Published online May 21, 2015. doi: 10.3748/wjg.v21.i19.5867
Peer-review started: December 16, 2014
First decision: January 8, 2015
Revised: January 15, 2015
Accepted: February 13, 2015
Article in press: February 13, 2015
Published online: May 21, 2015
Processing time: 155 Days and 6.9 Hours
AIM: To investigate the underlying molecular mechanisms of miR-451 to inhibit proliferation of esophageal carcinoma cell line EC9706.
METHODS: Assays for cell growth, apoptosis and invasion were used to evaluate the effects of miR-451 expression on EC cells. Luciferase reporter and Western blot assays were used to test whether cyclin-dependent kinase inhibitor 2D (CDKN2D) and MAP3K1 act as major targets of miR-451.
RESULTS: The results showed that CDKN2D and MAP3K1 are direct targets of miR-451. CDKN2D and MAP3K1 overexpression reversed the effect of miR-451. MiR-451 inhibited the proliferation of EC9706 by targeting CDKN2D and MAP3K1.
CONCLUSION: These findings suggest that miR-451 might be a novel prognostic biomarker and a potential target for the treatment of esophageal squamous cell carcinoma in the future.
Core tip: Recently miR-451 has been reported to be tumor suppressor in human cancer cells. In the previous studies we have reported that miR-451 expression in esophageal squamous cell carcinoma (ESCC) tissues was significantly reduced, and that upregulated expression of miR-451 induced apoptosis and suppressed cell proliferation, invasion and metastasis in esophageal carcinoma. However, the underlying molecular mechanisms remain unclear. In this study, we supposed and showed that cyclin-dependent kinase inhibitor 2D (CDKN2D) and MAP3K1 are the targets of miR-451 by the bioinformatics algorithms (TargetScan and miRBase). Moreover, we found that CDKN2D and MAP3K1 contributed to ESCC malignancy.
- Citation: Zang WQ, Yang X, Wang T, Wang YY, Du YW, Chen XN, Li M, Zhao GQ. MiR-451 inhibits proliferation of esophageal carcinoma cell line EC9706 by targeting CDKN2D and MAP3K1. World J Gastroenterol 2015; 21(19): 5867-5876
- URL: https://www.wjgnet.com/1007-9327/full/v21/i19/5867.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i19.5867
Esophageal squamous cell carcinoma (ESCC) is one of the most lethal malignancies worldwide[1,2]. ESCC is the 8th most common cancer and the 6th leading cause of cancer-related death. The traditional treatment for ESCC includes chemotherapy and radiation therapy[3,4]. However, many patients who are treated with such traditional therapy still experience disease progression, which suggests that ESCC is resistant to traditional therapy. New treatment choices are critically required and the mechanism of tumorigenesis is to be further clarified.
MicroRNAs (miRNAs) are small, endogenous noncoding RNAs that have been identified as post-transcriptional regulators of gene expression. MiRNAs exert their functions through imperfect base-pairing with the 3’-untranslated region (3’-UTR) of target mRNAs[5-8]. In human cancer, miRNAs can act as oncogenes or tumour suppressor genes during tumourigenesis. Recently, miR-451 has been reported to be induced during zebrafish, mouse, and human erythroid maturation as a key factor involved in regulating erythrocyte differentiation[9-11]. It was also reported that miR-451 might function as a tumor suppressor and modulate MDR1/P-glycoprotein expression in human cancer cells[12]. In previous studies we have reported that miR-451 expression in ESCC tissues was significantly reduced, and that upregulated expression of miR-451 induced apoptosis and suppressed cell proliferation, invasion and metastasis in esophageal carcinoma[13,14]. However, the underlying molecular mechanisms remain unclear. In this study, we supposed and showed that cyclin-dependent kinase inhibitor 2D (CDKN2D) and MAP3K1 are the targets of miR-451 by the bioinformatics algorithms (TargetScan and miRBase). Moreover, we found that CDKN2D and MAP3K1 contributed to ESCC malignancy. Our data demonstrate that miR-451 has potential values as a prognostic marker and a therapeutic target for ESCC.
EC9706 and KYSE150 cell lines were purchased from the Chinese Academy of Sciences Cell Bank. All cells were cultured in RPMI-1640 (Gibco, United States) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, United States) and grown in humidified 5% CO2 at 37 °C.
The miR-451 mimics used in this study was synthesized by Shanghai GenePharma Co. Ltd. For transfection, 2 × 105 cells were seeded into each well of six well plates and grown overnight until they were 50%-80% confluent. Cells were washed, placed in serum-free medium, and transfected using LipofectamineTM2000 according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, United States). After 6 h, the medium was changed to complete medium, and cells were cultured at 37 °C in 5% CO2.
The different experimental groups of EC9706 and KYSE150 cells were plated in 96-well plates at 1 × 104 cells per well and incubated for 48 h after transfection. The viability of cells was determined using Cell Counting Kit-8 (CCK-8; Dojindo, Japan) according to the manufacturer’s instructions. Viable cell numbers were estimated by measurement of optical density (OD) at 450 nm. All experiments were performed in triplicate.
Cells were suspended in RPMI-1640 containing 0.35% low melting agarose, and plated onto solidified 0.6% agarose containing RPMI-1640 in six-well culture plates at a density of 1 × 105 cells per dish. The plates were incubated for 2 wk at 37 °C in a 5% CO2 incubator, and the number of colonies was counted after staining with 0.1% crystal violet solution. All experiments were performed in triplicate.
The experimental groups of EC9706 cells were adjusted at 2 × 105/mL in each group 48 h after transfection. The upper chamber of 24-well Transwell Permeable Supports with 8 μm pores (Corning Cat. No. 3422) was loaded with 200 μL of cell suspension, and the lower chamber was loaded with 500 μL of medium containing 10% serum for incubation in an atmosphere of 5% CO2 at 37 °C for 48 h. Five wells were set for each group. The number of cells invading the matrigel was counted from 5 randomly selected visual fields using an inverted microscope. All experiments were performed in triplicate.
EC9706 cells were harvested 48 h after transfection and cell concentration was adjusted to 1 × 106 cells. Annexin V-FITC/PI Apoptosis Detetion Kit Ι (BestBio, Shanghai, China) was used to detect Annexin V. Results were obtained using FACScan Flow Cytometer (BD Biosciences, San Jose, CA, United States). Tests were repeated in triplicate. Data were analyzed with Cell Quest software. All experiments were performed in triplicate.
For cell cycle analysis by flow cytometry, cells in the logarithmic phase of growth were harvested by trypsinization, washed with PBS, fixed with 75% ethanol overnight at 4 °C and incubated with RNase at 37 °C for 30 min. Nuclei were stained with propidium iodide for 30 min. A total of 104 nuclei were examined in a FACSCalibur Flow Cytometer (BD Biosciences, San Jose, CA, United States). All experiments were performed in triplicate.
The experimental groups of EC9706 and KYSE150 cells in each group were lysed in lysis buffer for total protein extration. Protein concentrations were measured using the BCA method (KeyGEN, China), and 30 μg of protein was separated by 12% SDS-PAGE and electroblotted onto a nitrocellulose membrane (Whatman, United States). The membrane was blotted overnight at 4 °C with primary antibodies (mouse anti- CDKN2D and anti- MAP3K1, 1:1000) in Tris-buffered saline with 5% non-fat milk. A secondary antibody (HRP-conjugated goat anti-mouse IgG) was incubated with the membrane for 1 h after three washes with TBST. The protein band density was determined with Kodak Digital ID Image Analysis Software and was normalized with the density of β-actin. All experiments were performed in triplicate.
The human CDKN2D and MAP3K1 fragments containing putative binding sites for miR-451 were amplified by PCR from human genomic DNA. The mutant CDKN2D and MAP3K1 3’-UTRs were obtained by overlap extension PCR. The fragments were cloned into a pmirGLO reporter vector (Promega), downstream of the luciferase gene, to generate the recombinant vectors pmirGLO-CDKN2D-wt, pmirGLO-CDKN2D-mut, pmirGLO-MAP3K1-wt and pmirGLO-MAP3K1-mut. For the luciferase reporter assay, cells were transiently co-transfected with miRNA (miR-451 mimics or scrambled-miR-451 negative control) and reporter vectors (wild-type reporter vectors or mutant-type reporter vectors), using LipofectamineTM2000. Luciferase activities were measured using a Dual-Luciferase assay kit (Promega) according to the manufacturer’s instructions at 48 h post-transfection. All experiments were performed in triplicate.
Statistical testing was conducted with the assistance of SPSS 17.0 software. All data are expressed as mean ± SD. One-way analysis of variance (ANOVA) was used to analyze data. Results were considered significant when P values were < 0.05.
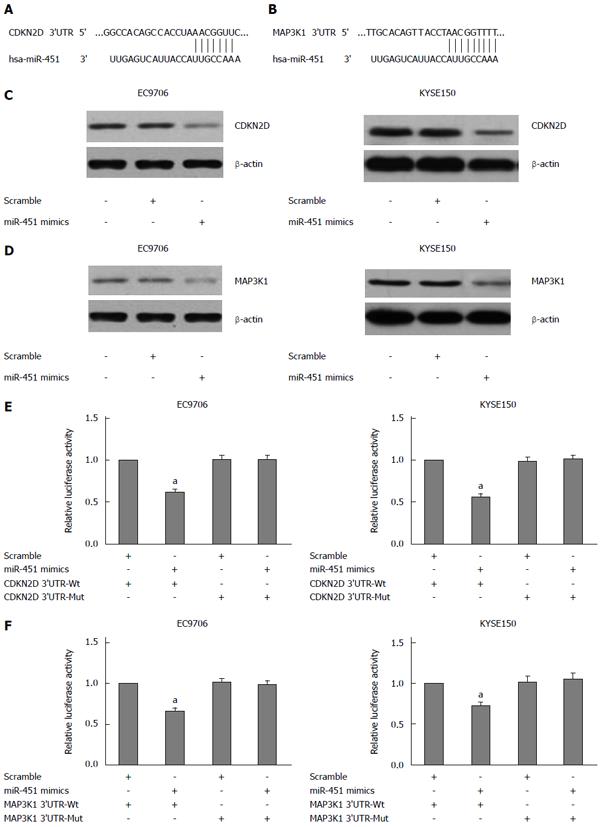
We based on the following criteria to search for the direct target of miR-451: the target should have oncogenic property and regulate the cell migration and invasion. Among these targets of miR-451 predicted by the bioinformatics algorithms (TargetScan and miRBase), we selected CDKN2D and MAP3K1. The 3’-UTR of CDKN2D contains the seed regions for miR-451 at the position of base 240 nt - 246 nt (Figure 1A). Similarly, the 3’-UTR of MAP3K1 contains the seed regions for miR-451 at the position of base 6270 nt - 6278 nt (Figure 1B).
Subsequent Western blot analysis indeed showed that CDKN2D and MAP3K1 expression was down-regulated in EC9706 and KYSE150 cells following transfection with the miR-451 mimics (Figure 1C and D). In order to test the specific regulation through the seed region, we constructed a reporter vector which consists of the luciferase coding sequence followed by the 3’-UTR of CDKN2D and MAP3K1. Wild type (pmirGLO-CDKN2D-3’-UTR, pmirGLO-MAP3K1-3’-UTR) or mutated sequences (pmirGLO-CDKN2D-mut 3’-UTR, pmirGLO-MAP3K1-mut 3’-UTR) within the seed region sites were cloned into the pmirGLO reporter vector. We used a Dual-Luciferase reporter system containing either wild-type or mutant 3’-UTRs of CDKN2D and MAP3K1, respectively. Co-transfection experiments showed that miR-451 significantly decreased the luciferase activity of wild type in EC9706 and KYSE150 cells (P < 0.05; Figure 1E and F), but this was not observed in mutant type (P > 0.05; Figure 1E and F). These data indicate that miR-451 negatively regulates CDKN2D and MAP3K1 expression by directly binding to putative binding sites in the 3’-UTR. Our results thus demonstrated that CDKN2D and MAP3K1 are direct targets of miR-451.
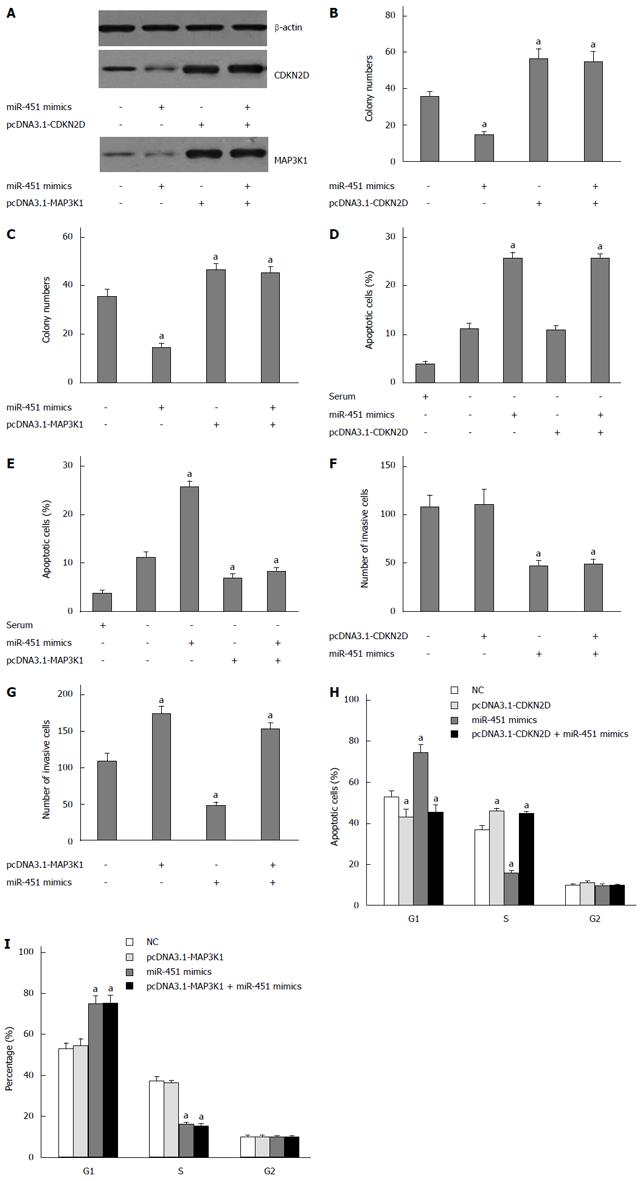
To explore the function of CDKN2D and MAP3K1 in EC9706 cells, we constructed pcDNA3.1-CDKN2D and pcDNA3.1-MAP3K1 lacking the 3’-UTR, and then they were transfected into EC9706 cells. Western blot assay showed that transfection of miR-451 mimics inhibited the expression of CDKN2D and MAP3K1, respectively (Figure 2A). Co-transfection of pcDNA3.1-CDKN2D and miR-451 abrogated the effects of miR-451 on CDKN2D expression (Figure 2A). Similarly, co-transfection of pcDNA3.1-MAP3K1 and miR-451 abrogated the effects of miR-451 on MAP3K1 expression (Figure 2A).
In the colony formation assays we found that exogenous expression of miR-451 decreased cell colony formation numbers (Figure 2B and C). Subsequently, we exogenously expressed recombinant CDKN2D lacking the 3’-UTR sequence (pcDNA3.1-CDKN2D) or MAP3K1 lacking the 3’-UTR sequence (pcDNA3.1-MAP3K1) in EC9706 cells. Cells transfected with pcDNA3.1-CDKN2D or pcDNA3.1-MAP3K1 alone showed significantly increased cell colony formation numbers (Figure 2B and C). When we, however, co-transfected cells with pcDNA3.1-CDKN2D or pcDNA3.1-MAP3K1 and miR-451, the expression of CDKN2D and pcDNA3.1-MAP3K1 lacking the 3’-UTR sequence were found to reverse the anti-proliferation of miR-451 (Figure 2B and C). From these results we conclude that expression of CDKN2D and MAP3K1 could partially reverse the anti-proliferation function of miR-451.
Our apoptosis assay indicated that exogenous expression of miR-451 increased cell apoptosis induced by serum starvation (Figure 2D and E). Subsequently, we exogenously expressed recombinant CDKN2D lacking the 3’-UTR sequence (pcDNA3.1-CDKN2D) or MAP3K1 lacking the 3’-UTR sequence (pcDNA3.1-MAP3K1) in EC9706 cells. Cells transfected with pcDNA3.1-CDKN2D alone did not show significantly decreased levels of apoptosis (Figure 2D). However, cells transfected with pcDNA3.1-MAP3K1 alone showed significantly decreased levels of apoptosis compared to the blank control (Figure 2E), and that when we co-transfected cells with pcDNA3.1-MAP3K1 and miR-451, the expression of MAP3K1 lacking the 3’-UTR sequence was found to reverse the pro-apoptotic functions of miR-451 (Figure 2E).
In the transwell assays we found that exogenous expression of miR-451 decreased cell invasiveness (Figure 2F and G). Subsequently, we exogenously expressed recombinant CDKN2D lacking the 3’-UTR sequence (pcDNA3.1-CDKN2D) or MAP3K1 lacking the 3’-UTR sequence (pcDNA3.1-MAP3K1) in EC9706 cells. Cells transfected with pcDNA3.1-MAP3K1 alone showed significantly increased cell invasiveness (Figure 2G). When we, however, co-transfected cells with pcDNA3.1-CDKN2D and miR-451, the expression of CDKN2D lacking the 3’-UTR sequence was found to reverse the anti-migration functions of miR-451 (Figure 2F). Similarly, when we co-transfected cells with pcDNA3.1-MAP3K1 and miR-451, the expression of MAP3K1 lacking the 3’-UTR sequence was found to reverse the anti- migration functions of miR-451 (Figure 2G).
Cell cycle analysis showed that administration of miR-451 mimic oligonucleotides significantly increased the percentage of cells in the G1 phase and decreased the percentage of cells in the S phase (Figure 2H and I). When we co-transfected cells with pcDNA3.1-CDKN2D and miR-451, the expression of CDKN2D lacking the 3’-UTR sequence was found to reverse G1 arrest of miR-451 (Figure 2H). When we co-transfected cells with pcDNA3.1-MAP3K1 and miR-451, the expression of MAP3K1 lacking the 3’-UTR sequence was not found to reverse G1 arrest of miR-451 (Figure 2I).
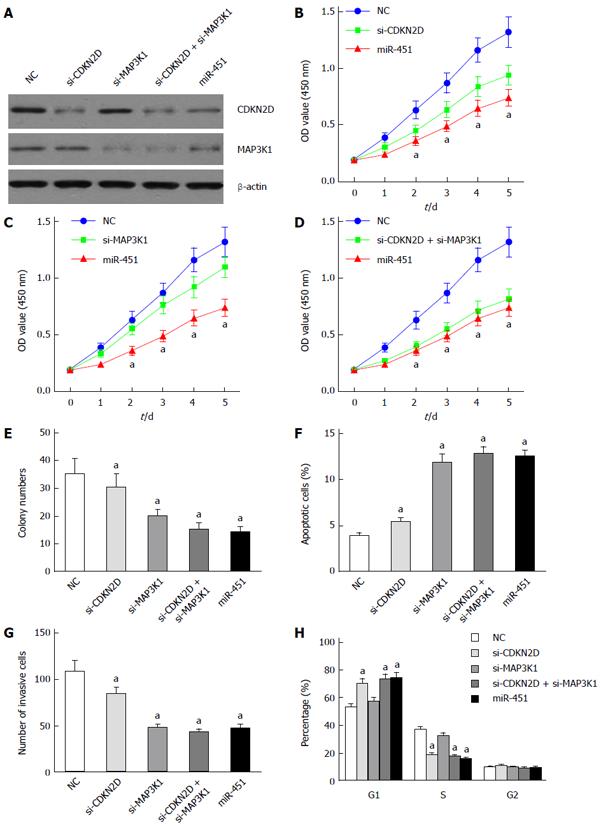
To further explore the biological significance of CDKN2D, MAP3K1 and miR-451 in EC9706 cells, CDKN2D-siRNAs, MAP3K1-siRNAs and miR-451 mimics were transfected into EC9706 cells. Western blot assay showed that transfection of CDKN2D-siRNAs, MAP3K1-siRNAs and miR-451 mimics inhibited the expression of CDKN2D and MAP3K1, respectively (Figure 3A).
CCK8 assay showed that CDKN2D, MAP3K1 silencing and miR-451 overexpression inhibited the cell proliferation (Figure 3B and C). For EC9706 cells transfected with si-CDKN2D, the inhibition was more obvious than cells transfected with si-MAP3K1 (Figure 3B and C). Compared to the NC group, co-transfection of si-CDKN2D and si-MAP3K1 also significantly inhibited the proliferation of EC9706 cells (Figure 3D). In addition, for co-transfected cells with si-CDKN2D and si-MAP3K1, the inhibitory effects were similar to those for cells overexpressing miR-451 (Figure 3D). Furthermore, colony formation assay obtained the similar results that CDKN2D, MAP3K1 silencing and miR-451 overexpression reduced EC9706 cell colony numbers (Figure 3E).
Invasion assay showed that knockdown of MAP3K1 and the overexpression of miR-451 repressed the invasion capacities of EC9706 cells (Figure 3G). For cells transfected with si-MAP3K1, the numbers of invasive cells were less than cells transfected with si-CDKN2D (Figure 3G). To investigate the effect of si-CDKN2D, si-MAP3K1 and miR-451 on apoptosis, we performed apoptosis assay. As showed in Figure 3F, CDKN2D, MAP3K1 silencing and the overexpression of miR-451 induced EC9706 cell apoptosis significantly compared to the blank control. For EC9706 cells transfected with si-MAP3K1, the apoptotic cells were more than those for cells transfected with si-CDKN2D. For co-transfected cells with si-CDKN2D and si-MAP3K1, the apoptosis effects were similar to those for cells overexpressing miR-451.
Cell cycle analysis showed that knockdown of CDKN2D and the overexpression of miR-451 significantly increased the percentage of cells in the G1 phase and decreased the percentage of cells in the S phase (Figure 3H). Altogether, these results confirm that miR-451 inhibits the proliferation of EC9706 by targeting CDKN2D and MAP3K1.
CDKN2D (p19INK4d), a negative regulator of the cell cycle, is located on chromosome 19p13. The protein encoded by this gene is a member of the INK4 family of cyclin-dependent kinase inhibitors. This protein has been shown to form a stable complex with CDK4 or CDK6, and prevent the activation of the CDK kinases, thus functioning as a cell growth regulator that controls cell cycle G1 progression. The abundance of the transcript of this gene was found to oscillate in a cell-cycle dependent manner with the lowest expression at mid G1 and a maximal expression during S phase. The negative regulation of the cell cycle involving this protein was shown to participate in repressing neuronal proliferation, as well as spermatogenesis[15-20]. Little is known of its role in cancer development and prognosis. CDKN2D expression in cancers has been examined in only a few studies and, to date, it has not been linked to cancer development.
Mitogen-activated protein kinases (MAPKs) are key mediators of evolutionarily conserved signaling networks that play an essential role in multiple aspects of cell physiology[21,22]. MAP3K1 or MEKK1 (MEK kinase 1) is a 196-kDa serine-threonine kinase that belongs to the MAP3K family and the STE superfamily[22,23]. MAP3K1 was originally identified as the mammalian homolog of the yeast MAP3Ks Ste11 and Byr2 that function in pheromone responsive signaling. Studies have demonstrated that MAP3K1 functions in cell survival, apoptosis, and cell motility/migration in multiple normal and tumor cell types[24,25].
In previous studies we have reported that miR-451 expression in ESCC tissues were significantly reduced, and that upregulated expression of miR-451 induced apoptosis and suppressed cell proliferation, invasion and metastasis in esophageal carcinoma[13,14]. In this study, we identified CDKN2D and MAP3K1 as the direct and functional targets of miR-451, which facilitated our understanding of the mechanisms underlying ESCC progression. Additionally, a further study indicated that CDKN2D and MAP3K1 overexpression reversed the effect of miR-451, and that miR-451 inhibited the proliferation of EC9706 by targeting CDKN2D and MAP3K1. The study demonstrates that miR-451 prefers to act as a potential target for the treatment of ESCC in the future.
MiRNAs have been shown to be important in the development and maintenance of normal cellular function, and an alteration in expression of miRNAs can result in human cancer initiation and tumor progression. MiRNAs can regulate target genes by increasing mRNA decay or by repressing translation. Each miRNA has the potential to target hundreds of genes that harbor in their 3’-UTR sequences complementary to the seed region of the miRNA[26-29]. In the study, for co-transfected cells with si-CDKN2D and si-MAP3K1, the inhibitory effects are similar to those for cells overexpressing miR-451.
However, miRNAs may function according to a combinatorial circuits model, in which a single miRNA may target multiple mRNAs, and several coexpressed miRNAs may target a single mRNA. Recent studies have suggested that the biological concept of “one hit-multiple targets” could be used in clinical therapeutics[30]. If the primary molecular defect of a disease is in the expression of a miRNA, the expression of several critical protein targets could be deregulated. In that case, one might recover the normal phenotype of the cells by normalizing the miRNA expression. Although individual targets responsible for observed phenotypes have been proposed for many miRNAs, it is likely that a specific miRNA may function through cooperative down-regulation of multiple targets. Thus, other target genes of miR-451 may also contribute to tumorigenesis.
In conclusion, we have identified that miR-451 inhibited the proliferation, invasion and induced the apoptosis of ESCC cells in vitro and in vivo by directly targeting CDKN2D and MAP3K1. MiR-451 might be a novel prognostic biomarker and a potential target for the treatment of ESCC in the future.
Esophageal squamous cell carcinoma (ESCC) is one of the most lethal malignancies worldwide. ESCC is the 8th most common cancer and the 6th leading cause of cancer-related death. The traditional treatments for ESCC include chemotherapy and radiation therapy. However, many patients who are treated with such traditional therapy still experience disease progression, which suggests that ESCC is resistant to traditional therapy. In human cancer, microRNAs (miRNAs) can act as oncogenes or tumour suppressor genes during tumourigenesis.
Recently miR-451 has been reported to be induced during zebrafish, mouse, and human erythroid maturation as a key factor involved in regulating erythrocyte differentiation. It was also reported that miR-451 might function as a tumor suppressor and modulate MDR1/P-glycoprotein expression in human cancer cells.
In previous studies the authors have reported that miR-451 expression in ESCC tissues were significantly reduced, and that upregulated expression of miR-451 induced apoptosis and suppressed cell proliferation, invasion and metastasis in esophageal carcinoma. However, the underlying molecular mechanisms remain unclear. In this study, the authors supposed and showed that CDKN2D and MAP3K1 are the targets of miR-451 by the bioinformatics algorithms (TargetScan and miRBase). Moreover, they found that CDKN2D and MAP3K1 contributed to ESCC malignancy.
The data suggest that miR-451 has potential values as a prognostic marker and a therapeutic target for ESCC.
MiRNAs are small, endogenous noncoding RNAs that have been identified as post-transcriptional regulators of gene expression. MiRNAs exert their functions through imperfect base-pairing with the 3’-untranslated region of target mRNAs.
The manuscript is basically good. This is an in vitro study that addressed the mechanism of tumor suppressive functions of a miRNA, miR-451. The authors authentically conducted the required experiments using esophageal cancer cells and revealed that miR-451 targeted CDKN2D and MAP3K1 and worked tumor-suppressively through the inhibition of the two kinases. The findings are expected to contribute to the development of molecularly targeted therapy for esophageal cancer.
P- Reviewer: Lee HC, Kanda T, Stanojevic GZ S- Editor: Qi Y L- Editor: Wang TQ E- Editor: Ma S
| 1. | Hongo M, Nagasaki Y, Shoji T. Epidemiology of esophageal cancer: Orient to Occident. Effects of chronology, geography and ethnicity. J Gastroenterol Hepatol. 2009;24:729-735. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 271] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 2. | Dawsey SP, Tonui S, Parker RK, Fitzwater JW, Dawsey SM, White RE, Abnet CC. Esophageal cancer in young people: a case series of 109 cases and review of the literature. PLoS One. 2010;5:e14080. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 66] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 3. | Baxi SH, Burmeister B, Harvey JA, Smithers M, Thomas J. Salvage definitive chemo-radiotherapy for locally recurrent oesophageal carcinoma after primary surgery: retrospective review. J Med Imaging Radiat Oncol. 2008;52:583-587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 4. | Matsushima K, Isomoto H, Kohno S, Nakao K. MicroRNAs and esophageal squamous cell carcinoma. Digestion. 2010;82:138-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 74] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 5. | Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857-866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5705] [Cited by in RCA: 6021] [Article Influence: 316.9] [Reference Citation Analysis (0)] |
| 6. | Esteller M. Cancer Epigenetics for the 21st Century: What’s Next? Genes Cancer. 2011;2:604-606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 46] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 7. | Chen WX, Ren LH, Shi RH. Implication of miRNAs for inflammatory bowel disease treatment: Systematic review. World J Gastrointest Pathophysiol. 2014;5:63-70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 71] [Cited by in RCA: 80] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 8. | Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152-D157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2680] [Cited by in RCA: 2783] [Article Influence: 185.5] [Reference Citation Analysis (0)] |
| 9. | Masaki S, Ohtsuka R, Abe Y, Muta K, Umemura T. Expression patterns of microRNAs 155 and 451 during normal human erythropoiesis. Biochem Biophys Res Commun. 2007;364:509-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 152] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 10. | Pase L, Layton JE, Kloosterman WP, Carradice D, Waterhouse PM, Lieschke GJ. miR-451 regulates zebrafish erythroid maturation in vivo via its target gata2. Blood. 2009;113:1794-1804. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 162] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 11. | Patrick DM, Zhang CC, Tao Y, Yao H, Qi X, Schwartz RJ, Jun-Shen Huang L, Olson EN. Defective erythroid differentiation in miR-451 mutant mice mediated by 14-3-3zeta. Genes Dev. 2010;24:1614-1619. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 147] [Cited by in RCA: 151] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 12. | Zhu H, Wu H, Liu X, Evans BR, Medina DJ, Liu CG, Yang JM. Role of MicroRNA miR-27a and miR-451 in the regulation of MDR1/P-glycoprotein expression in human cancer cells. Biochem Pharmacol. 2008;76:582-588. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 373] [Cited by in RCA: 365] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 13. | Wang T, Zang WQ, Li M, Wang N, Zheng YL, Zhao GQ. Effect of miR-451 on the biological behavior of the esophageal carcinoma cell line EC9706. Dig Dis Sci. 2013;58:706-714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 14. | Zang W, Wang Y, Du Y, Xuan X, Wang T, Li M, Ma Y, Li P, Chen X, Dong Z. Differential expression profiling of microRNAs and their potential involvement in esophageal squamous cell carcinoma. Tumor Biol. 2014;35:3295-3304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 15. | Roussel MF. The INK4 family of cell cycle inhibitors in cancer. Oncogene. 1999;18:5311-5317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 218] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 16. | Ortega S, Malumbres M, Barbacid M. Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochim Biophys Acta. 2002;1602:73-87. [PubMed] |
| 17. | Felisiak-Golabek A, Dansonka-Mieszkowska A, Rzepecka IK, Szafron L, Kwiatkowska E, Konopka B, Podgorska A, Rembiszewska A, Kupryjanczyk J. p19(INK4d) mRNA and protein expression as new prognostic factors in ovarian cancer patients. Cancer Biol Ther. 2013;14:973-981. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 18. | Gluick T, Yuan Z, Libutti SK, Marx SJ. Mutations in CDKN2C (p18) and CDKN2D (p19) may cause sporadic parathyroid adenoma. Endocr Relat Cancer. 2013;20:L27-L29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 19. | Bartkova J, Thullberg M, Rajpert-De Meyts E, Skakkebaek NE, Bartek J. Lack of p19INK4d in human testicular germ-cell tumours contrasts with high expression during normal spermatogenesis. Oncogene. 2000;19:4146-4150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 42] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 20. | Bartkova J, Rajpert-De Meyts E, Skakkebaek NE, Lukas J, Bartek J. Deregulation of the G1/S-phase control in human testicular germ cell tumours. APMIS. 2003;111:252-265; discussion 265-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 47] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 21. | Kyriakis JM, Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol Rev. 2001;81:807-869. [PubMed] |
| 22. | Uhlik MT, Abell AN, Cuevas BD, Nakamura K, Johnson GL. Wiring diagrams of MAPK regulation by MEKK1, 2, and 3. Biochem Cell Biol. 2004;82:658-663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 77] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 23. | Cuevas BD, Abell AN, Johnson GL. Role of mitogen-activated protein kinase kinase kinases in signal integration. Oncogene. 2007;26:3159-3171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 228] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 24. | Hagemann C, Blank JL. The ups and downs of MEK kinase interactions. Cell Signal. 2001;13:863-875. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 214] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 25. | Lange-Carter CA, Pleiman CM, Gardner AM, Blumer KJ, Johnson GL. A divergence in the MAP kinase regulatory network defined by MEK kinase and Raf. Science. 1993;260:315-319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 800] [Cited by in RCA: 853] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 26. | Pham TT, Angus SP, Johnson GL. MAP3K1: Genomic Alterations in Cancer and Function in Promoting Cell Survival or Apoptosis. Genes Cancer. 2013;4:419-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 100] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 27. | Ambros V. The functions of animal microRNAs. Nature. 2004;431:350-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7919] [Cited by in RCA: 8586] [Article Influence: 408.9] [Reference Citation Analysis (0)] |
| 28. | Engels BM, Hutvagner G. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene. 2006;25:6163-6169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 343] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 29. | Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769-773. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3585] [Cited by in RCA: 3739] [Article Influence: 187.0] [Reference Citation Analysis (0)] |
| 30. | Wurdinger T, Costa FF. Molecular therapy in the microRNA era. Pharmacogenomics J. 2007;7:297-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 58] [Article Influence: 3.1] [Reference Citation Analysis (0)] |