Published online Dec 28, 2014. doi: 10.3748/wjg.v20.i48.18177
Revised: June 21, 2014
Accepted: September 5, 2014
Published online: December 28, 2014
Processing time: 245 Days and 16 Hours
Inflammatory bowel disease (IBD) is a chronic inflammatory disease thought to be mediated by the microbiota of the intestinal lumen and inappropriate immune responses. Aberrant immune responses can cause secretion of harmful cytokines that destroy the epithelium of the gastrointestinal tract, leading to further inflammation. Interleukin (IL)-22 is a member of the IL-10 family of cytokines that was recently discovered to be mainly produced by both adaptive and innate immune cells. Several cytokines and many of the transcriptional factors and T regulatory cells are known to regulate IL-22 expression through activation of signal transducer and activator of transcription 3 signaling cascades. This cytokine induces antimicrobial molecules and proliferative and antiapoptotic pathways, which help prevent tissue damage and aid in its repair. All of these processes play a beneficial role in IBD by enhancing intestinal barrier integrity and epithelial innate immunity. In this review, we discuss recent progress in the involvement of IL-22 in the pathogenesis of IBD, as well as its therapeutic potential.
Core tip: Interleukin (IL)-22 is expressed by adaptive immune system cells and innate lymphocytes. Several cytokines and many transcriptional factors and T regulatory cells can regulate IL-22 expression. Through activation of signal transducer and activator of transcription 3 signaling cascades, IL-22 induces antimicrobial, proliferative and antiapoptotic pathways, which can help fix damaged tissue and promote tissue repair mechanisms. IL-22 is also associated with inflammatory bowel disease (IBD) susceptibility genes that regulate inflammatory responses in tissues. All of these processes play crucial roles in IBD pathogenesis and collectively provide an important rationale for the development of novel therapeutic measures for this disease.
- Citation: Li LJ, Gong C, Zhao MH, Feng BS. Role of interleukin-22 in inflammatory bowel disease. World J Gastroenterol 2014; 20(48): 18177-18188
- URL: https://www.wjgnet.com/1007-9327/full/v20/i48/18177.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i48.18177
Inflammatory bowel disease (IBD) is a group of inflammatory conditions of the small intestine and colon, and includes Crohn’s disease (CD) and ulcerative colitis (UC). Despite extensive research efforts, however, the etiology of IBD remains unclear. The current opinion about IBD pathogenesis is that the disease results from interactions between environmental factors, mainly microbes of the intestinal lumen and their products, and dysregulation of immune responses in genetically susceptible individuals[1]. Certain harsh environments that may affect barrier integrity (to increase barrier permeability to luminal macromolecular substances, such as protein antigens and microbial products) and over-absorption of luminal microbial products (which has been ascribed to a number of mucosal pathologies) can lead to an over-activation of immune system, thus resulting in mucosal inflammation[2].
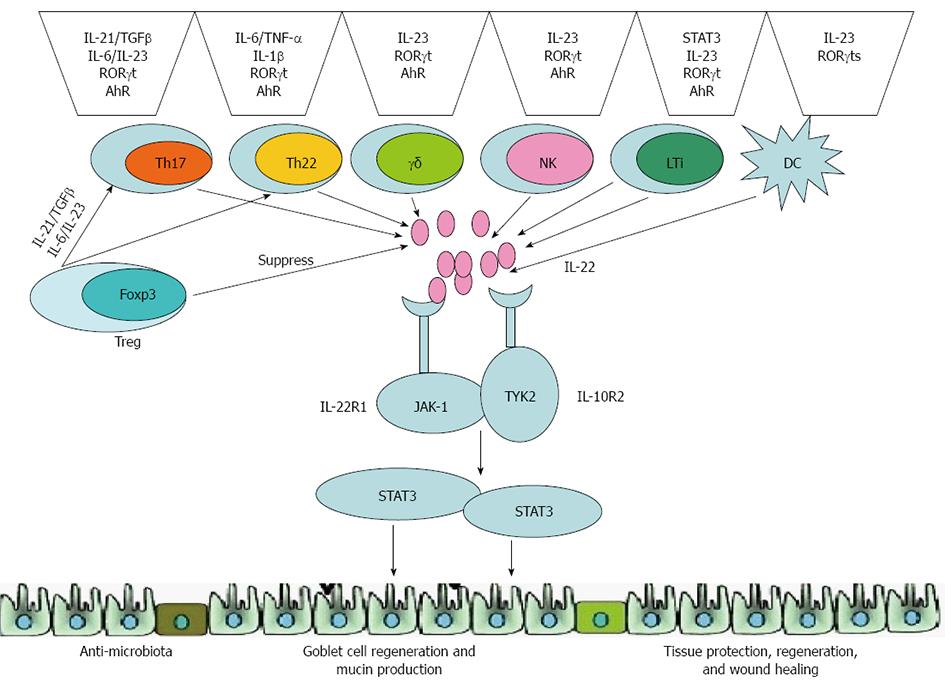
Interleukin (IL)-22, a member of the IL-10 cytokine family which is composed of IL-10, IL-19, IL-20, IL-24 and IL-26[3], is expressed by both the cells of the innate immune system [such as dendritic cells (DCs), lymphoid tissue inducer (LTi)-like cells and natural killer (NK) cells) as well as on the surface of adaptive lymphocytes (including CD4+ T cell subsets, CD8+ T cells and so on)[4]. Several cytokines [such as IL-23, IL-6, tumor necrosis factor (TNF) α, IL-1β, transforming growth factor (TGF) β and IL-17), many of the transcriptional factors (signal transducer and activator of transcription (STAT) 3, RAR-related orphan receptor (ROR) γt and aryl hydrocarbon receptor (AhR)][5] and T regulatory cells (Tregs) are known for their regulation of IL-22 expression[6]. Through activation of the Jak-STAT signal transduction pathway, IL-22 induces proliferative and anti-apoptotic pathways, as well as the production of antimicrobial peptides, which help prevent tissue destruction and assist in its repair and restoration[7]. IL-22 is also associated with IBD susceptibility genes that are crucial for regulating tissue responses during inflammation[8]. All of these processes play critical roles in the pathogenesis of IBD. In recent years, it was demonstrated that treatment with recombinant cytokine or gene therapy involving IL-22 can suppress the inflammatory response and alleviate tissue injury[8,9]. Thus, these findings suggest that further research focused on IL-22 may elucidate the underlying mechanisms of IBD and facilitate the development of novel effective, targeted therapeutic approaches for IBD. This review focuses on IL-22 and its functional role in IBD.
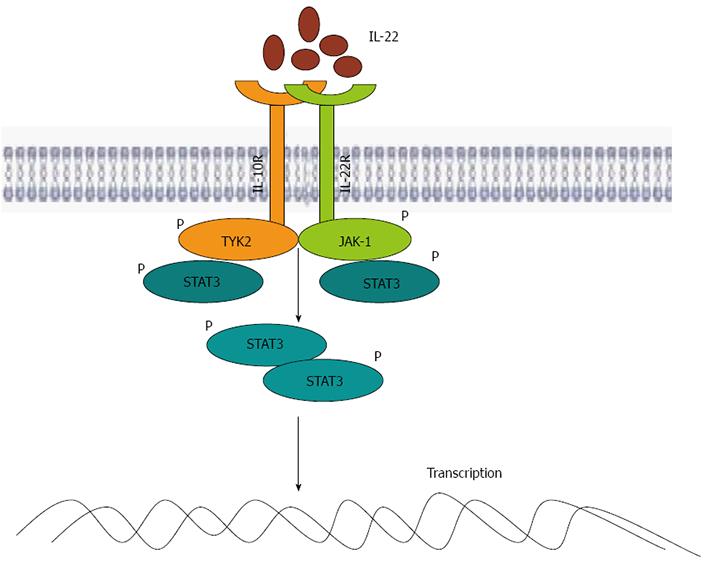
The IL-22 receptor is a heterodimer composed of IL-22 receptor 1 (IL-22R1) and IL-10 receptor 2 (IL-10R2)[10]. IL-10R2 is ubiquitously expressed by most cell types, while the expression of IL-22R1 is limited to nonhematopoietic cells (such as hepatic cells, pancreatic cells, kidney cells, epithelial cells, and skin keratinocytes)[10,11]. Therefore, the expression profile of IL-22R1 determines how IL-22 specifically targets innate cell populations, and not adaptive immune cells[12].
STAT3, STAT1 (in a relatively small number of cells) and STAT5 (in certain cells) were shown to be activated after IL-22 stimulation[13]. Further analysis has also demonstrated that IL-22 signaling propagates downstream phosphorylation signals, including several of the mitogen-activated protein kinase (MAPK) pathways (extracellular signal-regulated kinase (ERK)1/2, MEK1/2, C-Jun N-terminal kinase (JNK), and p38 kinase), and STAT1, STAT3 and STAT5 by utilizing Janus kinase (JAK)1 and tyrosine protein kinase (TYK)2[14] (Figure 1). The capacity of IL-22 to activate JNK, ERK1/2 and p38 MAPK pathways has been implicated in liver diseases[14]. Moreover, the strong activation of IL-22 to stimulate STAT3 has been confirmed in human colon cancer cell lines, human colonic biopsy, as well as the primary mouse colonic epithelial cells[15,16]. In fact, a recent study has shown that, compared with IL-6, IL-22 has a stronger ability to activate STAT3[17]. Pickert et al[18] have demonstrated that in dextran sulfate sodium (DSS)-induced colitis, the activation of epithelial STAT3 is more dependent on IL-22 than on IL-6, a well known activator of STAT3. This is due to IL-22R1 utilizing its constituent C-terminal tail to interact with the coiled-coil domain of STAT3, which has been to conformed in a recent discovery as a novel mechanism to activate STAT3[19]. Similar to other IL-10 family cytokines, IL-22 primarily relies on STAT3 to mediate its functions. Binding of cytokines to this receptor results in the activation of STAT3 signaling pathways, which in turn leads to the induction and production of various tissue-specific genes, including serum amyloid A (SAA), antimicrobial proteins (β-defensin, Reg3c and lipocalin-2) and mucins. Meanwhile, IL-22 also induces proliferative and antiapoptotic pathways in some responsive cells of certain tissues[10,20].
Basu et al[21] have suggested that both innate lymphoid cells (ILCs) and T cells produce IL-22. They showed that IL-22 produced by ILCs was strictly IL-23-dependent, and that the development of IL-22 induced by CD4+ T cells was via an IL-6-dependent mechanism that was augmented by IL-23 and was dependent on both transcription factors T-bet and AhR. At the same time, Wolk et al[22] confirmed that activation of murine T cells, especially T helper (Th) 1 cells, mainly express IL-22. A novel Th subset - the Th17 cells - was identified in 2005[23,24]. IL-17 (or IL-17A), a hallmark cytokine preferentially expressed by Th17 cells, distinguishes these cells from other Th subsets, such as Tregs, Th1 and Th2 cells[25]. Th17 cells play an essential role in host defense, especially against extracellular bacteria and other infectious bacteria, and are involved in the pathogenesis of various autoimmune diseases[26,27].
The level of IL-22 produced by Th17 cells is much higher than that of the production from undifferentiated Th0 cells or Th1 cells. However, the expression and regulation of IL-22 and IL-17 produced in T cells are unparalleled. Researchers have discovered that IL-6 and TGFβ are both required for inducing IL-17 expression in naïve T cells, yet IL-6 alone can sufficiently promote the expression of IL-22[28-30]. In fact, TGFβ has been shown to suppresses IL-22 production in a dose-dependent manner[28]. Through activation by anti-CD3 or concanavalin A (ConA), human T cells can produce IL-22[31]. Based on studies using the lineage marker chemokine CC receptor (CCR) 6 and CCR4, human Th17 cells produced in vitro or purified ex vivo from blood were shown to preferentially express IL-22[32,33]. Moreover, IL-17- and IL-22-expressing cells in human peripheral blood mononuclear cells (PBMCs) are defined by another surface maker, CD161[34]. The above-mentioned Th17 cells can produce IL-22 and IL-17; in addition, CD161+ human CD8+ T cells can also generate these two cytokines[35].
Recent studies have also demonstrated a unique cell subset, designated as the IL-22-producing CD4+ Th subset, in human peripheral blood, which expresses neither IL-17 nor interferon (IFN)-γ[36-38]. In skin, these cells mainly express CCR10. Moreover, the human IL-22-producing T cells can also be generated from naïve CD4+ T cells in the presence of IL-6, rather than TGFβ, which is consistent with what has been reported in the mouse system[36]. Human Langerhans cells are able to differentiate T cells into the only IL-22-producing Th cells in vitro[39].
The human innate immune cell types, such as NK and LTi cells, can also produce IL-22[40,41]. In addition to CD4+ T cells, the Th17 cells, CD8+ T cells and NK T cells also express high levels of IL-22 upon activation, especially when activation occurs along with IL-23 intervention[28,42]. Recently, LTi cells and developmentally-related NK-like cells (NK22), which express the NK marker NKp46, were demonstrated to be the main innate sources of IL-22 expression, especially in the intestinal tract[43,44]. Treatment of NK cells with IL-2 and IL-12 was shown to lead to expression of IL-22[45]. Human immature NK cells, defined as CD161+CD117+CD34-CD94- cells, express both IL-22 and AhR[46]. The equivalent NKp46+ NK-like cells in mice have been found to be developmentally linked to LTi cells[47,48].
Finally, subsets of myeloid cells express the IL-23 receptor (IL-23R) and combine with IL-23 to release lower levels of IL-22[49,50]; those cells that produce high levels of IL-22 may be the major cells of IL-22 origin in mucosal immunity. In contrast to the IL-22 produced by leukocytes, such IL-22 targets mainly tissue epithelial cells rather than immune cells[51]. Although expression of IL-10R2 is widespread, IL-22R1 expression has only been detected on epithelial cells. Upon binding to its receptors on the surface of these epithelial cells, IL-22 produces an accelerating effect on the proliferation and differentiation of these cells, and induces these cells to express genes involved in host defense and wound-healing responses[52].
These cellular functions of IL-22 underlie its crucial role in epithelial barrier defense, especially against invading extracellular bacteria. In fact, in a preclinical model of mucosal immune responses to Gram-negative bacteria, such as Klebsiella pneumoniae and Citrobacter rodentium, IL-22 played an indispensable role[53]. Moreover, IL-22 is associated with the development of various human autoimmune diseases[25]. The expression of IL-22 is unregulated in autoimmune diseases, such as IBD, rheumatoid arthritis and psoriasis. IL-23 appears to be a principal inducer of IL-22 in Th17, NK or NK-like cells, suggesting that IL-22 acts a pivotal mediator in IL-23-dependent immune reactions in skin and mucosal epithelia by stimulating innate antimicrobial responses as well as promoting tissue repair[54].
IL-23 is a member of the IL-12 cytokine family, and its stimulation of activated T cells induces IL-22 expression[54]. Research has found that il23a-/- and il22-/- mice are both highly susceptible to infection with extracellular Gram-negative bacteria, suggesting that a critical function of IL-23 in infection is to induce IL-22 expression[55,56]. Additionally, IL-23 has been found to be important in the terminal differentiation of Th17 cells, assisting in their proliferation and effector functions[56]. Therefore, the ability of IL-23 to enhance Th17 cell proliferation appears to be linked to IL-22 expression.
In addition to IL-23, other cytokines have been found to regulate the expression of IL-22. In cultures of purified naïve murine CD4+ T cells, IL-6 and T cell receptor (TCR) stimulation, or IL-6, TNFα, IL-1β and TCR stimulation, was sufficient to induce IL-22 expression[57]. Increasing concentrations of TGFβ dose-dependently inhibited IL-22 expression while maintaining stable IL-17A expression. It has recently been demonstrated that IL-17A can partially inhibit the expression of IL-22 from Th17 cells in vitro and in vivo, indicating that Th17 cell-associated IL-17A can also negatively regulate IL-22 expression. IL-22 expression in γδT cells can also be induced independently of IL-23 and TCR stimulation by IL-1β, as well as Toll-like receptor (TLR) 1, TLR2, and dectin-1 ligands[54,58].
Similar to cytokine-mediated regulation of IL-22, many of the transcriptional factors are known for regulation of IL-22 expression. STAT3 is critically involved in the induction of IL-22 expression in T cells[59]. Similarly, RORγt, a lineage-specifying transcription factor for the differentiation of Th17 cells, is also required for optimal expression of IL-22. STAT3 and RORγt both control expression of IL-23R, and this regulation may account for their ability to promote IL-22 production in Th17 cells. Therefore, many of the same transcription factors involved in Th17 cell differentiation are also required for IL-22 expression in CD4+ T cells[60]. In addition, AhR is a ligand-dependent transcription factor that is best known for its role in mediating toxicity to the organic compound dioxin. AhR also partially contributes to the differentiation of Th17 cells and is required for expression of IL-22, thus linking IL-22 and Th17 cells to toxicity following exposure to different environmental compounds[61]. A number of IL-22-producing innate cell populations have also been found to express STAT3, RORγt and AhR, yet the involvement of these transcription factors in regulating IL-22 expression in innate cell populations has yet to be examined[62] (Figure 2).
Recent studies have demonstrated a close relationship between CD4+Foxp3+ Tregs and proinflammatory IL-17-producing Th17 cells expressing the lineage-specific transcription factor RORγt. It has been shown that IL-17-secreting Foxp3+ T cells that express RORγt share features of conventional RORγt+ Th17 cells. However, RORγt+Foxp3+ Tregs mostly fail to secrete IL-22 after phorbol 12-myristate 13-acetate/ionomycin stimulation[63]. Foxp3 transcription factor binding sites (TFBSs) in the IL-22 promoter restrain RORγt+Foxp3+ T cells to produce IL-22 at the transcriptional level[64]. Despite the decreased expression of IL-22 in Foxp3+ Tregs, it has been found that Tregs can promote naïve T cell differentiation. In a mouse model of infection with oral Candida albicans, Foxp3+ Tregs were shown to powerfully promote the transition of naïve CD4+ T cells to responding CD4+ cells (Tresps)[65]. Tresps markedly produce IL-22. Therefore, there is the possibility that Tregs can regulate the expression of IL-22.
The IL-22 signaling pathway is activated through a heterogeneous receptor complex composed of two subunits, IL-22R1 and IL-10R2[66]. Although IL-10R2 is widely expressed on almost all of the cell types, the expression of IL-22R1 is restricted to the surfaces of nonhematopoietic cells such as epithelial cells, hepatocytes and keratinocytes[67]. This limited expression of IL-22R1 on nonhematopoietic cells allows IL-22 to specifically target innate cell populations within such tissues as the skin, kidney, digestive tract and respiratory systems[68]. A wide variety of innate and adaptive immune cells, including CD4+ T cells, and most notably Th17 and Th22 cells, CD8+ T cells, LTi cells, NK cells and DCs, can produce IL-22[69]. Upon binding to the IL-22R1 and IL-10R2 receptor complex, these cells produce IL-22 to activate receptor-associated JAK1 and TYK2, resulting in tyrosine phosphorylation of STAT3[70]. This in turn allows IL-22 to induce different kinds of tissue-specific genes, including those encoding proteins involved in antimicrobial defense, cellular differentiation, and expression of mucins; a large, heavily glycosylated family of proteins in the gastrointestinal tract forms a protective layer, which serves to separate commensal bacteria from pathogenic bacteria in the epithelium layer, thereby minimizing the immune response[71]. Through the production of antimicrobial peptides, enhancement of epithelial regeneration, and regulation of wound healing, IL-22 plays a particularly vital role in regulating intestinal inflammatory responses[72]. Furthermore, a direct effect of IL-22 on colonic epithelium is proliferation of epithelial cells, which maintains the integrity of the intestinal epithelium.
Recent studies have focused on possible protective effects of IL-22 in IBD, and have used several DSS-induced as well as Th1- and Th2-mediated colitis mouse models[73,74]. In the DSS-induced colitis model, feeding mice DSS causes disruption of the intestinal epithelial barrier, leading to colitis within 1 wk. In IL-22 knockout mice or wild-type (WT) mice, administration of neutralizing anti-IL-22 antibodies leads to more extensive epithelial destruction and inflammation in the colon, more severe weight loss, and more impaired recovery compared to the DSS-induced acute colitis model. In addition, T cells from IL-22-/- mice or IL-22-deficient mice cause a more severe colitis in the T cell transfer model of IBD[75]. In a Th1-cytokine-mediated model of colitis, expression of IL-22 by CD4+ T cells is crucial for relief of disease severity. Sugimoto et al[76] showed that receipt of supplemental IL-22 leads to rapid amelioration of local intestinal inflammation in the colons of Th2-mediated chronic colitis. IL-22 knockout mice showed delayed recovery from DSS-induced acute colonic injury. Treatment with neutralizing anti-IL-22 antibodies also impaired the recovery of WT mice. Finally, in IL-22-deficient and RAG1-deficient double knockout mice, lacking both T and B cells, no recovery was observed[77].
IL-22 gene delivery mediates STAT3 activation specifically within colonic epithelial cells and enhances reconstitution of goblet cells and production of mucus, thereby reinforcing the mucus barrier function within the gastroenterology tract[78]. In DSS-induced acute colonic injury, recovery is significantly impaired and delayed in IL-23R-deficient and RAG2-deficient double knockout mice lacking of IL-22 expression, and treatment with recombinant IL-22 rescues the recovery in these mice[79]. Pancreatic cells produce TGFβ and IL-10 upon IL-22 stimulation, which can inhibit IFN-γ production, facilitating relief of intestinal injury. These mouse models of colitis suggest that IL-22 plays a protective role in IBD through its ability to improve the integrity of the mucosal barrier and enhance the inherent epithelial defense function.
In addition to maintaining the mucosal barrier function in the gastrointestinal tract, IL-22 induces genes to encode anti-microbial proteins involved in bacterial defense and protection of intestinal mucosa, suggesting a role for IL-22 against extracellular bacteria in the innate immune system. CD and UC are thought to be driven by an abnormal immune response to the intestinal flora[80]. However, since intestinal dysbacteriosis is also a characteristic of IBD pathogenesis, it is difficult to determine whether there is an inflammatory response to abnormal flora or if an abnormal inflammatory response is altering the microbial communities[81]. Intestinal flora, as an environmental factor, may be associated with genetic susceptibility that alters the interactions between ourselves and our microbiome. The first major susceptibility gene discovered for CD is NOD2 (or CARD15), which is known as a receptor for bacterial peptidoglycan (PGN)[82]; another susceptibility gene, ATG16L1, has been shown to be critical for autophagy[83]. The intestinal flora may also lead to disorders of intestinal lymphoid cell subsets, such as Th17 cells and innate lymphocytes, which are important for regulating mucosal immunity[83]. Although there have been numerous studies investigating stool samples of and mucosa-associated bacteria in IBD patients, there has been a lack of consensus between the associations observed in these studies[83].
Although extensive changes have been reported, such as expansion of the Proteobacteria phylum in IBD patients[84], only few specific associations have been reproducibly identified. Although the causes of changes in microbiota species that can trigger IBD remain unclear, and studies on this subject are continuing, the general theme observed so far is that the diversity of microbial communities is significantly decreased in IBD[85]. There have also been repeated observations of the microbiota composition being disrupted during inflammation, resulting in dysbiosis that may induce or perpetuate the inflammatory condition. However, both host genotype and the environment have major impacts on the shape of such dysbiosis, as well as upon which members of the microbiota can stimulate pathogenic immune responses[86].
By promoting the maintenance of intestinal epithelial barrier function, IL-22 can prevent the spread of pathogenic microorganisms in the gut, such as enteropathogens, including Citrobacter rodentium and Salmonella typhimurium (enteric ecotype) in the gastrointestinal tract, thereby limiting bacterial growth. Tregs promote IL-22-dependent clearance of fungi during acute Candida albicans infection[87,88]. In addition, IL-22 can help to eliminate pathogenic microorganisms by inducing various anti-microbial proteins (Figure 2). IL-22 has already been confirmed as a regulator of the expression of antimicrobial proteins such as the S100 family proteins (S100A7, S100A8 and S100A9), β-defensin family proteins (β-defensins BD2 and BD3), Reg family proteins (RegIIIa, RegIIIb and RegIIIc) and lipocalin-2[83,89-91]. These proteins may be important in the control of gut pathogens. IL-22 plays a protective role in the host inflammatory response to microbial infections or in promoting the release of inflammatory mediators, depending on the type of pathogenic microorganisms causing the infection.
Song et al[92] showed that IL-22 plays an crucial role in host defense immunity against infection with the Gram-negative enteric bacteria Citrobacter rodentium, as an inducer of the expression of antibacterial peptides in colonic epithelial cells. The protective effect of IL-22 in systemic infections caused by Salmonella enterica has been demonstrated. IL-23-dependent IL-22 was required for both liver cells’ survival and pathogen defense against systemic Salmonella infection in mice, especially when accompanied by decreased production of IL-12[93].
IL-22 is not only able to protect our intestine against bacterial pathogens, but also plays a protective role in intestinal fungal infections with Candida albicans. Compared with infected WT mice, IL-22 knockout mice infected with Candida albicans hyphae intragastrically had a higher fungal burden and showed signs of more severe mucosal inflammatory hyperplasia in the stomach and colon[94,35]. These results indicate that IL-22 serves as a protective guardian in regulating inflammatory responses and maintaining mucosal barrier integrity in a variety of intestinal infections. However, IL-22 has also been shown to promote intestinal inflammation in parasite infection[95]. Toxoplasma gondii-infected IL-22 knockout mice and mice whose IL-22 was neutralized with an anti-IL-22 monoclonal antibody developed significantly less intestinal pathology and had less weight loss and mortality, despite having similar parasite burdens to infected WT mice. Perhaps the strongly skewed Th1 immune response caused by the Toxoplasma gondii infection may explain this difference.
As mentioned above, IL-22 produced by Th17 cells can be regulated by the gut microbiota. Different from the neutrophil induction response of IL-17, IL-22 serves an important role in tissue repair during mucosal immune system response[96]. Regardless, the relationship between the intestinal microbiota and IL-22-producing cells is extremely close. Most notably, it was recently shown that IL-22-producing innate lymphocytes play a crucial role in preventing systemic inflammation by inhibiting systemic dissemination of commensal bacteria[83]. Sonnenberg et al[97] administrated Rag1-/- mice a neutralizing anti-IL-22 monoclonal antibody, and found that the signs of systemic inflammation increased as did levels of lipopolysaccharide (LPS); in addition, bacteria could be cultured from the spleens and livers of these mice. The disseminated commensal bacteria were subsequently identified as Alcaligenes sp.
Therefore, together with the protective role against IBD, IL-22 also serves as a mucosal protector and plays a critical role in separating our intestinal tract from our gut flora. Under gut homeostatic conditions, viable bacterial pathogens are sampled by DCs that carry them to the mesenteric lymph nodes and these microbes do not disseminate systemically to secondary lymphoid tissues, indicating that a mesenteric guardian may act in concert with a mucosal firewall to distinguish intestinal bacteria[98].
In addition to its antibacterial activity, IL-22 can enhance the survival and proliferation of epithelial cells for tissue differentiation and healing[99]. IL-22 induces the expression of antiapoptotic proteins, including Bcl-xL, Bcl-2 and Mcl-1, as well as proteins directly involved in cell cycle and proliferation, such as c-Myc, cyclin D1, Rb2 and CDK4, and anti-inflammatory or protective proteins, such as IL-11 and follistatin[100-102]. Moreover, IL-22 has been shown to be capable of stimulating a colonic cancer cell line to express a molecule termed deleted in malignant brain tumor 1 (DMBT1), which may play a vital role in the differentiation of epithelial cells[103]. IL-22 has also been shown to induce RegIα, which serves as a trophic and antiapoptotic factor in the inflamed colon of UC patients. Recent research has determined that, through the activation of STAT3, IL-22 can induce the proliferation and reconstruction of mucosal epithelial cells in the intestinal tract[104]. This increased healing response can further prevent the penetration of microorganisms into the intestinal epithelial layers.
An attractive biological feature of IL-22 is its functional association with some major IBD susceptibility genes. Interaction of IL-23 with IL-23R has been implicated in the development of IL-22-producing innate cells, including ILCs, LTi cells and NK cells[4,33,34], and in the maintenance of IL-22-producing Th17 cells[105,106]. Functional polymorphisms of the IL-23R gene have been negatively correlated with development of both CD and UC[105,106]. IL-22 is located within a UC-risk locus on chromosome 12q15[107]. IL-22 can be combined with its receptors that are composed of IL-10R2 and IL-22R1. Polymorphisms of il110r2 are positively associated with both CD and UC[108].
Binding of IL-22 with its cognate receptor induces rapid activation of STAT3 through JAK1 and TYK2. Stat3, jak1 and tyk2 are all well-defined susceptibility genes of CD and, to a lesser extent, of UC[95,96]. STAT3 activation stimulates epithelial cells to produce Muc1. A recent genome-wide association study proposed muc1 as a potential candidate gene associated with CD[109]. In addition, genome-wide association analysis of IBD patients has identified gene mutations involved in encoding IL-22 and the IL-10R2 subunit of the IL-22R complex[110,111].
Due to its crucial roles in regulating barrier immunity and antimicrobiota, IL-22 may have therapeutic potential for IBD. Understanding the various mechanisms of IL-22 in regulating immunity, together with development of immunosuppressive drugs, may open up a new path for the future treatment of IBD.
Treatment with recombinant cytokine or gene therapy delivery of IL-22 may alleviate tissue damage during inflammatory responses. Suppressing the immune system via anti-inflammatory treatments, such as TNFα inhibition, can lead to unwanted dampening of the immune response, impairing the ability of its response to infection. However, IL-22 is an ideal therapeutic candidate because it specifically affects tissue responses and does not have direct effects on the immune response. IL-22 has produced the expected results in an experimental animal model of IBD. Administration of a more specific targeting agent of IL-22 via microinjection of an IL-22 DNA vaccine into already inflamed colonic tissues of mice with IBD has been shown to lead to reduce infiltration of inflammatory cells as well as to increase number of goblet cells[112]. This enhances the production of mucin, thereby buffering the colonic epithelium from commensal bacteria that may otherwise initiate an immune response. Andoh et al[113] did not find IL-22 expression in the gut mucosa of patients with infectious colitis. It seems that IL-22 plays a protective systemic role in CD[114] and a protective local role in UC[115,116]. It should be mentioned at this point that recently, Leppkes et al[117] demonstrated that the adoptive transfer of IL-22-deficient T cells into RAG1-deficient mice caused severe colitis that was indistinguishable from that caused by transferred WT cells.
Genome-wide linkage analysis of IBD patients has identified gene mutations involved in encoding IL-22 and the subunit complex of IL-10R2 and IL-22R1[111]. The IL-22R complex is highly expressed within the gastrointestinal tract and in the inflamed colon; IL-22 is expressed by CD4+ T cells, likely Th17 cells, and innate lymphocytes, such as NK cells and LTi-like cells. Using different experimental models of IBD - DSS-induced colitis, which is thought to be mainly driven by innate immune response cells, and CD4+CD45RBhigh T cell-mediated colitis, in which naive T cells devoid of Tregs are transferred into T cell-deficient mice where they proliferate unimpeded leading to colitis - IL-22 has been shown to be protective[118,119]. Furthermore, Strengell et al[118] showed that IL-22 can be therapeutic in IBD; gene therapy transfer of the IL-22 gene into the colons of already inflamed mice resulted in amelioration of inflammation. In addition, the authors reported that in vivo gene delivery of IL-22 attenuates Th2-mediated colitis and regulates the expression of genes related to mucus layer formation[120].
Some existing biologic therapies are also able to mediate effects on IL-22 expression in patients. Anti-TNFα antibodies (such as infliximab) and the anti-IL-6 antibody toclizumab have been used to treat IBD. Th22 cells depend on TNFα for differentiation; therefore, both Th17 and Th22 cells depend on IL-6 for differentiation, and they can indirectly decrease IL-22 expression in patients to treat IBD[121,122]. Lastly, ustekinumab is able to target both IL-12 and IL-23 and therefore prevent the differentiation of Th1, Th17 and Th22 cells, eliminating several sources of IL-22; this drug is currently being studied in Phase III clinical trials of CD[123]. The treatments mentioned above suppress inflammation by indirect inhibition of IL-22.
IL-22 plays a critical role in the regeneration of damaged epithelial monolayers and stimulates antimicrobial peptide generation. Importantly, the ability of IL-22 to promote intestinal wound healing and proliferation of intestinal epithelial cells in mice and humans has been reproducibly demonstrated by independent groups using different experimental methods, and recent advances in genome-wide association studies have led to results suggesting that the IL-22 pathway is closely related to some major IBD susceptibility genes. These collective findings clearly highlight IL-22 as a promising target for IBD therapy. Therefore, further extensive research on IL-22 is necessary to bring about novel and practical interventions for improving the quality of life of patients with IBD in a safe and effective way. Further understanding of the regulation and function of IL-22 would certainly play a favorable role in the future treatment of IBD.
P- Reviewer: Fujimori S, Rao VS, Torres MI S- Editor: Ma YJ L- Editor: AmEditor E- Editor: Zhang DN
| 1. | Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2693] [Cited by in RCA: 2747] [Article Influence: 119.4] [Reference Citation Analysis (2)] |
| 2. | Salim SY, Söderholm JD. Importance of disrupted intestinal barrier in inflammatory bowel diseases. Inflamm Bowel Dis. 2011;17:362-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 357] [Cited by in RCA: 450] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 3. | Sonnenberg GF, Fouser LA, Artis D. Functional biology of the IL-22-IL-22R pathway in regulating immunity and inflammation at barrier surfaces. Adv Immunol. 2010;107:1-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 4. | Ouyang W. Distinct roles of IL-22 in human psoriasis and inflammatory bowel disease. Cytokine Growth Factor Rev. 2010;21:435-441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 88] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 5. | Aujla SJ, Kolls JK. IL-22: a critical mediator in mucosal host defense. J Mol Med (Berl). 2009;87:451-454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 130] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 6. | Lin S, Yang X, Liang D, Zheng SG. Treg cells: a potential regulator for IL-22 expression? Int J Clin Exp Pathol. 2014;7:474-480. [PubMed] |
| 7. | Kim K, Kim G, Kim JY, Yun HJ, Lim SC, Choi HS. Interleukin-22 promotes epithelial cell transformation and breast tumorigenesis via MAP3K8 activation. Carcinogenesis. 2014;35:1352-1361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 90] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 8. | Mizoguchi A. Healing of intestinal inflammation by IL-22. Inflamm Bowel Dis. 2012;18:1777-1784. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 106] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 9. | Seiderer J, Brand S. IL-22: a two-headed cytokine in IBD? Inflamm Bowel Dis. 2009;15:473-474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 10. | Zenewicz LA, Flavell RA. Recent advances in IL-22 biology. Int Immunol. 2011;23:159-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 258] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 11. | Sonnenberg GF, Fouser LA, Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol. 2011;12:383-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 794] [Cited by in RCA: 822] [Article Influence: 58.7] [Reference Citation Analysis (0)] |
| 12. | Wolk K, Kunz S, Witte E, Friedrich M, Asadullah K, Sabat R. IL-22 increases the innate immunity of tissues. Immunity. 2004;21:241-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1080] [Cited by in RCA: 1118] [Article Influence: 53.2] [Reference Citation Analysis (0)] |
| 13. | Pestka S, Krause CD, Sarkar D, Walter MR, Shi Y, Fisher PB. Interleukin-10 and related cytokines and receptors. Annu Rev Immunol. 2004;22:929-979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 14. | Pan CX, Tang J, Wang XY, Wu FR, Ge JF, Chen FH. Role of interleukin-22 in liver diseases. Inflamm Res. 2014;63:519-525. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 15. | Bishop JL, Roberts ME, Beer JL, Huang M, Chehal MK, Fan X, Fouser LA, Ma HL, Bacani JT, Harder KW. Lyn activity protects mice from DSS colitis and regulates the production of IL-22 from innate lymphoid cells. Mucosal Immunol. 2014;7:405-416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 35] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 16. | Nishida A, Lau CW, Zhang M, Andoh A, Shi HN, Mizoguchi E, Mizoguchi A. The membrane-bound mucin Muc1 regulates T helper 17-cell responses and colitis in mice. Gastroenterology. 2012;142:865-874.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 61] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 17. | Begue B, Verdier J, Rieux-Laucat F, Goulet O, Morali A, Canioni D, Hugot JP, Daussy C, Verkarre V, Pigneur B. Defective IL10 signaling defining a subgroup of patients with inflammatory bowel disease. Am J Gastroenterol. 2011;106:1544-1555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 184] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 18. | Pickert G, Neufert C, Leppkes M, Zheng Y, Wittkopf N, Warntjen M, Lehr HA, Hirth S, Weigmann B, Wirtz S. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med. 2009;206:1465-1472. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 791] [Cited by in RCA: 858] [Article Influence: 53.6] [Reference Citation Analysis (0)] |
| 19. | Dumoutier L, de Meester C, Tavernier J, Renauld JC. New activation modus of STAT3: a tyrosine-less region of the interleukin-22 receptor recruits STAT3 by interacting with its coiled-coil domain. J Biol Chem. 2009;284:26377-26384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 58] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 20. | Neufert C, Pickert G, Zheng Y, Wittkopf N, Warntjen M, Nikolaev A, Ouyang W, Neurath MF, Becker C. Activation of epithelial STAT3 regulates intestinal homeostasis. Cell Cycle. 2010;9:652-655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 21. | Basu R, O’Quinn DB, Silberger DJ, Schoeb TR, Fouser L, Ouyang W, Hatton RD, Weaver CT. Th22 cells are an important source of IL-22 for host protection against enteropathogenic bacteria. Immunity. 2012;37:1061-1075. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 370] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 22. | Wolk K, Kunz S, Asadullah K, Sabat R. Cutting edge: immune cells as sources and targets of the IL-10 family members? J Immunol. 2002;168:5397-5402. [PubMed] |
| 23. | Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123-1132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3407] [Cited by in RCA: 3592] [Article Influence: 179.6] [Reference Citation Analysis (0)] |
| 24. | Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133-1141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3074] [Cited by in RCA: 3358] [Article Influence: 167.9] [Reference Citation Analysis (0)] |
| 25. | Ouyang W, Kolls JK, Zheng Y. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity. 2008;28:454-467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1353] [Cited by in RCA: 1338] [Article Influence: 78.7] [Reference Citation Analysis (0)] |
| 26. | Guo X, Qiu J, Tu T, Yang X, Deng L, Anders RA, Zhou L, Fu YX. Induction of innate lymphoid cell-derived interleukin-22 by the transcription factor STAT3 mediates protection against intestinal infection. Immunity. 2014;40:25-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 217] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 27. | Zheng Y, Danilenko DM, Valdez P, Kasman I, Eastham-Anderson J, Wu J, Ouyang W. Interleukin-22, a T(H)17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature. 2007;445:648-651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1483] [Cited by in RCA: 1500] [Article Influence: 83.3] [Reference Citation Analysis (0)] |
| 28. | Chung Y, Yang X, Chang SH, Ma L, Tian Q, Dong C. Expression and regulation of IL-22 in the IL-17-producing CD4+ T lymphocytes. Cell Res. 2006;16:902-907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 183] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 29. | Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179-189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2768] [Cited by in RCA: 2953] [Article Influence: 155.4] [Reference Citation Analysis (0)] |
| 30. | Mangan PR, Harrington LE, O’Quinn DB, Helms WS, Bullard DC, Elson CO, Hatton RD, Wahl SM, Schoeb TR, Weaver CT. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2372] [Cited by in RCA: 2489] [Article Influence: 131.0] [Reference Citation Analysis (0)] |
| 31. | Xie MH, Aggarwal S, Ho WH, Foster J, Zhang Z, Stinson J, Wood WI, Goddard AD, Gurney AL. Interleukin (IL)-22, a novel human cytokine that signals through the interferon receptor-related proteins CRF2-4 and IL-22R. J Biol Chem. 2000;275:31335-31339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 421] [Cited by in RCA: 429] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 32. | Acosta-Rodriguez EV, Rivino L, Geginat J, Jarrossay D, Gattorno M, Lanzavecchia A, Sallusto F, Napolitani G. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat Immunol. 2007;8:639-646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1373] [Cited by in RCA: 1457] [Article Influence: 80.9] [Reference Citation Analysis (0)] |
| 33. | Wilson NJ, Boniface K, Chan JR, McKenzie BS, Blumenschein WM, Mattson JD, Basham B, Smith K, Chen T, Morel F. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat Immunol. 2007;8:950-957. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1490] [Cited by in RCA: 1566] [Article Influence: 87.0] [Reference Citation Analysis (0)] |
| 34. | Kleinschek MA, Boniface K, Sadekova S, Grein J, Murphy EE, Turner SP, Raskin L, Desai B, Faubion WA, de Waal Malefyt R. Circulating and gut-resident human Th17 cells express CD161 and promote intestinal inflammation. J Exp Med. 2009;206:525-534. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 356] [Cited by in RCA: 376] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 35. | Billerbeck E, Kang YH, Walker L, Lockstone H, Grafmueller S, Fleming V, Flint J, Willberg CB, Bengsch B, Seigel B. Analysis of CD161 expression on human CD8+ T cells defines a distinct functional subset with tissue-homing properties. Proc Natl Acad Sci USA. 2010;107:3006-3011. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 292] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 36. | Duhen T, Geiger R, Jarrossay D, Lanzavecchia A, Sallusto F. Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells. Nat Immunol. 2009;10:857-863. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 822] [Cited by in RCA: 828] [Article Influence: 51.8] [Reference Citation Analysis (0)] |
| 37. | Eyerich S, Eyerich K, Pennino D, Carbone T, Nasorri F, Pallotta S, Cianfarani F, Odorisio T, Traidl-Hoffmann C, Behrendt H. Th22 cells represent a distinct human T cell subset involved in epidermal immunity and remodeling. J Clin Invest. 2009;119:3573-3585. [PubMed] |
| 38. | Trifari S, Kaplan CD, Tran EH, Crellin NK, Spits H. Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from T(H)-17, T(H)1 and T(H)2 cells. Nat Immunol. 2009;10:864-871. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 756] [Cited by in RCA: 788] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 39. | Fujita H, Nograles KE, Kikuchi T, Gonzalez J, Carucci JA, Krueger JG. Human Langerhans cells induce distinct IL-22-producing CD4+ T cells lacking IL-17 production. Proc Natl Acad Sci USA. 2009;106:21795-21800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 194] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 40. | Vivier E, Spits H, Cupedo T. Interleukin-22-producing innate immune cells: new players in mucosal immunity and tissue repair? Nat Rev Immunol. 2009;9:229-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 140] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 41. | Colonna M. Interleukin-22-producing natural killer cells and lymphoid tissue inducer-like cells in mucosal immunity. Immunity. 2009;31:15-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 244] [Cited by in RCA: 249] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 42. | Goto M, Murakawa M, Kadoshima-Yamaoka K, Tanaka Y, Nagahira K, Fukuda Y, Nishimura T. Murine NKT cells produce Th17 cytokine interleukin-22. Cell Immunol. 2009;254:81-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 82] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 43. | Hughes T, Becknell B, McClory S, Briercheck E, Freud AG, Zhang X, Mao H, Nuovo G, Yu J, Caligiuri MA. Stage 3 immature human natural killer cells found in secondary lymphoid tissue constitutively and selectively express the TH 17 cytokine interleukin-22. Blood. 2009;113:4008-4010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 102] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 44. | de Moura PR, Watanabe L, Bleicher L, Colau D, Dumoutier L, Lemaire MM, Renauld JC, Polikarpov I. Crystal structure of a soluble decoy receptor IL-22BP bound to interleukin-22. FEBS Lett. 2009;583:1072-1077. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 47] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 45. | Cella M, Fuchs A, Vermi W, Facchetti F, Otero K, Lennerz JK, Doherty JM, Mills JC, Colonna M. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722-725. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1031] [Cited by in RCA: 1050] [Article Influence: 65.6] [Reference Citation Analysis (0)] |
| 46. | Luci C, Reynders A, Ivanov II, Cognet C, Chiche L, Chasson L, Hardwigsen J, Anguiano E, Banchereau J, Chaussabel D. Influence of the transcription factor RORgammat on the development of NKp46+ cell populations in gut and skin. Nat Immunol. 2009;10:75-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 463] [Cited by in RCA: 468] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 47. | Crellin NK, Trifari S, Kaplan CD, Cupedo T, Spits H. Human NKp44+IL-22+ cells and LTi-like cells constitute a stable RORC+ lineage distinct from conventional natural killer cells. J Exp Med. 2010;207:281-290. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 210] [Cited by in RCA: 220] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 48. | Cupedo T, Crellin NK, Papazian N, Rombouts EJ, Weijer K, Grogan JL, Fibbe WE, Cornelissen JJ, Spits H. Human fetal lymphoid tissue-inducer cells are interleukin 17-producing precursors to RORC+ CD127+ natural killer-like cells. Nat Immunol. 2009;10:66-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 534] [Cited by in RCA: 549] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 49. | De Luca A, Zelante T, D’Angelo C, Zagarella S, Fallarino F, Spreca A, Iannitti RG, Bonifazi P, Renauld JC, Bistoni F. IL-22 defines a novel immune pathway of antifungal resistance. Mucosal Immunol. 2010;3:361-373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 220] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 50. | Alam MS, Maekawa Y, Kitamura A, Tanigaki K, Yoshimoto T, Kishihara K, Yasutomo K. Notch signaling drives IL-22 secretion in CD4+ T cells by stimulating the aryl hydrocarbon receptor. Proc Natl Acad Sci USA. 2010;107:5943-5948. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 135] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 51. | Takatori H, Kanno Y, Watford WT, Tato CM, Weiss G, Ivanov II, Littman DR, O’Shea JJ. Lymphoid tissue inducer-like cells are an innate source of IL-17 and IL-22. J Exp Med. 2009;206:35-41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 577] [Cited by in RCA: 602] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 52. | Ouyang W, Valdez P. IL-22 in mucosal immunity. Mucosal Immunol. 2008;1:335-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 53] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 53. | Aujla SJ, Chan YR, Zheng M, Fei M, Askew DJ, Pociask DA, Reinhart TA, McAllister F, Edeal J, Gaus K. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat Med. 2008;14:275-281. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 956] [Cited by in RCA: 941] [Article Influence: 55.4] [Reference Citation Analysis (0)] |
| 54. | Molle C, Zhang T, Ysebrant de Lendonck L, Gueydan C, Andrianne M, Sherer F, Van Simaeys G, Blackshear PJ, Leo O, Goriely S. Tristetraprolin regulation of interleukin 23 mRNA stability prevents a spontaneous inflammatory disease. J Exp Med. 2013;210:1675-1684. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 83] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 55. | Aujla SJ, Dubin PJ, Kolls JK. Th17 cells and mucosal host defense. Semin Immunol. 2007;19:377-382. [PubMed] |
| 56. | McGeachy MJ, Chen Y, Tato CM, Laurence A, Joyce-Shaikh B, Blumenschein WM, McClanahan TK, O’Shea JJ, Cua DJ. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat Immunol. 2009;10:314-324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 891] [Cited by in RCA: 838] [Article Influence: 52.4] [Reference Citation Analysis (0)] |
| 57. | Huber M, Heink S, Grothe H, Guralnik A, Reinhard K, Elflein K, Hünig T, Mittrücker HW, Brüstle A, Kamradt T. A Th17-like developmental process leads to CD8(+) Tc17 cells with reduced cytotoxic activity. Eur J Immunol. 2009;39:1716-1725. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 189] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 58. | Singh B, Schwartz JA, Sandrock C, Bellemore SM, Nikoopour E. Modulation of autoimmune diseases by interleukin (IL)-17 producing regulatory T helper (Th17) cells. Indian J Med Res. 2013;138:591-594. [PubMed] |
| 59. | Lim C, Savan R. The role of the IL-22/IL-22R1 axis in cancer. Cytokine Growth Factor Rev. 2014;25:257-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 60. | Ramirez JM, Brembilla NC, Sorg O, Chicheportiche R, Matthes T, Dayer JM, Saurat JH, Roosnek E, Chizzolini C. Activation of the aryl hydrocarbon receptor reveals distinct requirements for IL-22 and IL-17 production by human T helper cells. Eur J Immunol. 2010;40:2450-2459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 140] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 61. | Kang N, Tang L, Li X, Wu D, Li W, Chen X, Cui L, Ba D, He W. Identification and characterization of Foxp3(+) gammadelta T cells in mouse and human. Immunol Lett. 2009;125:105-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 62. | Molinero LL, Cubre A, Mora-Solano C, Wang Y, Alegre ML. T cell receptor/CARMA1/NF-κB signaling controls T-helper (Th) 17 differentiation. Proc Natl Acad Sci USA. 2012;109:18529-18534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 50] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 63. | Jeron A, Hansen W, Ewert F, Buer J, Geffers R, Bruder D. ChIP-on-chip analysis identifies IL-22 as direct target gene of ectopically expressed FOXP3 transcription factor in human T cells. BMC Genomics. 2012;13:705. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 64. | Wang X, Zheng XY, Ma C, Wang XK, Wu J, Adem A, Zhu J, Zhang HL. Mitigated Tregs and augmented Th17 cells and cytokines are associated with severity of experimental autoimmune neuritis. Scand J Immunol. 2014;80:180-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 65. | Pandiyan P, Conti HR, Zheng L, Peterson AC, Mathern DR, Hernández-Santos N, Edgerton M, Gaffen SL, Lenardo MJ. CD4(+)CD25(+)Foxp3(+) regulatory T cells promote Th17 cells in vitro and enhance host resistance in mouse Candida albicans Th17 cell infection model. Immunity. 2011;34:422-434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 215] [Cited by in RCA: 218] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 66. | Kumar P, Thakar MS, Ouyang W, Malarkannan S. IL-22 from conventional NK cells is epithelial regenerative and inflammation protective during influenza infection. Mucosal Immunol. 2013;6:69-82. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 144] [Cited by in RCA: 152] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 67. | Bleicher L, de Moura PR, Watanabe L, Colau D, Dumoutier L, Renauld JC, Polikarpov I. Crystal structure of the IL-22/IL-22R1 complex and its implications for the IL-22 signaling mechanism. FEBS Lett. 2008;582:2985-2992. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 76] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 68. | Wolk K, Sabat R. Interleukin-22: a novel T- and NK-cell derived cytokine that regulates the biology of tissue cells. Cytokine Growth Factor Rev. 2006;17:367-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 238] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 69. | Yang X, Zheng SG. Interleukin-22: a likely target for treatment of autoimmune diseases. Autoimmun Rev. 2014;13:615-620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 70. | Geremia A, Arancibia-Cárcamo CV, Fleming MP, Rust N, Singh B, Mortensen NJ, Travis SP, Powrie F. IL-23-responsive innate lymphoid cells are increased in inflammatory bowel disease. J Exp Med. 2011;208:1127-1133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 479] [Cited by in RCA: 521] [Article Influence: 37.2] [Reference Citation Analysis (0)] |
| 71. | Akdis M, Palomares O, van de Veen W, van Splunter M, Akdis CA. TH17 and TH22 cells: a confusion of antimicrobial response with tissue inflammation versus protection. J Allergy Clin Immunol. 2012;129:1438-149; 1438-149;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 127] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 72. | Zhang N, Pan HF, Ye DQ. Th22 in inflammatory and autoimmune disease: prospects for therapeutic intervention. Mol Cell Biochem. 2011;353:41-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 85] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 73. | Bhan AK, Mizoguchi E, Smith RN, Mizoguchi A. Colitis in transgenic and knockout animals as models of human inflammatory bowel disease. Immunol Rev. 1999;169:195-207. [PubMed] |
| 74. | Yeste A, Mascanfroni ID, Nadeau M, Burns EJ, Tukpah AM, Santiago A, Wu C, Patel B, Kumar D, Quintana FJ. IL-21 induces IL-22 production in CD4+ T cells. Nat Commun. 2014;5:3753. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 130] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 75. | Zenewicz LA, Yancopoulos GD, Valenzuela DM, Murphy AJ, Stevens S, Flavell RA. Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity. 2008;29:947-957. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 76. | Sugimoto K, Ogawa A, Mizoguchi E, Shimomura Y, Andoh A, Bhan AK, Blumberg RS, Xavier RJ, Mizoguchi A. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J Clin Invest. 2008;118:534-544. [PubMed] |
| 77. | Zindl CL, Lai JF, Lee YK, Maynard CL, Harbour SN, Ouyang W, Chaplin DD, Weaver CT. IL-22-producing neutrophils contribute to antimicrobial defense and restitution of colonic epithelial integrity during colitis. Proc Natl Acad Sci USA. 2013;110:12768-12773. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 265] [Cited by in RCA: 284] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 78. | Willson TA, Jurickova I, Collins M, Denson LA. Deletion of intestinal epithelial cell STAT3 promotes T-lymphocyte STAT3 activation and chronic colitis following acute dextran sodium sulfate injury in mice. Inflamm Bowel Dis. 2013;19:512-525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 79. | Monteleone I, Rizzo A, Sarra M, Sica G, Sileri P, Biancone L, MacDonald TT, Pallone F, Monteleone G. Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology. 2011;141:237-48, 248.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 501] [Cited by in RCA: 505] [Article Influence: 36.1] [Reference Citation Analysis (0)] |
| 80. | Rath E, Haller D. Unfolded protein responses in the intestinal epithelium: sensors for the microbial and metabolic environment. J Clin Gastroenterol. 2012;46 Suppl:S3-S5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 81. | Leung JM, Loke P. A role for IL-22 in the relationship between intestinal helminths, gut microbiota and mucosal immunity. Int J Parasitol. 2013;43:253-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 82. | Hugot JP, Chamaillard M, Zouali H, Lesage S, Cézard JP, Belaiche J, Almer S, Tysk C, O’Morain CA, Gassull M. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599-603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4223] [Cited by in RCA: 3903] [Article Influence: 162.6] [Reference Citation Analysis (0)] |
| 83. | Kaser A, Blumberg RS. ATG16L1 Crohn’s disease risk stresses the endoplasmic reticulum of Paneth cells. Gut. 2014;63:1038-1039. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 84. | Mukhopadhya I, Hansen R, El-Omar EM, Hold GL. IBD-what role do Proteobacteria play? Nat Rev Gastroenterol Hepatol. 2012;9:219-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 417] [Cited by in RCA: 574] [Article Influence: 44.2] [Reference Citation Analysis (1)] |
| 85. | Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260-270. [PubMed] |
| 86. | Elson CO, Cong Y. Host-microbiota interactions in inflammatory bowel disease. Gut Microbes. 2012;3:332-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 94] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 87. | Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1508] [Cited by in RCA: 1549] [Article Influence: 91.1] [Reference Citation Analysis (0)] |
| 88. | Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio SP, Paixao TA, Butler BP, Chu H, Santos RL, Berger T. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe. 2009;5:476-486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 444] [Cited by in RCA: 428] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 89. | Kato-Kogoe N, Nishioka T, Kawabe M, Kataoka F, Yamanegi K, Yamada N, Hata M, Yamamoto T, Nakasho K, Urade M. The promotional effect of IL-22 on mineralization activity of periodontal ligament cells. Cytokine. 2012;59:41-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 90. | Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, Fouser LA. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203:2271-2279. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1807] [Cited by in RCA: 1830] [Article Influence: 96.3] [Reference Citation Analysis (0)] |
| 91. | Sabat R, Wolk K. Research in practice: IL-22 and IL-20: significance for epithelial homeostasis and psoriasis pathogenesis. J Dtsch Dermatol Ges. 2011;9:518-523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 92. | Song X, Zhu S, Shi P, Liu Y, Shi Y, Levin SD, Qian Y. IL-17RE is the functional receptor for IL-17C and mediates mucosal immunity to infection with intestinal pathogens. Nat Immunol. 2011;12:1151-1158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 243] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 93. | Schulz SM, Köhler G, Schütze N, Knauer J, Straubinger RK, Chackerian AA, Witte E, Wolk K, Sabat R, Iwakura Y. Protective immunity to systemic infection with attenuated Salmonella enterica serovar enteritidis in the absence of IL-12 is associated with IL-23-dependent IL-22, but not IL-17. J Immunol. 2008;181:7891-7901. [PubMed] |
| 94. | Stange J, Hepworth MR, Rausch S, Zajic L, Kühl AA, Uyttenhove C, Renauld JC, Hartmann S, Lucius R. IL-22 mediates host defense against an intestinal intracellular parasite in the absence of IFN-γ at the cost of Th17-driven immunopathology. J Immunol. 2012;188:2410-2418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 95. | Wilson MS, Feng CG, Barber DL, Yarovinsky F, Cheever AW, Sher A, Grigg M, Collins M, Fouser L, Wynn TA. Redundant and pathogenic roles for IL-22 in mycobacterial, protozoan, and helminth infections. J Immunol. 2010;184:4378-4390. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 108] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 96. | Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485-517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3754] [Cited by in RCA: 3787] [Article Influence: 236.7] [Reference Citation Analysis (0)] |
| 97. | Sonnenberg GF, Monticelli LA, Alenghat T, Fung TC, Hutnick NA, Kunisawa J, Shibata N, Grunberg S, Sinha R, Zahm AM. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321-1325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 593] [Cited by in RCA: 584] [Article Influence: 44.9] [Reference Citation Analysis (0)] |
| 98. | Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. 2012;12:503-516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 627] [Cited by in RCA: 663] [Article Influence: 51.0] [Reference Citation Analysis (0)] |
| 99. | Ki SH, Park O, Zheng M, Morales-Ibanez O, Kolls JK, Bataller R, Gao B. Interleukin-22 treatment ameliorates alcoholic liver injury in a murine model of chronic-binge ethanol feeding: role of signal transducer and activator of transcription 3. Hepatology. 2010;52:1291-1300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 364] [Cited by in RCA: 367] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 100. | Dudakov JA, Hanash AM, Jenq RR, Young LF, Ghosh A, Singer NV, West ML, Smith OM, Holland AM, Tsai JJ. Interleukin-22 drives endogenous thymic regeneration in mice. Science. 2012;336:91-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 299] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 101. | Zenewicz LA, Yancopoulos GD, Valenzuela DM, Murphy AJ, Karow M, Flavell RA. Interleukin-22 but not interleukin-17 provides protection to hepatocytes during acute liver inflammation. Immunity. 2007;27:647-659. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 527] [Cited by in RCA: 547] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 102. | Radaeva S, Sun R, Pan HN, Hong F, Gao B. Interleukin 22 (IL-22) plays a protective role in T cell-mediated murine hepatitis: IL-22 is a survival factor for hepatocytes via STAT3 activation. Hepatology. 2004;39:1332-1342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 465] [Cited by in RCA: 503] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 103. | Fukui H, Sekikawa A, Tanaka H, Fujimori Y, Katake Y, Fujii S, Ichikawa K, Tomita S, Imura J, Chiba T. DMBT1 is a novel gene induced by IL-22 in ulcerative colitis. Inflamm Bowel Dis. 2011;17:1177-1188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 47] [Article Influence: 3.4] [Reference Citation Analysis (1)] |
| 104. | Zwiers A, Kraal L, van de Pouw Kraan TC, Wurdinger T, Bouma G, Kraal G. Cutting edge: a variant of the IL-23R gene associated with inflammatory bowel disease induces loss of microRNA regulation and enhanced protein production. J Immunol. 2012;188:1573-1577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 86] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 105. | Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066-2078. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1967] [Cited by in RCA: 2200] [Article Influence: 137.5] [Reference Citation Analysis (6)] |
| 106. | Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307-317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2019] [Cited by in RCA: 1881] [Article Influence: 134.4] [Reference Citation Analysis (2)] |
| 107. | Silverberg MS, Cho JH, Rioux JD, McGovern DP, Wu J, Annese V, Achkar JP, Goyette P, Scott R, Xu W. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet. 2009;41:216-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 108. | Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schäffer AA, Noyan F, Perro M, Diestelhorst J, Allroth A, Murugan D. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033-2045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1149] [Cited by in RCA: 1092] [Article Influence: 68.3] [Reference Citation Analysis (0)] |
| 109. | Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118-1125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2110] [Cited by in RCA: 2005] [Article Influence: 133.7] [Reference Citation Analysis (0)] |
| 110. | Kryczek I, Lin Y, Nagarsheth N, Peng D, Zhao L, Zhao E, Vatan L, Szeliga W, Dou Y, Owens S. IL-22(+)CD4(+) T cells promote colorectal cancer stemness via STAT3 transcription factor activation and induction of the methyltransferase DOT1L. Immunity. 2014;40:772-784. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 313] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 111. | Floss DM, Mrotzek S, Klöcker T, Schröder J, Grötzinger J, Rose-John S, Scheller J. Identification of canonical tyrosine-dependent and non-canonical tyrosine-independent STAT3 activation sites in the intracellular domain of the interleukin 23 receptor. J Biol Chem. 2013;288:19386-19400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 53] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 112. | Shimauchi T, Sasada K, Kito Y, Mori T, Hata M, Fujiyama T, Ito T, Hirakawa S, Tokura Y. CD8+ Sézary syndrome with interleukin-22 production modulated by bacterial sepsis. Br J Dermatol. 2013;168:881-883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 113. | Andoh A, Zhang Z, Inatomi O, Fujino S, Deguchi Y, Araki Y, Tsujikawa T, Kitoh K, Kim-Mitsuyama S, Takayanagi A. Interleukin-22, a member of the IL-10 subfamily, induces inflammatory responses in colonic subepithelial myofibroblasts. Gastroenterology. 2005;129:969-984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 379] [Cited by in RCA: 397] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 114. | Wolk K, Witte E, Hoffmann U, Doecke WD, Endesfelder S, Asadullah K, Sterry W, Volk HD, Wittig BM, Sabat R. IL-22 induces lipopolysaccharide-binding protein in hepatocytes: a potential systemic role of IL-22 in Crohn’s disease. J Immunol. 2007;178:5973-5981. [PubMed] |
| 115. | Sabat R, Ouyang W, Wolk K. Therapeutic opportunities of the IL-22-IL-22R1 system. Nat Rev Drug Discov. 2014;13:21-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 427] [Cited by in RCA: 463] [Article Influence: 42.1] [Reference Citation Analysis (0)] |
| 116. | Kreymborg K, Etzensperger R, Dumoutier L, Haak S, Rebollo A, Buch T, Heppner FL, Renauld JC, Becher B. IL-22 is expressed by Th17 cells in an IL-23-dependent fashion, but not required for the development of autoimmune encephalomyelitis. J Immunol. 2007;179:8098-8104. [PubMed] |
| 117. | Leppkes M, Becker C, Ivanov II, Hirth S, Wirtz S, Neufert C, Pouly S, Murphy AJ, Valenzuela DM, Yancopoulos GD. RORgamma-expressing Th17 cells induce murine chronic intestinal inflammation via redundant effects of IL-17A and IL-17F. Gastroenterology. 2009;136:257-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 363] [Cited by in RCA: 378] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 118. | Strengell M, Lehtonen A, Matikainen S, Julkunen I. IL-21 enhances SOCS gene expression and inhibits LPS-induced cytokine production in human monocyte-derived dendritic cells. J Leukoc Biol. 2006;79:1279-1285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 69] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 119. | Sheikh SZ, Hegazi RA, Kobayashi T, Onyiah JC, Russo SM, Matsuoka K, Sepulveda AR, Li F, Otterbein LE, Plevy SE. An anti-inflammatory role for carbon monoxide and heme oxygenase-1 in chronic Th2-mediated murine colitis. J Immunol. 2011;186:5506-5513. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 112] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 120. | Kumar P, Rajasekaran K, Palmer JM, Thakar MS, Malarkannan S. IL-22: An Evolutionary Missing-Link Authenticating the Role of the Immune System in Tissue Regeneration. J Cancer. 2013;4:57-65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 121. | Caproni M, Antiga E, Melani L, Volpi W, Del Bianco E, Fabbri P. Serum levels of IL-17 and IL-22 are reduced by etanercept, but not by acitretin, in patients with psoriasis: a randomized-controlled trial. J Clin Immunol. 2009;29:210-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 142] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 122. | Yao X, Huang J, Zhong H, Shen N, Faggioni R, Fung M, Yao Y. Targeting interleukin-6 in inflammatory autoimmune diseases and cancers. Pharmacol Ther. 2014;141:125-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 123. | Tuskey A, Behm BW. Profile of ustekinumab and its potential in patients with moderate-to-severe Crohn’s disease. Clin Exp Gastroenterol. 2014;7:173-179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |