strategy.
Secretion of transmitters and hormones is regarded as a hallmark of neuroendocrine cells and tumors. Synaptic-like microvesicle proteins, such as amphiphysin, synaptic vesicle protein, SV2, and synapsin 1, are found in a majority of GISTs[9]. Expression of these proteins enables GISTs to secrete neurotransmitters or hormones, suggesting that GISTs adopt a neuroendocrine phenotype. SLITRK3 is one of the six isoforms of SLIT and neurotropic tyrosine receptor kinase (NTRK)-like family member (Slitrk1-6), which are neuronal transmembrane proteins that control neurite growth[10]. Recently, SLITRK3 has been identified as a post-synaptic adhesion molecule that selectively regulates inhibitory synapse development and is important for normal functional GABAergic synapse development[11]. GABA is a key inhibitory neurotransmitter and mediates synaptic transmission, neural network development[12], and is involved in digestive diseases such as esophageal reflux and gastric cancer[13-15]. GISTs may originate from the interstitial cells of Cajal (ICCs), with pacemaker potentials suggesting that mutations in genes involved in synapse or neural development may underlie GIST behavior[9]. In agreement with this, we have previously found that the expression of SLITRK3 was increased in a high-risk group compared to a low-risk group (unpublished data), and Milde et al[16] showed higher SLITRK3 expression levels in lymphoma.
The aim of this study was to assess the influence of SLITRK3 on the prognosis of GIST and determine whether SLITRK3 can help improve current risk stratification systems. We hypothesized that up-regulation of SLITRK3 is strongly-associated with high recurrence risk and poor prognosis in GIST patients. We tested this by using qRT-PCR and immunohistochemistry on GIST samples and examining the relationship to patient outcome.
Formalin-fixed paraffin-embedded (FFPE) tissue sections were collected from GIST patients who underwent surgery at Renji Hospital, Shanghai Jiaotong University School of Medicine, China from 2004 to 2012. The inclusion criteria for this study were as follows: (1) primary GIST cases with definite pathologic diagnosis, as previously described[17]; (2) all cases received surgical resection; and (3) no reoccurrence or metastasis was detected. The exclusion criteria were: (1) chemotherapy, radiotherapy, or other anti-tumor therapy before surgery; and (2) incomplete clinicopathologic data. A total of 417 tumor tissue samples, with tumor adjacent normal tissue available for 139, were collected.
All cases were divided into four groups according to the risk table published by the National Institutes of Health (NIH) (Table 1)[8]. Tissue microarray and immunohistochemical staining were performed to access the expression levels of SLITRK3 in these samples. The follow-up data, including survival, reoccurrence, and metastasis as re-examination results, were obtained from outpatient medical records or from patients and their relatives by telephone interview using a follow-up questionnaire.
| Risk level | Tumor size (cm) | Mitotic count (50/HPF) | Primary tumor location |
| Very low | ≤ 2.0 | ≤ 5 | Any |
| Low | 2.1-5.0 | ≤ 5 | Any |
| Medium | 2.1-5.0 | > 5 | Stomach |
| < 5.0 | 6-10 | Any | |
| 5.1-10.0 | ≤ 5 | Stomach | |
| High | Any | Any | Tumor rupture |
| > 10.0 | Any | Any | |
| Any | > 10 | Any | |
| > 5.0 | > 5 | Any | |
| 2.1-5.0 | > 5 | Non stomach | |
| 5.1-10.0 | ≤ 5 | Non stomach |
Additionally, 35 fresh frozen GIST specimens were obtained between 2010 and 2012 from GIST patients who received surgical resection at Renji Hospital, Shanghai Jiaotong University School of Medicine, China. The samples were used for qRT-PCR detection of SLITRK3 expression.
Tissue microarrays were constructed by Suzhou Xinxin Biotechnology Co., Ltd (Xinxin Biotechnology Co, Suzhou, China). First, 139 GIST tissues with paired tumor adjacent normal tissues were used to construct 3 microarrays, while the other 278 GIST tissues were used to construct another 4 microarrays. Tissue paraffin blocks of GIST samples were stained with hematoxylin-eosin to confirm the diagnoses, and were marked at fixed points with most typical histological characteristics under a microscope. Two 1.6 mm cores per donor block were transferred into a recipient block tissue microarray, with each dot array containing fewer than 160 dots. Three-micron-thick sections were cut from the recipient block and transferred to glass slides with an adhesive tape transfer system for ultraviolet cross linkage.
The slides were baked at 56 °C for 1 h, de-paraffinized in xylene for 20 min, and rehydrated through a graded series of ethanol concentrations (5 min in 100% ethanol followed by 5 min in 70% ethanol). Antigen retrieval was performed in a pressure cooker for 10 min with 0.01 mol/L sodium citrate buffer (pH 6.0). Endogenous peroxidase activity was quenched with 0.3% hydrogen peroxide in methanol at 37 °C for 30 min. Next, an SLITRK3 antibody (NBP1-93619, Novus Biologicals, Colorado, United States; concentration: 1:100) was applied to cover the specimens overnight at 4 °C, which was followed by incubation with a labeled polymer-HRP anti-rabbit secondary antibody (Dako, CA, United States) for 30 min at room temperature. Staining was detected with diaminobenzidine (Thermo, MA, United States) as chromogen and counterstained with hematoxylin prior to coverslipping. The staining intensity and percentage of positive cells were recorded by two pathologists of Renji Hospital (Liu Q and Shen YY) and a consensus score was obtained for each slide. Immunohistochemical scoring was categorized as follows: (1) staining intensity was scored from 0 to 3: 0 for no staining, 1 for weak staining, 2 for moderate staining, and 3 for strong staining; (2) staining area was graded into 0-3 levels: 0 for no staining area, 1 for extent to less than 1/3, 3 for more than 2/3, and 2 for in-between; and (3) immunohistochemical classification was based on the sum of intensity and extent score: 0 as negative (-), 1-2 as weakly positive (+), 3-4 as positive (++), and 5-6 as strongly positive (+++). We further ranked the protein level into two classes as (-) or (+) for SLITRK3 low expression while (++) or (+++) was for SLITRK3 high expression.
Total RNA was extracted from fresh frozen GIST specimens and GIST cells using RNAiso Plus (Takara, Dalian, China) according to the manufacturer’s instructions. RNA quantity and quality were measured by NanoDrop 2000 (NanoDrop, DE, United States). RNA integrity was assessed by standard denaturing agarose gel electrophoresis. Reverse-transcription reactions were performed with Prime Script® RT Master Mix (Takara, Dalian, China) according to the manufacturer’s instructions. First-strand cDNA was synthesized from 2 μg of total RNA.
All qRT-PCR primer sequences were obtained from the Primer Bank database (http://pga.mgh.harvard.edu/primerbank/) (Table 2). Relative quantification of cDNA samples were measured by the SYBR-Green method in a final volume of 20 μL with Power SYBR® Green PCR Master Mix (Applied Biosystems, NY, USA) according to the manufacturer’s instructions. All reactions were performed on ABI ViiA™ 7 Real-Time PCR System (Applied Biosystems, NY, United States) in triplicate and the results were analyzed by ViiA™ 7 software. The 2-△Ct method was used to quantify the relative gene expression levels and 18S was used for normalization.
| Gene name | Primer | Sequence (5'-3') | Tm (°C) | Amplicon size (bp) |
| SLITRK3 | Forward | TTCCATAGCTGAGATGCTTCACA | 61.4 | 87 |
| Reverse | GGAATCGGGGTAGTCCATCC | 61.2 | ||
| 18S | Forward | GTAACCCGTTGAACCCCATT | 60.4 | 151 |
| Reverse | CCATCCAATCGGTAGTAGCG | 61.7 |
For comparisons, one-way analyses of variance, Wilcoxon signed rank test, and chi-squared tests were performed where appropriate. Kaplan-Meier curves were used to visualize biomarker expression, and NIH risk stage with respect to overall survival (OS) and disease-free survival (DFS) data. Univariate and multivariate analyses were based on the Cox proportional hazards regression model. Only those factors statistically significant (P < 0.05) in univariate analysis had access to the next multivariate analyses. Statistical analyses were all performed using SPSS 19.0 software (Chicago, IL, United States). All statistical tests were 2-sided, and P-value differences < 0.05 were considered statistically significant. All statistical analysis was reviewed and confirmed by Zhi-Gang Zhang.
Detailed clinicopathological data is shown in Table 3. Of the 417 paraffin-embedded GIST tissue samples, the predominant cell types were spindle cell (n = 271; 65.0%), epithelioid cell (n = 54; 12.9%), and mixed (n = 92; 22.1%). The maximum tumor diameter detected in GIST patients ranged from 0.5 to 30 cm (median: 5.5 cm). Risk stratification was performed according to the NIH risk classification, and suggested that there were 33 (7.9%) very low-risk cases, 154 (36.9%) low-risk cases, 67 (16.1%) intermediate-risk cases, and 163 (39.1%) high-risk cases.
| Clinicopathological factors | n (%) | |
| Gender | Male | 226 (54.2) |
| Female | 191 (45.8) | |
| Age (yr) | ≤ 60 | 223 (53.5) |
| > 60 | 194 (46.5) | |
| Median | 60 | |
| Gastrointestinal bleeding | No | 315 (75.5) |
| Yes | 102 (24.5) | |
| Primary tumor site | Stomach | 229 (54.9) |
| Small bowel | 123 (29.5) | |
| Colon | 21 (5.0) | |
| Others | 44 (10.6) | |
| Predominant cell type | Spindle | 271 (65.0) |
| Epithelioid | 54 (12.9) | |
| Mixed | 92 (22.1) | |
| Primary tumor size (cm) | 0-5 | 202 (48.4) |
| 5.1-10 | 138 (33.1) | |
| > 10 | 77 (18.5) | |
| Median | 5.5 | |
| Mitotic index (per 50 HPFs) | 0-5 | 309 (74.1) |
| 6-10 | 60 (14.4) | |
| > 10 | 48 (11.5) | |
| NIH stage | Very low risk | 33 (7.9) |
| Low risk | 154 (36.5) | |
| Intermediate risk | 67 (16.1) | |
| High risk | 163 (39.1) | |
| Recurrence | No | 331 (79.4) |
| Yes | 71 (17.0) | |
| Insufficient data | 15 (3.6) | |
| Death from illness | No | 376 (90.2) |
| Yes | 26 (6.2) | |
| Insufficient data | 15 (3.6) |
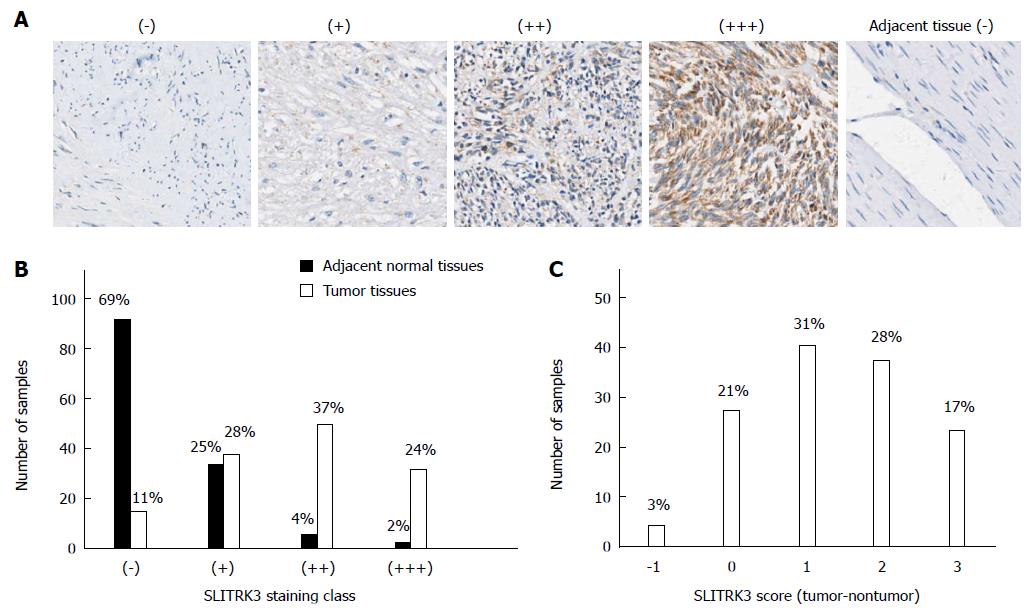
We performed immunohistochemistry in 139 GIST tissue samples which had both tumor (T) and adjacent non-tumor (N) tissue to determine if expression levels of SLITRK3 differed between tumor and non-tumor tissue. The results showed that SLITRK3 protein was expressed at different levels in different tissue samples and was divided into four classes, as described in materials and methods (Figure 1A). Most of the adjacent non-tumor tissues were (-) or (+), while most tumor samples ranged from (+) to (+++) (Figure 1B and Table 4), indicating higher SLITRK3 protein levels in tumor samples. The difference between tumor and paired adjacent normal tissues (T-N), ranging from -1 to 3 [(-) for 0 and (+++) for 3], revealed that SLITRK3 expression was increased in 76.3% (100/131) of GIST tumors where T-N > 0 (Figure 1C). Wilcoxon signed rank test further confirmed that GIST tumors have a significantly higher expression of SLITRK3 protein than adjacent normal tissue tumor samples (P < 0.001).
| Tissue | SLITRK3 level | |||
| - | + | ++ | +++ | |
| Tumor | 14 (11) | 37 (28) | 49 (37) | 31 (24) |
| Non-tumor tissue | 91 (69) | 33 (25) | 5 (4) | 2 (2) |
In order to better understand the significance of SLITRK3 expression in GIST tumor tissues, we expanded the tissue microarray sample size to 417 cases (4 cases were off-chip and not included in the statistics). Among the 413 GIST tumor tissues, SLITRK3 staining was strongly positive (+++) in 85 cases (20.6%), positive (++) in 142 cases (34.4%), weakly positive (+) in 112 cases (27.1%), and negative (-) in 74 cases (17.9%). We then ranked the protein level into two classes: (-) or (+) for low expression and (++) or (+++) for high expression, in order to further investigate the relationship between SLITRK3 and clinicopathological factors in GIST. Chi-square test revealed that the SLITRK3 protein level was not associated with gender, age, or predominant cell type, but was closely related with gastrointestinal bleeding, primary tumor site, primary tumor size, mitotic index, and NIH classification (Table 5).
| Clinicopathological factors1 | SLITRK3 expression | Number of patients | χ2 | P value2 | ||
| Low | High | |||||
| Gender | Male | 100 | 123 | 224 | 0.001 | 1.000 |
| Female | 85 | 104 | 189 | |||
| Age (yr) | ≤ 60 | 99 | 121 | 220 | 0.000 | 1.000 |
| > 60 | 87 | 106 | 193 | |||
| Gastrointestinal bleeding | No | 153 | 160 | 313 | 7.722 | 0.006 |
| Yes | 33 | 67 | 100 | |||
| Primary tumor site | Stomach | 120 | 106 | 226 | 24.730 | < 0.0013 |
| Small bowel | 34 | 88 | 122 | |||
| Colon | 14 | 7 | 21 | |||
| Others | 18 | 26 | 44 | |||
| Predominant cell type | Spindle | 126 | 144 | 270 | 2.552 | 0.279 |
| Epithelioid | 26 | 27 | 53 | |||
| Mixed | 34 | 56 | 90 | |||
| Primary tumor size | ≤ 5 cm | 116 | 83 | 199 | 27.260 | < 0.0013 |
| > 5 cm | 70 | 144 | 214 | |||
| Mitotic index | ≤ 5/50 HPF | 157 | 149 | 306 | 18.763 | < 0.0013 |
| > 5/50 HPF | 29 | 78 | 107 | |||
| NIH stage | Very low risk | 28 | 3 | 31 | 53.340 | < 0.0013 |
| Low risk | 87 | 67 | 154 | |||
| Intermediate risk | 25 | 41 | 66 | |||
| High risk | 46 | 116 | 162 | |||
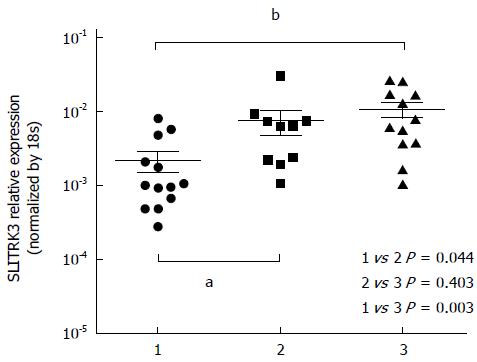
To further confirm SLITRK3 expression in GIST, the relative expression levels of SLITRK3 mRNAs were analyzed by qRT-PCR in 35 fresh GISTs samples. The relative expression level of SLITRK3 mRNA in the low risk group (n = 13), intermediate risk group (n = 10), and high risk group (n = 12) were 0.002 ± 0.002, 0.008 ± 0.009, and 0.011 ± 0.009, respectively, indicating a gradually increasing trend. SLITRK3 expression in GIST tumor tissues of the intermediate and high risk groups were significantly higher than those of the low-risk group (P = 0.003 and P = 0.044) (Figure 2).
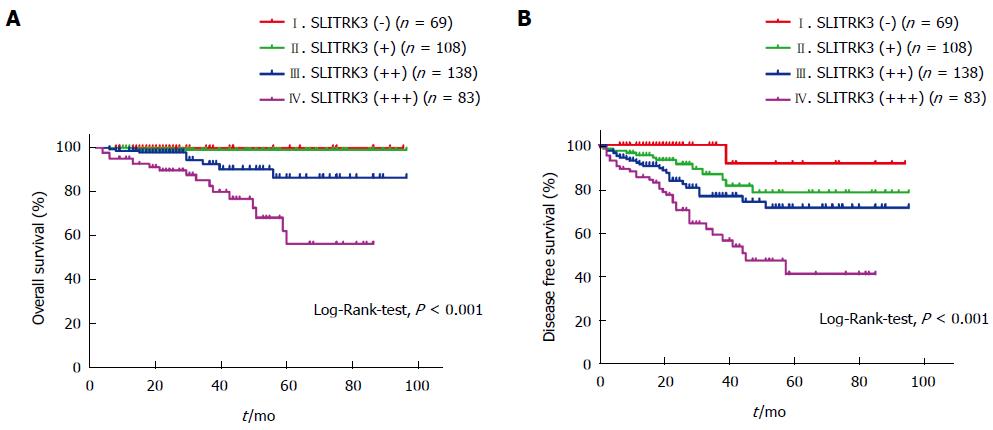
The relationship between SLITRK3 expression and overall survival (OS) or disease-free survival (DFS) in GIST patients was investigated. All 398 cases with complete follow-up data were classified into four classes according to SLITRK3 expression levels and calculated according to the Kaplan-Meier method. The 5-year OS rates decreased successively from 100% in (-), 99.0% in (+), 86.7% in (++), and 57.4% in (+++) (Log-Rank-test, P < 0.001) (Figure 3A). Meanwhile, the 5-year DFS rates also decreased successively form 91.7% in (-), 78.6% in (+), 71.6% in (++), and 41.8% in (+++) (Log-Rank-test, P < 0.001) (Figure 3B).
To further investigate whether SLITRK3 can be used as an independent predictor associated with poor prognosis in GIST, univariate and multivariate analysis were performed. Univariate analysis revealed that primary tumor size, mitotic index, NIH stage, and SLITRK3 expression were significantly associated with OS (Table 6). Univariate analysis revealed that primary tumor site, primary tumor size, mitotic index, NIH stage, and SLITRK3 expression were significantly associated with DFS (Table 7).
| Factor | 5-yr OS rate (%) | OS HR (95%CI) | P value | |
| Gender | Male: Female | 81.3: 89.2 | 0.508 (0.221-1.169) | 0.111 |
| Age (yr) | ≤ 60: > 60 | 88.9: 80.0 | 2.186 (0.989-4.832) | 0.053 |
| Gastrointestinal bleeding | No: Yes | 86.8: 81.2 | 1.259 (0.559-2.837) | 0.578 |
| Primary tumor site | Gastric: Non-gastric | 83.0: 86.8 | 1.143 (0.529-2.470) | 0.733 |
| Predominant cell type | Spindle: Epithelioid: Mixed | 87.7: 82.4: 88.8 | 0.935 (0.586-1.491) | 0.778 |
| Primary tumor size (cm) | ≤ 5: > 5 | 99.2: 74.0 | 22.726 (3.079-167.742) | 0.0021 |
| Mitotic index (HPFs) | ≤ 5/50: 6-10/50: > 10/50 | 96.5: 83.0: 39.7 | 4.727 (2.887-7.740) | < 0.0011 |
| NIH stage | Very low: Low: Mid: High | 100.0: 100.0: 88.6: 70.3 | 8.005 (2.365-27.098) | 0.0011 |
| SLITRK3 expression | (-): (+): (++): (+++) | 100.0: 99.0: 86.7: 57.4 | 4.164 (2.227-7.786) | < 0.0011 |
| Factor | 5-yr DFS rate (%) | DFS HR (95%CI) | P value | |
| Gender | Male: Female | 69.0: 69.7 | 0.643 (0.398-1.038) | 0.071 |
| Age (yr) | ≤ 60: > 60 | 72.1: 65.6 | 1.285 (0.806-2.049) | 0.292 |
| Gastrointestinal bleeding | No: Yes | 71.6: 65.7 | 1.425 (0.874-2.322) | 0.155 |
| Primary tumor site | Gastric: Non-gastric | 81.5: 59.4 | 2.669 (1.623-4.388) | 0.0011 |
| Predominant cell type | Spindle: Epithelioid: Mixed | 72.0: 67.1: 71.3 | 0.877 (0.661-1.164) | 0.365 |
| Primary tumor size (cm) | ≤ 5: > 5 | 92.2: 50.8 | 8.183 (3.921-17.079) | < 0.0011 |
| Mitotic index (HPFs) | ≤ 5/50: 6-10/50: > 10/50 | 84.2: 50.7: 19.0 | 3.289 (2.545-4.253) | < 0.0011 |
| NIH stage | Very low: Low: Mid: High | 100.0: 93.3: 87.1: 40.0 | 5.421 (3.249-9.045) | < 0.0011 |
| SLITRK3 expression | (-): (+): (++): (+++) | 91.7: 78.6: 71.6: 41.8 | 2.082 (1.575-2.753) | < 0.0011 |
Only those factors with statistically significant relationships with DFS in the univariate analysis were entered in the Cox’s proportional-hazard model for multivariate analysis (Table 8). The NIH stage is based on primary tumor site, tumor size, and mitotic index, and correlated with each of them strongly, so we developed 3 models for multivariate analysis. Analysis included NIH stage without SLITRK3 expression in model A, SLITRK3 expression instead of NIH stage in model B, and both in model C. In model A, primary tumor site, mitotic index, and NIH stage were statistically significant indicators of poor DFS, and model B showed that SLITRK3 expression is also a significant indicator of poor DFS. Importantly, in model C, SLITRK3 expression (but not NIH stage) was an independent risk factor for GIST recurrence.
| Factor | Model A | Model B | Model C | ||||
| DFS HR(95%CI) | P value | DFS HR(95%CI) | P value | DFS HR(95%CI) | P value | ||
| Primary tumor site | Gastric: Non-gastric | 1.762 (1.036-2.999) | 0.0371 | 2.046 (1.232-3.401) | 0.0062 | 1.593 (0.928-2.733) | 0.091 |
| Primary tumor size (cm) | ≤ 5: > 5 | 1.388 (0.567-3.398) | 0.473 | 3.370 (1.545-7.351) | 0.0022 | 1.183 (0.477-2.934) | 0.717 |
| Mitotic index (HPFs) | ≤ 5/50: 6-10/50: > 10/50 | 2.027 (1.494-2.749) | < 0.0012 | 2.549 (1.930-3.366) | < 0.0012 | 2.032 (1.496-2.760) | < 0.0012 |
| NIH stage | Very low: Low: Mid: High | 2.753 (1.426-5.314) | 0.0032 | 2.707 (1.387-5.283) | 0.0042 | ||
| SLITRK3 expression | (-): (+): (++): (+++) | 1.508 (1.151-1.976) | 0.0032 | 1.465 (1.114-1.928) | 0.0062 | ||
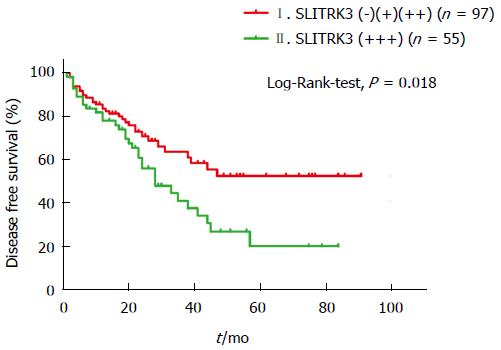
In order to find out whether SLITRK3 can help improve the NIH stage, we further investigated a subgroup of 152 high risk cases. The Kaplan-Meier analysis revealed that the 5-year DFS rate in the SLITRK3 (+++) group was significantly lower than the others (20.3% vs 52.7%, Log-Rank-test, P = 0.018) (Figure 4).
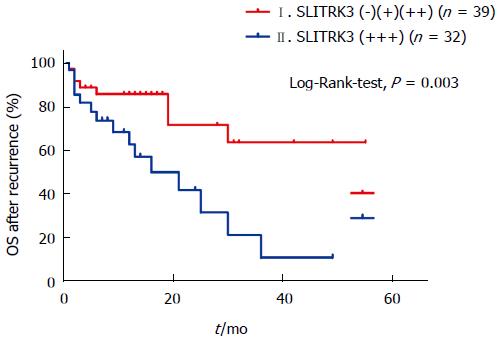
Furthermore, we found that 71 patients suffered from disease recurrence, mostly with an original high NIH risk rating. Kaplan-Meier curve analysis based on SLITRK3 expression of these patients showed that 1-year and 3-year OS rates in the SLITRK3 (-), (+), (++) group was 83.3% and 61.7%, respectively; the SLITRK3 (+++) group was 58.5% and 9.7%, respectively (Log-Rank-test, P =0.003) (Figure 5).
In the current study, we examined the correlation between SLITRK3 and GLIST behavior and survival. We found that SLITKR3 was expressed more highly in tumor tissue, correlated well with clinicopathological features, and predicted poor survival in patients. Most importantly, increasing SLITKR3 expression correlated with decreased overall survival and disease-free survival.
Currently, risk stratification schemes for operable GIST, such as the NIH consensus criteria, modified consensus criteria, and AFIP-Miettinen criteria, all depend on tumor site, tumor size, and mitosis index[8,18-20]. Mitosis count is one of the most valuable prognostic factors in GIST, but has limitations and controversial reliability[21]. Observation of mitosis can be subjective, time consuming, and affected by the high power field (HPF) area of the microscope and tissue fixation time. A previous study using 16 different pathologists and different microscopes resulted in a wide counting range from the same sample[22]. Moreover, according to current risk stratification, abrupt changes can occur in estimating risk of recurrence when the tumor size or mitosis index is close to a cutoff value. This is especially important due to the existence of small and mitotically inactive malignant GISTs[1,23,24]. Together, these factors suggest that the current risk criteria can be improved significantly.
We found that the clinical measurements in our study, including age, sex, tumor size, and mitoses index, were all similar to previously reported studies[21,25-27]. Patients who suffered from disease recurrence or death were mainly from the high-risk group. The 5-year OS and DFS rate of our database were 85.0% and 69.4%, respectively, compared with 72.3% and 70.5%, respectively, from a large multicenter, retrospective analysis of clinical data published in Lancet Oncology 2012[21]. The better 5-year OS rate of our study might reflect differences in standardized treatment and the use of IM adjuvant therapy.
SLITRK3 is expressed predominantly in neural tissues and has neurite-modulating activity[28]. However, the function of SLITRK3 in solid tumors is poorly studied. In our previous study (unpublished data), the expression of SLITRK3 monotonically increased from the low-risk group to the high-risk group. We found that SLITRK3 was also up-regulated in tumor compared to non-tumor tissue by using immunohistochemistry and qRT-PCR. This finding is in agreement with Milde et al[16], who demonstrated that SLITRK3 was up-regulated in lymphoma. SLITRK3 expression was also associated with a higher incidence of GI bleeding, a common symptom of GIST and a good indicator for high-risk patients[29,30]. Furthermore, SLITRK3 expression correlated with NIH risk classification, and the reduced overall survival and disease-free progression suggests that elevated expression of SLITRK3 is a good tumor biomarker candidate, particularly for aggressive GISTs.
The function and mechanism of SLITRK3 protein in the malignant processes of GIST it still unclear, necessitating the need for experiments both in vitro and in vivo. However, it is very likely that there are other potential GIST risk-related genes, as suggested by our previous gene microarray (unpublished data). In agreement with this, the potassium channel tetramerization domain containing protein 10 (KCTD10) has been shown to correlate with GIST prognosis[31]. Future studies and identification of novel prognostic biomarkers will help further stratify risk groups and direct treatment strategies for GIST.
Our detailed analysis showed that SLITRK3 mRNA expression level increased according to NIH risk classification. We found that SLIRTK3 protein level was closely associated with tumor site, tumor size, and mitotic index. As current risk stratification schemes are mainly based on these three features, it is not surprising that up-regulation of SLIRTK3 is strongly associated with a high-risk NIH grade. However, we found that NIH stage was a significant unfavorable factor for OS in univariate analysis, but not multivariate analysis. This suggests that the current NIH criteria is very likely not an optimal prognostic tool. We found that under all circumstances that SLITRK3 expression was a significant predictor of poor prognosis. Therefore, we believe that the combination of SLITRK3 expression and NIH criteria will better stratify post-operative GIST patients. Many patients with operable GIST can be cured by surgery alone and may not benefit from IM adjuvant therapy. Given the expense of IM adjuvant therapy and its associated side effects[32], an improved selection of patients for adjuvant therapy will be of clinical benefit. Due to the poor prognosis and reduced OS, we strongly suggest that patients with high SLITRK3 expression, especially those who also are in the NIH high-risk groups, receive IM adjuvant therapy and close follow-up management after surgery.
In conclusion, we have identified SLITRK3 as a novel prognostic molecular biomarker that may help guide treatment of GIST.
Recent studies have shown that adjuvant therapy with imatinib, a small molecule tyrosine kinase inhibitor, can prolong both survival and time to metastasis following surgery. However, most micro-gastro intestinal stromal tumors (GIST) (less than 1 cm in diameter) have little malignancy potential, despite the presence of a KIT or PDGFRA mutation. Furthermore, a 2002 risk assessment for aggressive GISTs showed that tumor growth rates can be affected by numerous factors.
In agreement with this, the authors have previously found that the expression of SLIT and NTRK-like family member 3 (SLITRK3) was increased in a high-risk group when compared to a low-risk group (unpublished data), and Milde et al showed higher SLITRK3 expression levels in lymphoma.
The authors hypothesized that up-regulation of SLITRK3 is strongly associated with high recurrence risk and poor prognosis in GIST patients. We tested this by using qRT-PCR and immunohistochemistry on GIST samples and examining the relationship to patient outcome.
The authors have identified SLITRK3 as a novel prognostic molecular biomarker that may help guide treatment of GIST.
In this study, the authors aimed to assess the influence of SLITRK3 on the prognosis of GIST, determine whether different degrees of SLITRK3 expression were significantly associated with overall survival, and assess whether this can help improve current risk stratification systems according to tables published by the National Institutes of Health for GIST.
P- Reviewer: Aseni P, Lu XF, Tang HH, Yang CH S- Editor: Qi Y L- Editor: Rutherford A E- Editor: Liu XM
| 1. | Nilsson B, Bümming P, Meis-Kindblom JM, Odén A, Dortok A, Gustavsson B, Sablinska K, Kindblom LG. Gastrointestinal stromal tumors: the incidence, prevalence, clinical course, and prognostication in the preimatinib mesylate era--a population-based study in western Sweden. Cancer. 2005;103:821-829. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 803] [Cited by in RCA: 875] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 2. | Tryggvason G, Gíslason HG, Magnússon MK, Jónasson JG. Gastrointestinal stromal tumors in Iceland, 1990-2003: the icelandic GIST study, a population-based incidence and pathologic risk stratification study. Int J Cancer. 2005;117:289-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 247] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 3. | Goettsch WG, Bos SD, Breekveldt-Postma N, Casparie M, Herings RM, Hogendoorn PC. Incidence of gastrointestinal stromal tumours is underestimated: results of a nation-wide study. Eur J Cancer. 2005;41:2868-2872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 207] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 4. | Rubió J, Marcos-Gragera R, Ortiz MR, Miró J, Vilardell L, Gironès J, Hernandez-Yagüe X, Codina-Cazador A, Bernadó L, Izquierdo A. Population-based incidence and survival of gastrointestinal stromal tumours (GIST) in Girona, Spain. Eur J Cancer. 2007;43:144-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 56] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 5. | Hirota S. Gastrointestinal stromal tumors: their origin and cause. Int J Clin Oncol. 2001;6:1-5. [PubMed] |
| 6. | Corless CL, Ballman KV, Antonescu CR, Kolesnikova V, Maki RG, Pisters PW, Blackstein ME, Blanke CD, Demetri GD, Heinrich MC. Pathologic and molecular features correlate with long-term outcome after adjuvant therapy of resected primary GI stromal tumor: the ACOSOG Z9001 trial. J Clin Oncol. 2014;32:1563-1570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 221] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 7. | Demetri GD, Benjamin RS, Blanke CD, Blay JY, Casali P, Choi H, Corless CL, Debiec-Rychter M, DeMatteo RP, Ettinger DS. NCCN Task Force report: management of patients with gastrointestinal stromal tumor (GIST)--update of the NCCN clinical practice guidelines. J Natl Compr Canc Netw. 2007;5 Suppl 2:S1-S29; quiz S30. [PubMed] |
| 8. | Fletcher CD, Berman JJ, Corless C, Gorstein F, Lasota J, Longley BJ, Miettinen M, O’Leary TJ, Remotti H, Rubin BP. Diagnosis of gastrointestinal stromal tumors: A consensus approach. Hum Pathol. 2002;33:459-465. [PubMed] |
| 9. | Bümming P, Nilsson O, Ahlman H, Welbencer A, Andersson MK, Sjölund K, Nilsson B. Gastrointestinal stromal tumors regularly express synaptic vesicle proteins: evidence of a neuroendocrine phenotype. Endocr Relat Cancer. 2007;14:853-863. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 10. | Aruga J, Mikoshiba K. Identification and characterization of Slitrk, a novel neuronal transmembrane protein family controlling neurite outgrowth. Mol Cell Neurosci. 2003;24:117-129. [PubMed] |
| 11. | Takahashi H, Katayama K, Sohya K, Miyamoto H, Prasad T, Matsumoto Y, Ota M, Yasuda H, Tsumoto T, Aruga J. Selective control of inhibitory synapse development by Slitrk3-PTPδ trans-synaptic interaction. Nat Neurosci. 2012;15:389-398, S1-S2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 155] [Cited by in RCA: 198] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 12. | Akerman CJ, Cline HT. Refining the roles of GABAergic signaling during neural circuit formation. Trends Neurosci. 2007;30:382-389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 125] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 13. | Herman MA, Alayan A, Sahibzada N, Bayer B, Verbalis J, Dretchen KL, Gillis RA. micro-Opioid receptor stimulation in the medial subnucleus of the tractus solitarius inhibits gastric tone and motility by reducing local GABA activity. Am J Physiol Gastrointest Liver Physiol. 2010;299:G494-G506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 14. | Ren LH, Chen WX, Qian LJ, Li S, Gu M, Shi RH. Addition of prokinetics to PPI therapy in gastroesophageal reflux disease: a meta-analysis. World J Gastroenterol. 2014;20:2412-2419. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 76] [Cited by in RCA: 65] [Article Influence: 5.9] [Reference Citation Analysis (1)] |
| 15. | Matuszek M, Jesipowicz M, Kleinrok Z. GABA content and GAD activity in gastric cancer. Med Sci Monit. 2001;7:377-381. [PubMed] |
| 16. | Milde T, Shmelkov SV, Jensen KK, Zlotchenko G, Petit I, Rafii S. A novel family of slitrk genes is expressed on hematopoietic stem cells and leukemias. Leukemia. 2007;21:824-827. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 17. | Poveda A, del Muro XG, López-Guerrero JA, Martínez V, Romero I, Valverde C, Cubedo R, Martín-Broto J. GEIS 2013 guidelines for gastrointestinal sarcomas (GIST). Cancer Chemother Pharmacol. 2014;74:883-898. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 18. | Miettinen M, Lasota J. Gastrointestinal stromal tumors: pathology and prognosis at different sites. Semin Diagn Pathol. 2006;23:70-83. [PubMed] |
| 19. | Joensuu H. Risk stratification of patients diagnosed with gastrointestinal stromal tumor. Hum Pathol. 2008;39:1411-1419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 699] [Cited by in RCA: 865] [Article Influence: 50.9] [Reference Citation Analysis (0)] |
| 20. | Gold JS, Gönen M, Gutiérrez A, Broto JM, García-del-Muro X, Smyrk TC, Maki RG, Singer S, Brennan MF, Antonescu CR. Development and validation of a prognostic nomogram for recurrence-free survival after complete surgical resection of localised primary gastrointestinal stromal tumour: a retrospective analysis. Lancet Oncol. 2009;10:1045-1052. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 373] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 21. | Joensuu H, Vehtari A, Riihimäki J, Nishida T, Steigen SE, Brabec P, Plank L, Nilsson B, Cirilli C, Braconi C. Risk of recurrence of gastrointestinal stromal tumour after surgery: an analysis of pooled population-based cohorts. Lancet Oncol. 2012;13:265-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 576] [Cited by in RCA: 671] [Article Influence: 47.9] [Reference Citation Analysis (0)] |
| 22. | Gal R, Rath-Wolfson L, Rosenblatt Y, Halpern M, Schwartz A, Koren R. An improved technique for mitosis counting. Int J Surg Pathol. 2005;13:161-165. [PubMed] |
| 23. | Tanaka J, Oshima T, Hori K, Tomita T, Kim Y, Watari J, Oh K, Hirota S, Matsumoto T, Miwa H. Small gastrointestinal stromal tumor of the stomach showing rapid growth and early metastasis to the liver. Dig Endosc. 2010;22:354-356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 24. | Miettinen M, El-Rifai W, H L Sobin L, Lasota J. Evaluation of malignancy and prognosis of gastrointestinal stromal tumors: a review. Hum Pathol. 2002;33:478-483. [PubMed] |
| 25. | Rosa F, Alfieri S, Tortorelli AP, Di Miceli D, Papa V, Ricci R, Doglietto GB. Gastrointestinal stromal tumors: prognostic factors and therapeutic implications. Tumori. 2012;98:351-356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 26. | Linhares E, Gonçalves R, Valadão M, Vilhena B, Herchenhorn D, Romano S, Ferreira MA, Ferreira CG, Ramos Cde A, de Jesus JP. Gastrointestinal stromal tumor: analysis of 146 cases of the center of reference of the National Cancer Institute--INCA. Rev Col Bras Cir. 2011;38:398-406. [PubMed] |
| 27. | Maor Y, Avidan B, Melzer E, Bar-Meir S. Long-term clinical outcome of patients with gastric gastrointestinal stromal tumors. Dig Dis Sci. 2010;55:2893-2898. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 28. | Aruga J, Yokota N, Mikoshiba K. Human SLITRK family genes: genomic organization and expression profiling in normal brain and brain tumor tissue. Gene. 2003;315:87-94. [PubMed] |
| 29. | Miettinen M, Sobin LH, Lasota J. Gastrointestinal stromal tumors of the stomach: a clinicopathologic, immunohistochemical, and molecular genetic study of 1765 cases with long-term follow-up. Am J Surg Pathol. 2005;29:52-68. [PubMed] |
| 30. | Lv A, Li Z, Tian X, Guan X, Zhao M, Dong B, Hao C. SKP2 high expression, KIT exon 11 deletions, and gastrointestinal bleeding as predictors of poor prognosis in primary gastrointestinal stromal tumors. PLoS One. 2013;8:e62951. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 31. | Kubota D, Yoshida A, Tsuda H, Suehara Y, Okubo T, Saito T, Orita H, Sato K, Taguchi T, Yao T. Gene expression network analysis of ETV1 reveals KCTD10 as a novel prognostic biomarker in gastrointestinal stromal tumor. PLoS One. 2013;8:e73896. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 32. | Wu L, Zhang Z, Yao H, Liu K, Wen Y, Xiong L. Clinical efficacy of second-generation tyrosine kinase inhibitors in imatinib-resistant gastrointestinal stromal tumors: a meta-analysis of recent clinical trials. Drug Des Devel Ther. 2014;8:2061-2067. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |