Published online Nov 28, 2014. doi: 10.3748/wjg.v20.i44.16489
Revised: July 4, 2014
Accepted: July 24, 2014
Published online: November 28, 2014
Processing time: 216 Days and 23.7 Hours
The intestinal microbiota is the collection of the living microorganisms (bacteria, fungi, protozoa, and viruses) inhabiting the gastrointestinal tract. Novel bacterial identification approaches have revealed that the gastrointestinal microbiota of dogs and cats is, similarly to humans, a highly complex ecosystem. Studies in dogs and cats have demonstrated that acute and chronic gastrointestinal diseases, including inflammatory bowel disease (IBD), are associated with alterations in the small intestinal and fecal microbial communities. Of interest is that these alterations are generally similar to the dysbiosis observed in humans with IBD or animal models of intestinal inflammation, suggesting that microbial responses to inflammatory conditions of the gut are conserved across mammalian host types. Studies have also revealed possible underlying susceptibilities in the innate immune system of dogs and cats with IBD, which further demonstrate the intricate relationship between gut microbiota and host health. Commonly identified microbiome changes in IBD are decreases in bacterial groups within the phyla Firmicutes and Bacteroidetes, and increases within Proteobacteia. Furthermore, a reduction in the diversity of Clostridium clusters XIVa and IV (i.e., Lachnospiraceae and Clostridium coccoides subgroups) are associated with IBD, suggesting that these bacterial groups may play an important role in maintenance of gastrointestinal health. Future studies are warranted to evaluate the functional changes associated with intestinal dysbiosis in dogs and cats.
Core tip: Several studies in dogs and cats have demonstrated that acute and chronic gastrointestinal diseases, including inflammatory bowel disease (IBD), are associated with alterations in the small intestinal and fecal microbial communities. Of interest is that these alterations are generally similar to the dysbiosis observed in humans with IBD or animal models of intestinal inflammation, suggesting that microbial responses in inflammatory conditions of the gut are conserved across mammalian host types, and dogs and cats may serve as models to study therapeutic approaches to spontaneous inflammatory conditions of the gastrointestinal tract.
- Citation: Honneffer JB, Minamoto Y, Suchodolski JS. Microbiota alterations in acute and chronic gastrointestinal inflammation of cats and dogs. World J Gastroenterol 2014; 20(44): 16489-16497
- URL: https://www.wjgnet.com/1007-9327/full/v20/i44/16489.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i44.16489
The intestinal microbiota is the collection of the living microorganisms (bacteria, fungi, protozoa, and viruses) inhabiting the gastrointestinal (GI) tract. Novel bacterial identification approaches have revealed that the gastrointestinal microbiota of dogs and cats is, similarly to humans, a highly complex ecosystem, comprising at least several hundred different bacterial phylotypes[1-3]. It has been suggested that the intestine of mammals is home to a total of 1010-1014 microbial cells, which is approximately 10 times more than the number of host cells. This complex microbial ecosystem and its interplay with eukaryotic host cells have a significant impact on health and disease of dogs and cats. The stimulation of the host immune system and the microbial metabolites produced by the resident microbiome are thought to be one of the most important driving forces behind the coevolution of gastrointestinal microbiota with their host. Gut microbes aid the host by acting as a defending barrier against enteropathogens. They also aid in digestion of complex fiber sources and produce various short-chain fatty acids and other metabolites that provide nutritional support for enterocytes, and which play an important role in the development and regulation of the host immune system[4,5].
Several studies in dogs and cats have demonstrated that acute and chronic gastrointestinal diseases, including inflammatory bowel disease (IBD), are associated with alterations in small intestinal and fecal microbial communities[6-14]. Of interest is that these alterations are generally similar to the dysbiosis observed in humans with IBD or animal models of intestinal inflammation[15-20], suggesting that microbial responses in inflammatory conditions of the gut are conserved across mammalian host types, and dogs and cats may serve as models to study therapeutic approaches to spontaneous inflammatory conditions of the gastrointestinal tract. Recent data support this model, as it has been shown that for example probiotic products (i.e., VSL#3 strains) show similar clinical benefits in dogs with IBD as have been previously demonstrated in humans[21].
Studies have also revealed possible underlying susceptibilities in the innate immune system of dogs and cats with IBD, which further demonstrates the intricate relationship between gut microbiota and host health[10,22-25]. Currently, a major hurdle for a more detailed understanding of host-microbe interactions in dogs and cats is the fact that to date most studies evaluating microbiota in GI diseases have examined only a single time point or have evaluated only a small number of diseased animals.
Yet the possibility to alter the microbiome holds promise as a therapeutic mean in veterinary medicine, and recent studies would confirm that direct or indirect manipulations of the intestinal microbiome via antibiotics, diet and/or probiotics may have beneficial effects in gastrointestinal diseases of dogs and cats[21,26-29].
Various studies have evaluated the bacterial communities in healthy dogs and cats using either traditional bacterial culture or novel next-generation sequencing approaches. Based on traditional bacterial culture, the small intestine of dogs and cats harbors generally low bacterial counts, ranging between 102 to 105 cfu/g of small intestinal content; however, some studies have identified much higher counts in healthy dogs and cats with up to 109 cfu/g[30,31]. Cats appear to have higher counts of anaerobic bacteria compared to dogs in the proximal small intestine[31]. The total bacterial count in the colon ranges between approximately 109 and 1011 cfu/g and the most abundant cultivable groups are Bacteroides, Clostridium, Lactobacillus, Bifidobacterium, and Enterobacteriaceae[32,33]. Next-generation sequencing studies of the 16S rRNA gene have described the canine and feline microbiome, which on higher phylogenetic levels resembles the microbiome of humans and other mammals. On average, 10 different bacterial phyla have been identified in the feline and canine gut, with Firmicutes, Bacteroidetes, Proteobacteria, Fusobacteria, and Actinobacteria making up the vast majority of all gut microbes[1,3,11,34-36]. Minor abundant members are the phyla Tenericutes, Verrucomicrobia, Cyanobacteria, and Chloroflexi. The Firmicutes contain various sequences affiliated with Clostridium cluster IV and Clostridium cluster XIVa and these are together with Bacteroides or Prevotella the predominant bacterial groups in fecal samples[3,35,37]. Helicobacter are the predominant group in the stomach (> 90% of sequencing reads)[38], while the duodenum is home to Enterobacteriaceae, Clostridiales, Bacteroidales, and Lactobacillales[36].
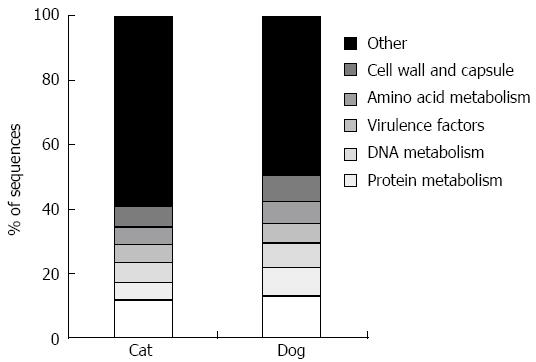
The canine[39] and feline[40,41] fecal metagenomes (i.e., shotgun sequencing of genomic DNA) have also been studied. This approach yields information regarding microbial genes present in a sample, and allows assessment of the functional capabilities of the microbiota, summarized in Figure 1. Despite variation in the microbial populations of cats and dogs, the functional capabilities are noted to be highly conserved.
More detailed overviews about the canine and feline microbiota in healthy animals have been reported previously[42-45].
In recent years, the GI microbiota has garnered strong interest due to the potential etiopathologic role in host health and disease. Many studies in humans and animal models have suggested that various GI disorders are associated with alterations of the GI microbiota. While specific enteropathogens have been recognized in cats and dogs (i.e., Campylobacter jejuni, Clostridium difficile, Clostridium perfringens, and Salmonella), most of them are found in similar frequency across healthy animals. Therefore, their cause-effect relations remain unclear[46,47]. It is now well recognized that more broad changes in the intestinal microbiome are associated with acute and chronic GI disease. Examples of recent studies in companion animals and their findings are summarized in Table 1. The cause-effect relationships of the alterations are still being elucidated, but especially in chronic enteropathies such as IBD there is now strong evidence that the gut microbiota plays an important part in the pathogenesis of the disease. Studies in humans have shown an association between IBD and microbial dysbiosis in the intestine. In these studies, a decrease in the bacterial phyla Firmicutes and Bacteroidetes, and an increase in Proteobacteria and Actinobacteria were associated with IBD[16]. Furthermore, a reduction in the diversity of Clostridium clusters XIVa and IV (i.e., Lachnospiraceae and C. coccoides subgroups) are associated with IBD, suggesting that this bacterial group may play an important role in maintenance of gastrointestinal health, possibly due to production of short chain fatty acids (SCFA). Similar studies have now been reported in dogs and cats with IBD, and a comparison of the observed microbial shifts for humans, dogs, and cats with IBD is provided in Table 2.
| Ref. | Species | Sampling location | Animal (sample size) | Method | Microbial changes in diseased animals |
| Suchodolski et al[], 2012 | Dog | Duodenal biopsies | IBD (n = 14) | 454-pyrosequencing | Increase in Proteobacteria (Diaphorobacter, Acinetobacter) |
| HC (n = 6) | (16S rRNA gene) | Reduction in Fusobacteria, Bacteroidaceae, Prevotellaceae, Clostridiales | |||
| Suchodolski et al[], 2010 | Dog | Duodenal biopsies | IBD (n = 7) | Gene clone libraries | Increase in Proteobacteria |
| HC (n = 7) | (16S rRNA gene) | Decrease in Clostridia | |||
| Allenspach et al[6], 2010 | Dog | Duodenal brushings | Chronic enteropathies (n = 13) | Gene clone libraries | Increase in Actinobacteria, Lactobacillales, Erysipelotrichales |
| HC (n = 8) | (16S rRNA gene) | ||||
| Xenoulis et al[9], 2008 | Dog | Duodenal brushings | IBD (n = 10) | Gene clone libraries | Increase in Enterobacteriaceae (E. coli); |
| HC (n = 9) | (16S rRNA gene) | Reduction in biodiversity | |||
| Suchodolski et al[36], 2008 | Dog | Duodenal brushings | Chronic enteropathies (n = 71) | Gene clone libraries | No significant differences in fungal communities |
| HC (n = 64) | (fungal ITS gene) | ||||
| Glanemann et al[52], 2008 | Dog | Stomach, duodenum, | Chronic GI disease (n = 42) | PCR | Presence of Mycobacterium avium subspecies paratuberculosis |
| Colon biopsies | HC (n = 14) | Detected in 8/42 (19%) of dogs with chronic GI disease | |||
| Manchester et al[59], 2013 | Dog | Colon biopsies | Granulomatous colitis (n = 6) | FISH | Presence of invasive E. coli |
| Simpson et al[58], 2006 | Dog | Colon biopsies | Granulomatous colitis (n = 13) | FISH | Intracellular translocation of adherent and invasive E. coli |
| HC (n = 38) | |||||
| Rossi et al[21], 2014 | Dog | Fecal samples | IBD (n = 20 ) | qPCR | Decreased in Faecalibacterium spp. And Turicibacter spp. |
| HC (n = 10) | (16S rRNA gene) | ||||
| Foster et al[62], 2013 | Dog | Fecal samples | Acute diarrhea (n = 7) | 454-pyrosequencing | No significant differences in fungal communities |
| HC (n = 12) | (18S rRNA gene) | ||||
| Suchodolski et al[14], 2012 | Dog | Fecal samples | IBD (n = 19) | 454-pyrosequencing | AHD: most profound alterations in their microbiome |
| AHD (n = 13) | (16S rRNA gene) | Increase in Sutterella, Clostridium perfringens | |||
| NHD (n = 12) | qPCR | Decrease in Blautia, Ruminococcaceae, Turicibacter | |||
| HC (n = 32) | (16S rRNA gene) | IBD: Decrease in Faecalibacterium spp., Fusobacteria | |||
| Markel et al[55], 2012 | Dog | Fecal samples | Chronic enteropathies (n = 87) | qPCR | Decrease in Faecalibacterium spp., Turicibacter spp., Ruminococcaceae |
| AHD (n = 48) | (16S rRNA gene) | Increase in C. perfringens and E. coli | |||
| HC (n = 180) | |||||
| Jia et al[65], 2010 | Dog | Fecal samples | Chronic diarrhea (n = 9) | FISH | Increase in Bacteroides |
| HC (n = 8) | |||||
| Glanemann et al[52], 2008 | Dog | Fecal samples | Diarrhea (n = 4) | T-RFLP | Increase in C. perfringens, E. faecalis, and E. faecium |
| HC (n = 9) | |||||
| Ghosh et al[13], 2013 | Cat | Ileum full-thick biopsies | Severe systemic ill (n = 50) | FISH | Increase in E. faecalis |
| HC (n = 50) | PCR | Attachemt of E. coli to intestinal epithelial cell | |||
| Janeczko et al[10], 2008 | Cat | Small intestine biopsies | IBD (n = 17) | FISH | Increase in Enterobacteriaceae |
| HC (n = 10) | |||||
| Abecia et al[54], 2010 | Cat | Fecal samples | IBD (n = 8) | FISH | No significant differences in specific bacterial populaltion |
| HC (n = 10) | |||||
| Inness et al[53], 2007 | Cat | Fecal samples | IBD (n = 11) | FISH | Decreased total bacteria, Bifidobacterium spp. and Bacteroides |
| HC (n = 34) | Increase in Desulfovibrio |
| Organism | Human (Crohn's disease) | Human (ulcerative colitis) | Dog | Cat |
| Firmicutes | 1245Decreased[15,18,56] | 124Decreased[15,18] | 16Decreased[7] | |
| Class Clostridia | 12467Decreased[7,8,14] | |||
| Family Ruminococcaceae (Clostridial cluster IV) | 24Decreased[15] | 24Decreased[15] | 12467Decreased[7,8,14] | |
| Family Lachnospiraceae (Clostridial cluster XIVa) | 1245Decreased[15,18] | 1245Decreased[15,18] | 167Decreased[7,8] | |
| Bacteroidetes | 14Decreased[18] | 14Decreased[18] | 167Decreased[7,9]; 17increased[8] | |
| Genus Bacteroides | 16Decreased[7] | 15Decreased[53] | ||
| Fusobacteria | 167Decreased[7,8] | |||
| Proteobacteria | 14Increased[18] | 14Increased[18] | 167Increased[7-9] | 15Increased[10] |
| Family Enterobacteriaceae | 14Unchanged[18]; 25increased [20] | 14Unchanged[18]; 25decreased[20] | 167Increased[7-9] | 15Increased[10] |
| E. coli | 24Unchanged[15]; 145Increased[19] | 17Increased[8] | ||
| Adherent-Invasive E. coli | 15Increased[19] | 135Increased[58] | ||
| Actinobacteria | 14Increased[18] | 14Increased[18] | 16Increased[7] | |
| Mycobacterium avium subspecies pseudotuberculosis | 1Controversial[18] | 14Increased[52] | ||
| Genus Bifidobacterium | 24Decreased[15] | 24Decreased[15] | 15Decreased[53] |
In veterinary medicine, chronic enteropathies with intestinal inflammation are commonly seen in dogs and cats. The response to treatment is used to allow for distinction of different types of enteropathies, such as food-responsive diarrhea, antibiotic-responsive diarrhea, and steroid-responsive diarrhea. Idiopathic IBD is a subgroup of enteropathies and it is defined as an inflammation of the GI tract with persistent or recurrent GI signs due to unknown cause[48]. To diagnose IBD, known causes for GI inflammation need to be excluded. Therefore, empirical treatments are applied sequentially, starting with a dietary trial, followed by antibiotic therapy if there is a lack of response to diet, and finally, treatment with anti-inflammatory drugs, if response to previous treatments was inadequate. Similarly to human IBD, the exact pathogenesis of canine/feline IBD is unknown, but is suspected to be the result of an abnormal interplay between an altered intestinal microbiota, an underlying genetic susceptibility of the host, and dietary and/or environmental factors[48]. Consequently, several studies have revealed possible underlying susceptibilities in the innate immune system of dogs and cats with chronic GI inflammation. These include altered differential expression of Toll-like receptors (TLR)-2 and 4[25,49], single nucleotide polymorphisms that lead to hyper-responsiveness of TLR-5 to flagellin in German Shepherd dogs (GSDs)[22], and decreased expression of CD11c(+) cells in dogs with IBD[50]. There is also well known anecdotal evidence that certain breeds are more prone to chronic GI inflammation. In addition to GSDs, which have been shown to possess polymorphisms in the TLR-4 and TLR-5 genes that are significantly associated with IBD[51], other dog breeds such as Rottweiler, Border Collie, Boxer dog, and Weimaraner have been shown to possess increased risks for developing IBD[23]. Of those breeds, breed specific studies evaluating the association between mucosa-adherent microbiota and intestinal inflammation were performed only in GSDs and Boxer dogs. In GSDs with chronic intestinal inflammation, the mucosa-adherent microbiota were analyzed in small intestinal brush samples and showed a significant over-representation of Bacilli and Erysipelotrichi when compared to healthy Greyhound dogs[6]. Interestingly, this is somewhat different to the results observed in other studies where more diverse populations of dogs with chronic intestinal inflammation were evaluated. In these studies, the most frequently observed changes in the mucosa-adherent microbiota in the small intestine were increases in members of the Proteobacteria, especially Escherichia coli-like organisms[9] or Pseudomonas[8], with concurrent decreases of members of Firmicutes and Bacteroidetes. In a more recent study evaluating mucosa-adherent microbiota in the duodenum of dogs with IBD by next-generation sequencing, the proportions of Fusobacteria, Bacteroidaceae, Prevotellaceae, and Clostridiales were significantly increased in healthy dogs. In contrast, specific bacterial genera within Proteobacteria, including Diaphorobacter and Acinetobacter, were either more abundant or more frequently identified in dogs with IBD[7]. One study evaluated specifically the presence of Mycobacterium avium subspecies paratuberculosis in duodenal biopsies of dogs with IBD or intestinal neoplasia by qPCR and reported that 19% of diseased dogs were PCR positive for this organism[52]. Less published information is available about the mucosa-adherent microbiota of cats with IBD. While sequencing methods have not yet been reported for the characterization of feline IBD, a study using fluorescent in situ hybridization (FISH) has revealed an increase in Enterobacteriaceae in duodenal biopsies of cats with IBD[10]. Furthermore, a relationship between increased bacterial numbers and the severity of histological inflammation was observed[10].
Several studies have evaluated the fecal microbiota in dogs and cats with chronic GI disease. In one study, cats with IBD had lower FISH counts for total bacteria, Bacteroides spp., and Bifidobacterium spp., but higher counts of Desulfovibrio spp. compared to healthy cats[53]. Desulfovibrio spp. are a sulfate-reducing bacterial group and able to produce hydrogen sulfides, which may be associated with the pathogenesis of feline IBD. However, another study did not identify significant differences in FISH counts between cats with IBD and controls, although the same bacterial groups were targeted[54]. A recent study utilized 454-pyrosequencing of 16S rRNA genes to describe changes in fecal microbiota in cats with chronic diarrhea and their response to dietary modifications[29]. Several bacterial groups correlated with improved fecal scores after therapeutic response to diet. Those included Slackia spp., Campylobacter upsaliensis, Enterobacteriaceae Raoultella spp., Collinsella spp., and unidentified genera within Clostridiales and Lachnospiraceae[29].
More data about the fecal microbiota are available in dogs. In one study, fecal samples from healthy dogs, dogs with acute non-hemorrhagic diarrhea, dogs with acute hemorrhagic diarrhea, and dogs with active or therapeutically controlled idiopathic IBD were analyzed by sequencing of the 16S rRNA gene[14]. Dogs with acute diarrhea, especially those with acute hemorrhagic diarrhea, had the most profound changes in bacterial groups in their microbiome. Dogs with acute hemorrhagic diarrhea had significant decreases in Blautia, Ruminococcaceae including Faecalibacterium, and Turicibacter spp., and significant increases in genus Sutterella and C. perfringens compared to healthy dogs. In another recent study, the fecal microbiome of healthy dogs, dogs with chronic enteropathies, and dogs with acute hemorrhagic diarrhea was evaluated by qPCR assays for selected bacterial groups[55]. The most pronounced changes were decreases in Faecalibacterium spp., Turicibacter spp., and Ruminococcaceae in CE and AHD. E. coli and C. perfringens were significantly increased in CE and AHD[55]. Especially Faecalibacterium spp. is an important group that frequently appears depleted in canine GI disease. This has been confirmed in another study evaluating the fecal microbiota of dogs with idiopathic IBD, in which Faecalibacterium spp. was the major bacterial group decreased in diseased dogs[21]. Noteworthy, Faecalibacterium spp. correlated with improvement in clinical activity index, suggesting that Faecalibacterium spp. may be important for canine GI health, and also may be useful as a monitoring marker for improvement of fecal dysbiosis[14,21].
While the above discussed studies have reported changes in microbial groups in GI disease of dogs and cats, only limited information is available about the metabolic consequences that are associated with this dysbiosis, as currently no comprehensive functional studies have been reported in dogs or cats. Alterations in the composition of intestinal microbiota are thought to be an important factor in the pathogenesis of chronic GI diseases. It can be hypothesized that the observed microbiome changes may lead to altered intestinal barrier function, damage to the intestinal brush border and enterocytes, an increased competition for nutrients and vitamins, and to an increased deconjugation of bile acids. Of interest is that commonly depleted groups in GI disease are Lachnospiraceae, Ruminococcaceae, and Faecalibacterium. These bacterial groups, important producers of SCFA, may play an important role in maintenance of gastrointestinal health, as their depletion leads to decreased production of SCFA (e.g., butyrate, acetate), which may impair the capability of the host to down-regulate aberrant intestinal immune response. The importance of some of these bacterial groups that are depleted in IBD have recently been demonstrated in humans. For example, Faecalibacterium prausnitzii is consistently reduced in human IBD and this bacterium has been shown to secrete metabolites with anti-inflammatory properties, thereby down-regulating interleukin (IL)-12 and interferon gamma and increasing IL-10 secretions[56]. Disturbances may result in a dysregulation of adaptive immune responses, and lead to inflammation and/or reduced activity against infection. Also, some bacteria produce various toxic agents such as ammonia, D-lactate, endotoxin (LPS), or exotoxin (enterotoxin), and compete for vitamins or other nutrients. Consequently, depletions in serum vitamin B12 concentrations and also increases in serum concentrations of D-lactate are potential consequences of intestinal dysbiosis in cats[57]. However, more comprehensive metabolomics studies are needed in companion animals to elucidate the consequences of the dysbiosis observed in GI disease.
A specific form of colitis occurs in Boxer dogs[58] and occasionally also in French Bulldogs[59]. This disease is termed granulomatous colitis. Microbiota analysis based on sequencing of 16S rRNA genes in combination with FISH has revealed invasive bacteria in the colonic mucosa of Boxer dogs with granulomatous colitis. Based on comparative 16S rRNA gene analysis, these bacteria have high phylogenetic similarity to Escherichia coli (E. coli) and Shigella. In situ analysis with 16S rRNA gene based FISH probes against E. coli showed multifocal clusters of invasive bacteria within macrophages in the colonic mucosa[58]. The eradication of these invasive E. coli in Boxer dogs and French Bulldogs with granulomatous colitis correlates with clinical remission, inferring a causal relationship between these bacteria and the disease[59]. Of interest is that these observed phylotypes of E. coli isolated from Boxer dogs have high phylogenetic resemblance to E. coli associated with Crohn’s disease in humans[16,59]. The breed specific predisposition of Boxer dogs and French bulldogs to E. coli associated granulomatous colitis highly suggests the presence of a genetic susceptibility that impairs their ability to fend off adherent and invasive E. coli.
Bacteria invading the intestinal mucosa may also be part of neutrophilic IBD in other dog breeds. Due to the recognized association of granulomatous and neutrophilic IBD with invasive bacteria, specialized testing based on FISH has been developed that allows localizing the bacteria in intestinal biopsies for better guidance of treatment decisions[59].
While bacteria are by far the most abundant constituents of the mammalian GI tract, it is now recognized that the gut harbors a highly diverse population of fungal organisms. FISH and shotgun sequencing studies of human and canine fecal DNA have estimated the abundance of fungal organisms and archaea as < 2% of total microbiota[39,60]. A recent metagenomic approach estimated that the feline GI microbiota constitutes 0.02% fungi, 0.09% archaea, and 0.09% viruses[41]. Fungi were described using pyrosequencing of the fungal 18S rRNA gene in pooled fecal samples of cats[3], with Aspergillus and Saccharomyces being the most abundant fungal genera. A study reported the prevalence and identification of fungal organisms in the small intestine of healthy dogs and dogs with chronic enteropathies[61]. The results indicated a high prevalence (up to 76.1% of dogs) and high diversity of fungal organisms in the canine duodenum. Furthermore, dogs with gastrointestinal disease harbored opportunistic fungal pathogens. A total of 51 different phylotypes were identified, with the most frequently observed phylotypes being Pichia spp., Cryptococcus spp., Candida spp., and Trichosporon spp.[61].
A recent study has characterized the fungal microbiome (mycobiome) of 19 dogs (12 healthy dogs and 7 dogs with acute diarrhea) using fungal tag-encoded FLX-Titanium amplicon pyrosequencing[62]. Five distinct fungal phyla were identified, with Ascomycota (median: 97.9% of obtained sequences) and Basidiomycota (median 1.0%) being the most abundant. A total of 219 fungal genera were identified across all 19 dogs with a median (range) of 28 (4-69) genera per sample. Candida was the most abundant genus found in dogs. However, no significant differences were observed in the relative proportions of fungal communities between healthy and diseased dogs. Therefore, additional studies are needed to elucidate the importance of fungi on intestinal health and disease of animals.
Studies using molecular approaches have provided clear evidence for alterations in microbial communities in the small and large intestine of dogs and cats with GI disorders. However, currently there is a lack of comprehensive studies evaluating the functional consequences of these alterations. A better understanding of these mechanisms will allow for the development of treatment modalities (e.g., prebiotics, probiotics, metabolites) aiming at modulating microbial communities and their produced metabolites. Anecdotal case reports have reported some success using fecal transplantation in dogs with chronic diarrhea. Results of initial studies suggest that the administration of probiotic strains can be useful in dogs with GI disease. For example, probiotic strains have shown benefits in dogs with IBD[21], puppies with acute parvoviral enteritis[63], and adult dogs with non-specific diarrhea[26,27]. In cats, probiotics strains have been shown to be beneficial in cats with chronic diarrhea[28] and stress-related diarrhea in a shelter environment[64]. However, future studies will need to evaluate how these microbial changes impact the immune and metabolic status of dogs and cats.
P- Reviewer: M’Koma A, Jovanovic I, Zhang L S- Editor: Gou SX L- Editor: A E- Editor: Zhang DN
| 1. | Suchodolski JS, Dowd SE, Westermarck E, Steiner JM, Wolcott RD, Spillmann T, Harmoinen JA. The effect of the macrolide antibiotic tylosin on microbial diversity in the canine small intestine as demonstrated by massive parallel 16S rRNA gene sequencing. BMC Microbiol. 2009;9:210. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 141] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 2. | Hand D, Wallis C, Colyer A, Penn CW. Pyrosequencing the canine faecal microbiota: breadth and depth of biodiversity. PLoS One. 2013;8:e53115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 71] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 3. | Handl S, Dowd SE, Garcia-Mazcorro JF, Steiner JM, Suchodolski JS. Massive parallel 16S rRNA gene pyrosequencing reveals highly diverse fecal bacterial and fungal communities in healthy dogs and cats. FEMS Microbiol Ecol. 2011;76:301-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 265] [Cited by in RCA: 291] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 4. | Sunvold GD, Fahey GC, Merchen NR, Titgemeyer EC, Bourquin LD, Bauer LL, Reinhart GA. Dietary fiber for dogs: IV. In vitro fermentation of selected fiber sources by dog fecal inoculum and in vivo digestion and metabolism of fiber-supplemented diets. J Anim Sci. 1995;73:1099-1109. [PubMed] |
| 5. | Sunvold GD, Fahey GC, Merchen NR, Bourquin LD, Titgemeyer EC, Bauer LL, Reinhart GA. Dietary fiber for cats: in vitro fermentation of selected fiber sources by cat fecal inoculum and in vivo utilization of diets containing selected fiber sources and their blends. J Anim Sci. 1995;73:2329-2339. [PubMed] |
| 6. | Allenspach K, House A, Smith K, McNeill FM, Hendricks A, Elson-Riggins J, Riddle A, Steiner JM, Werling D, Garden OA. Evaluation of mucosal bacteria and histopathology, clinical disease activity and expression of Toll-like receptors in German shepherd dogs with chronic enteropathies. Vet Microbiol. 2010;146:326-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 80] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 7. | Suchodolski JS, Dowd SE, Wilke V, Steiner JM, Jergens AE. 16S rRNA gene pyrosequencing reveals bacterial dysbiosis in the duodenum of dogs with idiopathic inflammatory bowel disease. PLoS One. 2012;7:e39333. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 185] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 8. | Suchodolski JS, Xenoulis PG, Paddock CG, Steiner JM, Jergens AE. Molecular analysis of the bacterial microbiota in duodenal biopsies from dogs with idiopathic inflammatory bowel disease. Vet Microbiol. 2010;142:394-400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 124] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 9. | Xenoulis PG, Palculict B, Allenspach K, Steiner JM, Van House AM, Suchodolski JS. Molecular-phylogenetic characterization of microbial communities imbalances in the small intestine of dogs with inflammatory bowel disease. FEMS Microbiol Ecol. 2008;66:579-589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 169] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 10. | Janeczko S, Atwater D, Bogel E, Greiter-Wilke A, Gerold A, Baumgart M, Bender H, McDonough PL, McDonough SP, Goldstein RE. The relationship of mucosal bacteria to duodenal histopathology, cytokine mRNA, and clinical disease activity in cats with inflammatory bowel disease. Vet Microbiol. 2008;128:178-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 136] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 11. | Desai AR, Musil KM, Carr AP, Hill JE. Characterization and quantification of feline fecal microbiota using cpn60 sequence-based methods and investigation of animal-to-animal variation in microbial population structure. Vet Microbiol. 2009;137:120-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 48] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 12. | Craven M, Mansfield CS, Simpson KW. Granulomatous colitis of boxer dogs. Vet Clin North Am Small Anim Pract. 2011;41:433-445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 13. | Ghosh A, Borst L, Stauffer SH, Suyemoto M, Moisan P, Zurek L, Gookin JL. Mortality in kittens is associated with a shift in ileum mucosa-associated enterococci from Enterococcus hirae to biofilm-forming Enterococcus faecalis and adherent Escherichia coli. J Clin Microbiol. 2013;51:3567-3578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 14. | Suchodolski JS, Markel ME, Garcia-Mazcorro JF, Unterer S, Heilmann RM, Dowd SE, Kachroo P, Ivanov I, Minamoto Y, Dillman EM. The fecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PLoS One. 2012;7:e51907. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 249] [Cited by in RCA: 333] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 15. | Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, Cosnes J, Corthier G, Marteau P, Doré J. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183-1189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 857] [Cited by in RCA: 911] [Article Influence: 56.9] [Reference Citation Analysis (0)] |
| 16. | Packey CD, Sartor RB. Commensal bacteria, traditional and opportunistic pathogens, dysbiosis and bacterial killing in inflammatory bowel diseases. Curr Opin Infect Dis. 2009;22:292-301. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 263] [Cited by in RCA: 251] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 17. | Craven M, Egan CE, Dowd SE, McDonough SP, Dogan B, Denkers EY, Bowman D, Scherl EJ, Simpson KW. Inflammation drives dysbiosis and bacterial invasion in murine models of ileal Crohn’s disease. PLoS One. 2012;7:e41594. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 153] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 18. | Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780-13785. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3731] [Cited by in RCA: 3421] [Article Influence: 190.1] [Reference Citation Analysis (1)] |
| 19. | Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R, Orsi RH, Wiedmann M, McDonough P, Kim SG. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J. 2007;1:403-418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 456] [Cited by in RCA: 480] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 20. | Swidsinski A, Loening-Baucke V, Vaneechoutte M, Doerffel Y. Active Crohn’s disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflamm Bowel Dis. 2008;14:147-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 207] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 21. | Rossi G, Pengo G, Caldin M, Palumbo Piccionello A, Steiner JM, Cohen ND, Jergens AE, Suchodolski JS. Comparison of microbiological, histological, and immunomodulatory parameters in response to treatment with either combination therapy with prednisone and metronidazole or probiotic VSL#3 strains in dogs with idiopathic inflammatory bowel disease. PLoS One. 2014;9:e94699. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 148] [Cited by in RCA: 184] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 22. | Kathrani A, Holder A, Catchpole B, Alvarez L, Simpson K, Werling D, Allenspach K. TLR5 risk-associated haplotype for canine inflammatory bowel disease confers hyper-responsiveness to flagellin. PLoS One. 2012;7:e30117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 23. | Kathrani A, Werling D, Allenspach K. Canine breeds at high risk of developing inflammatory bowel disease in the south-eastern UK. Vet Rec. 2011;169:635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 56] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 24. | Luckschander N, Hall JA, Gaschen F, Forster U, Wenzlow N, Hermann P, Allenspach K, Dobbelaere D, Burgener IA, Welle M. Activation of nuclear factor-kappaB in dogs with chronic enteropathies. Vet Immunol Immunopathol. 2010;133:228-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 35] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 25. | McMahon LA, House AK, Catchpole B, Elson-Riggins J, Riddle A, Smith K, Werling D, Burgener IA, Allenspach K. Expression of Toll-like receptor 2 in duodenal biopsies from dogs with inflammatory bowel disease is associated with severity of disease. Vet Immunol Immunopathol. 2010;135:158-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 50] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 26. | Kelley RL, Minikhiem D, Kiely B, O’Mahony L, O’Sullivan D, Boileau T, Park JS. Clinical benefits of probiotic canine-derived Bifidobacterium animalis strain AHC7 in dogs with acute idiopathic diarrhea. Vet Ther. 2009;10:121-130. [PubMed] |
| 27. | Herstad HK, Nesheim BB, L’Abée-Lund T, Larsen S, Skancke E. Effects of a probiotic intervention in acute canine gastroenteritis--a controlled clinical trial. J Small Anim Pract. 2010;51:34-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 61] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 28. | Hart ML, Suchodolski JS, Steiner JM, Webb CB. Open-label trial of a multi-strain synbiotic in cats with chronic diarrhea. J Feline Med Surg. 2012;14:240-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 29. | Ramadan Z, Xu H, Laflamme D, Czarnecki-Maulden G, Li QJ, Labuda J, Bourqui B. Fecal microbiota of cats with naturally occurring chronic diarrhea assessed using 16S rRNA gene 454-pyrosequencing before and after dietary treatment. J Vet Intern Med. 2014;28:59-65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 30. | German AJ, Day MJ, Ruaux CG, Steiner JM, Williams DA, Hall EJ. Comparison of direct and indirect tests for small intestinal bacterial overgrowth and antibiotic-responsive diarrhea in dogs. J Vet Intern Med. 2003;17:33-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 56] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 31. | Johnston K, Lamport A, Batt RM. An unexpected bacterial flora in the proximal small intestine of normal cats. Vet Rec. 1993;132:362-363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 48] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 32. | Benno Y, Nakao H, Uchida K, Mitsuoka T. Impact of the advances in age on the gastrointestinal microflora of beagle dogs. J Vet Med Sci. 1992;54:703-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 81] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 33. | Mentula S, Harmoinen J, Heikkilä M, Westermarck E, Rautio M, Huovinen P, Könönen E. Comparison between cultured small-intestinal and fecal microbiotas in beagle dogs. Appl Environ Microbiol. 2005;71:4169-4175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 85] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 34. | Middelbos IS, Vester Boler BM, Qu A, White BA, Swanson KS, Fahey GC. Phylogenetic characterization of fecal microbial communities of dogs fed diets with or without supplemental dietary fiber using 454 pyrosequencing. PLoS One. 2010;5:e9768. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 165] [Cited by in RCA: 206] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 35. | Ritchie LE, Steiner JM, Suchodolski JS. Assessment of microbial diversity along the feline intestinal tract using 16S rRNA gene analysis. FEMS Microbiol Ecol. 2008;66:590-598. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 120] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 36. | Suchodolski JS, Camacho J, Steiner JM. Analysis of bacterial diversity in the canine duodenum, jejunum, ileum, and colon by comparative 16S rRNA gene analysis. FEMS Microbiol Ecol. 2008;66:567-578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 170] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 37. | Ritchie LE, Burke KF, Garcia-Mazcorro JF, Steiner JM, Suchodolski JS. Characterization of fecal microbiota in cats using universal 16S rRNA gene and group-specific primers for Lactobacillus and Bifidobacterium spp. Vet Microbiol. 2010;144:140-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 65] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 38. | Garcia-Mazcorro JF, Suchodolski JS, Jones KR, Clark-Price SC, Dowd SE, Minamoto Y, Markel M, Steiner JM, Dossin O. Effect of the proton pump inhibitor omeprazole on the gastrointestinal bacterial microbiota of healthy dogs. FEMS Microbiol Ecol. 2012;80:624-636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 98] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 39. | Swanson KS, Dowd SE, Suchodolski JS, Middelbos IS, Vester BM, Barry KA, Nelson KE, Torralba M, Henrissat B, Coutinho PM. Phylogenetic and gene-centric metagenomics of the canine intestinal microbiome reveals similarities with humans and mice. ISME J. 2011;5:639-649. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 244] [Cited by in RCA: 256] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 40. | Barry KA, Middelbos IS, Vester Boler BM, Dowd SE, Suchodolski JS, Henrissat B, Coutinho PM, White BA, Fahey GC, Swanson KS. Effects of dietary fiber on the feline gastrointestinal metagenome. J Proteome Res. 2012;11:5924-5933. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 66] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 41. | Tun HM, Brar MS, Khin N, Jun L, Hui RK, Dowd SE, Leung FC. Gene-centric metagenomics analysis of feline intestinal microbiome using 454 junior pyrosequencing. J Microbiol Methods. 2012;88:369-376. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 71] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 42. | Minamoto Y, Hooda S, Swanson KS, Suchodolski JS. Feline gastrointestinal microbiota. Anim Health Res Rev. 2012;13:64-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 35] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 43. | Hooda S, Minamoto Y, Suchodolski JS, Swanson KS. Current state of knowledge: the canine gastrointestinal microbiome. Anim Health Res Rev. 2012;13:78-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 63] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 44. | Suchodolski JS. Companion animals symposium: microbes and gastrointestinal health of dogs and cats. J Anim Sci. 2011;89:1520-1530. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 111] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 45. | Suchodolski JS. Intestinal microbiota of dogs and cats: a bigger world than we thought. Vet Clin North Am Small Anim Pract. 2011;41:261-272. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 76] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 46. | Queen EV, Marks SL, Farver TB. Prevalence of selected bacterial and parasitic agents in feces from diarrheic and healthy control cats from Northern California. J Vet Intern Med. 2012;26:54-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 46] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 47. | Marks SL, Rankin SC, Byrne BA, Weese JS. Enteropathogenic bacteria in dogs and cats: diagnosis, epidemiology, treatment, and control. J Vet Intern Med. 2011;25:1195-1208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 178] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 48. | Simpson KW, Jergens AE. Pitfalls and progress in the diagnosis and management of canine inflammatory bowel disease. Vet Clin North Am Small Anim Pract. 2011;41:381-398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 116] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 49. | Burgener IA, Jungi TW. Dysregulation of toll-like receptors in inflammatory bowel disease and food-responsive diarrhea. J Vet Intern Med. 2007;21:612-612. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 50. | Kathrani A, Schmitz S, Priestnall SL, Smith KC, Werling D, Garden OA, Allenspach K. CD11c+ cells are significantly decreased in the duodenum, ileum and colon of dogs with inflammatory bowel disease. J Comp Pathol. 2011;145:359-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 51. | Kathrani A, House A, Catchpole B, Murphy A, German A, Werling D, Allenspach K. Polymorphisms in the TLR4 and TLR5 gene are significantly associated with inflammatory bowel disease in German shepherd dogs. PLoS One. 2010;5:e15740. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 94] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 52. | Glanemann B, Schönenbrücher H, Bridger N, Abdulmawjood A, Neiger R, Bülte M. Detection of Mycobacterium avium subspecies paratuberculosis-specific DNA by PCR in intestinal biopsies of dogs. J Vet Intern Med. 2008;22:1090-1094. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 53. | Inness VL, McCartney AL, Khoo C, Gross KL, Gibson GR. Molecular characterisation of the gut microflora of healthy and inflammatory bowel disease cats using fluorescence in situ hybridisation with special reference to Desulfovibrio spp. J Anim Physiol Anim Nutr (Berl). 2007;91:48-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 77] [Article Influence: 4.3] [Reference Citation Analysis (1)] |
| 54. | Abecia L, Hoyles L, Khoo C, Frantz N, McCartney AL. Effects of a novel galactooligosaccharide on the faecal microbiota of healthy and inflammatory bowel disease cats during a randomized, double-blind, cross-over feeding study. Int J Probiotics Prebiotics. 2010;5:61. |
| 55. | Markel M, Berghoff N, Unterer S, Oliveira-Barros LM, Grellet A, Allenspach K, Toresson L, Luckschander N, Suchodolski JS. Characterization of fecal dysbiosis in dogs with chronic enteropathies and acute hemorrhagic diarrhea. J Vet Internal Med. 2012;26:765-766. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 56. | Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731-16736. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2747] [Cited by in RCA: 3190] [Article Influence: 187.6] [Reference Citation Analysis (0)] |
| 57. | Packer RA, Moore GE, Chang CY, Zello GA, Abeysekara S, Naylor JM, Steiner JM, Suchodolski JS, O’Brien DP. Serum D-lactate concentrations in cats with gastrointestinal disease. J Vet Intern Med. 2012;26:905-910. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (1)] |
| 58. | Simpson KW, Dogan B, Rishniw M, Goldstein RE, Klaessig S, McDonough PL, German AJ, Yates RM, Russell DG, Johnson SE. Adherent and invasive Escherichia coli is associated with granulomatous colitis in boxer dogs. Infect Immun. 2006;74:4778-4792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 195] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 59. | Manchester AC, Hill S, Sabatino B, Armentano R, Carroll M, Kessler B, Miller M, Dogan B, McDonough SP, Simpson KW. Association between granulomatous colitis in French Bulldogs and invasive Escherichia coli and response to fluoroquinolone antimicrobials. J Vet Intern Med. 2013;27:56-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 60. | Scanlan PD, Marchesi JR. Micro-eukaryotic diversity of the human distal gut microbiota: qualitative assessment using culture-dependent and -independent analysis of faeces. ISME J. 2008;2:1183-1193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 229] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 61. | Suchodolski JS, Morris EK, Allenspach K, Jergens AE, Harmoinen JA, Westermarck E, Steiner JM. Prevalence and identification of fungal DNA in the small intestine of healthy dogs and dogs with chronic enteropathies. Vet Microbiol. 2008;132:379-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 35] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 62. | Foster ML, Dowd SE, Stephenson C, Steiner JM, Suchodolski JS. Characterization of the fungal microbiome (mycobiome) in fecal samples from dogs. Vet Med Int. 2013;2013:658373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 46] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 63. | Arslan HH, Aksu DS, Terzi G, Nisbet C. Therapeutic effects of probiotic bacteria in parvoviral enteritis in dogs. Rev Med Vet-Toulouse. 2012;163:55-59. |
| 64. | Bybee SN, Scorza AV, Lappin MR. Effect of the probiotic Enterococcus faecium SF68 on presence of diarrhea in cats and dogs housed in an animal shelter. J Vet Intern Med. 2011;25:856-860. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 91] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 65. | Jia J, Frantz N, Khoo C, Gibson GR, Rastall RA, McCartney AL. Investigation of the faecal microbiota associated with canine chronic diarrhoea. FEMS Microbiol Ecol. 2010;71:304-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 58] [Article Influence: 3.6] [Reference Citation Analysis (0)] |