Published online Sep 28, 2014. doi: 10.3748/wjg.v20.i36.13088
Revised: March 11, 2014
Accepted: April 30, 2014
Published online: September 28, 2014
Processing time: 271 Days and 7.5 Hours
AIM: To investigate over-expression of Osteopontin (OPN) pathway expression and mechanisms of action in human alcoholic liver disease (ALD), in vivo and in vitro acute alcohol models.
METHODS: OPN pathway was evaluated in livers from patients with progressive stages of human ALD and serum from drinkers with and without liver cirrhosis. In vitro stellate LX2 cells exposed to acute alcohol and in vivo in acute alcoholic steatosis mouse models were also investigated for OPN pathway expression and function. WT and OPN-/- mice were administered an acute dose of alcohol and extent of liver injury was examined by histopathology and liver biochemistry after 16-24 h. The causative role of OPN was studied in OPN knockout animals and in vitro in stellate LX2 cells, utilizing siRNA, aptamer and neutralizing antibodies to block OPN and OPN pathway. OPN pathway expression and downstream functional consequences were measured for signaling by Western blotting, plasmin activation by spectrophotometric assays and cell migration by confocal imaging and quantitation.
RESULTS: OPN expression positively correlated with disease severity in patients with progressive stages of ALD. In vivo, associated with alcoholic steatosis, a single dose of acute alcohol significantly increased hepatic OPN mRNA and protein, and a cleaved OPN form in a dose dependent manner. OPN mRNA and secreted OPN also increased in parallel with activation of LX2 stellate cells within 4 h of a single dose of alcohol. Expression of OPN receptors, αvβ3-integrin and CD44, increased in human ALD, and in vivo and in vitro with alcohol administration. This was accompanied by downstream phosphorylation of Akt and Erk, increased mRNA expression of several fibrogenesis, fibrinolysis and extracellular matrix pathway genes, plasmin activation and hepatic stellate cell (HSC) migration. Inhibition of OPN and OPN-receptor mediated signaling partially inhibited alcohol-induced HSC activation, plasmin activity and cell migration.
CONCLUSION: OPN is a key mediator of the alcohol-induced effects on hepatic stellate cell functions and liver fibrogenesis.
Core tip: The present study confirms a novel hypothesis that alcohol induced plasmin is mediated via Osteopontin (OPN) in hepatic stellate cell (HSC). We show that OPN has a key role in alcohol-induced HSC functions such as signalling, cell migration and activation of fibrinolysis, extracellular matrix and fibrogenic pathways. Identification of transcriptional isoform OPN-C in patients with alcoholic cirrhosis and LX2, and proteolytically cleaved cOPN in mice with a single dose of alcohol is novel. Importantly, we have defined novel mechanisms of OPN action in alcohol-induced liver injury that have a broader significance in other forms of liver injury.
-
Citation: Seth D, Duly A, Kuo PC, McCaughan GW, Haber PS. Osteopontin is an important mediator of alcoholic liver disease
via hepatic stellate cell activation. World J Gastroenterol 2014; 20(36): 13088-13104 - URL: https://www.wjgnet.com/1007-9327/full/v20/i36/13088.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i36.13088
Over-expression of Osteopontin (OPN) in human alcoholic liver disease (ALD) was first identified by our group[1,2]. We showed significantly up-regulated OPN at the portal-parenchymal interface in reactive biliary ductules and other liver cells in cirrhotic patients[1,2]. Since then, others have confirmed our findings[3] and have demonstrated increased OPN in alcoholic hepatitis patients[4]. Experimental administration of alcohol and lipopolysaccharide (LPS) in rats led to increased OPN in association with liver disease[5,6]. Up-regulated OPN was also associated with phosphorylated Akt (P-Akt)[7], collagen 1 (Col1) and TNF-α indicating activation of fibrotic events in non-alcoholic steatohepatitis (NASH)[8], liver necrosis in the carbon tetrachloride (CCl4) model[9], and elevated serum alanine aminotransferase (ALT) levels in a drug induced liver injury mouse model[10]. These data suggest that OPN has a pathogenic role in liver injury. Nonetheless, it is unclear whether over-expression of OPN is a cause or association of tissue injury. Furthermore, little is known about the respective contribution of the multiple OPN isoforms arising from transcriptional splicing and post-translational modifications[11]. Transcriptional isoforms are known to be associated with cancers, specifically OPN-C is linked to more aggressive tumors and poor prognosis[12,13], but its role remains controversial[14]. We were the first to observe differential expression and functions of OPN-C isoform in hepatocyte and stellate cell culture models of alcohol[15], indicating the importance of studying its role in alcoholic liver injury.
One of the intriguing effects of OPN in metastatic cancer cells is enhancement of plasmin activation by increasing urokinase plasminogen activator (μPA) secretion through Akt[16] and Erk-dependent pathways[17,18]. In liver injury, activation of plasmin contributes to both tissue remodelling during fibrogenesis and recovery from hepatic injury by promoting fibrinolysis and removing fibrin[19,20]. Hepatic stellate cell (HSC) mediated plasminogen (PLG) activation and extracellular matrix (ECM) remodeling are recognised components of fibrogenesis[19]. In alcoholic liver injury, increased plasminogen activators have been shown to regulate liver matrix remodelling through activation of plasminogen to plasmin[21,22]. We demonstrated that acute experimental administration of alcohol increased plasminogen, leading to altered plasmin and fibrinolysis, both in vivo and in vitro in liver cells, including HSCs[23]. However, the role of OPN in these actions of alcohol has not been clearly defined. This study examined the hypothesis that OPN drives alcohol induced plasmin regulation in liver cells and contributes to the process of fibrogenesis in ALD.
This work has been carried out in accordance with the Human Ethics Review Committee of Royal Prince Alfred Hospital (HREC/09/RPAH/148; HREC/11/RPAH/88) and The University of Sydney Animal Welfare Committee (K75-8-2009-3-5157 and K75-8-2009-3-4978).
Hepatic mRNAs from patients with progressive stages of ALD and non-diseased donor liver described previously[1] were used for expression analysis. Serum samples from drinkers consuming > 50 g/d (female) and > 80 g/d (male) with no liver disease (alcoholic control) and with liver cirrhosis (alcoholic cirrhosis) (Table 1) were tested for secreted OPN by enzyme-linked immunosorbent assay (ELISA).
All animals used in these experiments were 9-11 wk-old C57BL6 female mice. WT mice were administered a single dose of ethanol (2 g/4 g/6 g per kg body weight for 16 h) or equal volume of saline (n = 4/group) as described[23]. In another set of experiment, an additional subset of WT and OPN knockout (OPN-/-) mice (global OPN deficient in C57BL6 background)[24] were given 6 g/kg alcohol (OPN-/-n = 8; WT n = 6) or saline (Control: OPN-/-n = 7; WT n = 5). Blood and tissue samples were collected 24 h post-gavage. Serum was tested for liver functions (ALT, aspartate aminotransferase) as described[23] and liver cholesterol and triglyceride content were determined at the Sydney South West Pathology Services, RPAH. Liver tissue was used for histology, RNA and protein extraction.
Hepatic stellate LX2[23,25] cells were maintained in DMEM with 10% fetal calf serum unless otherwise specified. For alcohol and antibody inhibition studies, cells were seeded at a density of 3 × 106 cells/well in serum-free media in 6-well plates overnight prior to treatment. Cells were incubated ± 10 mmol/L ethanol (alcohol) for 1 h (signaling), 4 h (RNA, protein, migration, immunohistochemistry) and 16h (plasmin assay). For immunohistochemistry, cells (1 × 104 per well) were seeded in chamber slides with and without alcohol treatment. Treatments included 1 mmol/L 4-methylpyrazole (4-MP) to block alcohol metabolism, and antibodies (Table 2) against OPN, CD44v6, αvβ3-integrin (ITGAV), and OPN-R3 aptamer[26] to inhibit OPN pathway. For signaling experiments, insulin (100 nmol/L) was added 30 min prior to the end of the incubation period to induce baseline phosphorylation in LX2 cells.
| Specificity | Host | Isotype | Use | Dilution | Samples | Source (Catalog No.) |
| Primary antibodies | ||||||
| OPNK20 (SPP1) | Goat | Polyclonal IgG | WB | 1:1000 | Human, LX2 | Santa Cruz, Santa Cruz, CA, United States (sc-10591) |
| Blocking | 200 ìg/mL | |||||
| OPN (AF808) | Goat | Polyclonal IgG | WB | 1:1000 | Mice | RD (AF808) |
| Capture anti-OPN | Mouse | Monoclonal | ELISA | 3 ìg/mL | Human, LX2 | RD (MAB14331) |
| Detection biotynlated anti-OPN | Goat | Polyclonal IgG | ELISA | 200 ng/mL | Human, LX2 | RD (BAF1433) |
| OPN-R3 aptamer blocks SPP1 binding to receptors | 100 nmol/L | LX2 | Duke University, NC, United States | |||
| αvβ3-integrin (CD51/61) (ITGAV) | Mouse | Monoclonal | Blocking | 10 ìg/mL | LX2 | RD, Minneapolis, MN, United States (MAB3050) |
| CD44v6 | Mouse | Monoclonal IgG1 | Blcoking | 1:1000 | LX2 | RD (BBA13) |
| Vimentin | Rat | Monoclonal | IF | 1:200 | LX2 | RD (MAB2105) |
| Phospho-Akt Ser473 (P-Akt) | Rabbit | Polyclonal | WB | 1:1000 | LX2, Mice | Cell Signalling Technology (No. 9271) |
| Total-Akt (pan) | Rabbit | Polyclonal IgG | WB | 1:1000 | LX2, Mice | Cell Signalling Technology (No. 4691) |
| Phospho-Erk p44/42 (P-Erk) | Mouse | Monoclonal IgG | WB | 1:2000 | LX2, Mice | Cell Signalling Technology, Danvers, MA, United States (No. 9106) |
| Total-Erk (T-Erk) | Rabbit | Polyclonal | WB | 1:1000 | LX2, Mice | Cell Signalling Technology (No. 9102) |
| GAPDH | Mouse | Monoclonal | WB | 1:200000 | Mice | Ambion (AM4300) |
| Secondary antibodies | ||||||
| Anti-mouse HRP (P0161) | 1:10000 | Dako Australia Pty Ltd, Campbellfield, VIC, Australia | ||||
| Anti-rabbit HRP (P0449) | 1:7500 | |||||
| Anti-goat HRP (P0448) | 1:75000 | |||||
| AlexaFluor 488; AlexaFluor 594 | 1:400 | Molecular probes | ||||
Amount of secreted OPN in serum and culture media (CM) was determined by ELISA as described[27,28]. Briefly, 96-well plates were coated overnight with 3 μg/mL anti-human OPN antibody (Table 2) and blocked with 5% skim milk. 100 μL serum or CM from cells and recombinant human OPN (0-125 ng/mL) for standard curve, were added to wells. Streptavidin-HRP (1/10000 dilution, Dako, Carpinteria, CA) and SigmaFast were used for spectrophotometric detection of secreted OPN (ng/mL) at 450 nm.
Quantitative-polymerase chain reaction (Q-PCR) was carried out as described previously[1,2,23,29] using specific human and mouse primers (Table 3).
| Target gene | Forward (5’→3’) | Reverse (5’→3’) | Expected size (bps) | Ref. |
| OPN and OPN pathway | ||||
| Total h-OPN | AATGGTGCATACAAGGCCATC | TGTCCTTCCCACGGCTGT | 90 | NM_000582[1] |
| h-OPN-C | CTGAGGAAAAGCAGAATG | AATGGAGTCCTGGCTGT | 149 | [40] |
| h-CD44v6 | GCTTTCAATAGCACCTTGCC | GTTGCCAAACCACTGTTCCT | 867 | [41] |
| h-αvβ3-integrin | TTGTTGCTACTGGCTGTTTTG | TCCCTTTCTTGTTCTTCTTGAG | 89 | NM_002210 |
| m-OPN | CAGCCTGCAAGATCCTA | GCGCAAGGAGATTCTGCT TCT | 71 | [7] |
| m-αvβ3-integrin | GTAATCGAGATGCCCCAGAG | CTTCCATCCAGGGCAATATG | 142 | [42] |
| m-CD44 | TCTGCCATCTAGCACTAAGAGC | GTCTGGGTATTGAAAGGTGTAGC | 106 | [43] |
| Fibrogenesis and ECM | ||||
| h-TGFβ | CCACAACGAAATCTATGAC | CGGTGACATCAAAAGATAAC | 260 | NM_000660 |
| m-TGF-β | CCTTCCTGCTCCTCATGGCCA | GTCCTTCCTAAAGTCAATGTA | 149 | [44] |
| h-Collagen 1α1 | GATTCCCTGGACCTAAAGGTGC | AGCCTCTCCATCTTTGCCAGCA | 106 | NM_000088.3 |
| m-Collagen 1 (Col1) | GAGCGGAGAGTACTGGATCG | GCTTCTTTTCCTTGGGGTTC | 158 | [45] |
| h-Collagen 4α4 | CAGGCTCAACTGGTCTAAGAGG | AGGTGGACCAAAGTGACTGGCA | 157 | NM_000092.4 |
| h-MMP2 | ATGACAGCTGCACCACTGAG | ATTTGTTGCCCAGGAAAGTG | 164 | NM_004530.2 |
| h-MMP3 | GCAGTTTGCTCAGCCTATCC | AGTGTCGGAGTCCAGCTTC | 194 | NM_002422.3 |
| h-MMP9 | ACGCCGCTCACCTTCACT | AGGAGGAAAGGCGTGTGC | 111 | NM_004994 |
| m-MMP7 | GCGGAGATGCTCACTTTGACA | ATTCATGGGTGGCAGCAAAC | 87 | [46] |
| m-MMP9 | CCCACATTTGACGTCCAGAGAAGAA | GTTTTTGATGCTATTGCTGAGATCCA | 205 | [46] |
| m-α-sma | AACAGGAATACGACGAAG | CAGGAATGATTTGGAAAGGA | 134 | [47] |
| Plasmin-Plasminogen system | ||||
| h-μPA | CACACACTGCTTCATTGATTAC | TTGGTGGTGACTTCAGAGCCG | 409 | NM_002658 |
| h-PLG (Plasminogen) | GCAGTGGAGAAAACTATGACGGC | GAAGATGGTGGAGGTGTTGTGC | 244 | NM_000301[23] |
| h-PAI-1 | CTTTGGTGAAGGGTCTGCTGTG | TGGGTTTCTCCTCCTGTTGTCAG | 183 | M16006[23] |
| m-PLG | GCCCAACCTACCAATGTCTGAAAG | GGTGGAACTGAGGAATCTGACTGG | 292 | [23] |
| m-μPA | AGGGTGAGCGCCAATAGCAT | GATACATTCACGTGGAGCAT | 530 | [48] |
| m-PAI-1 | ATGCCATCTTTGTCCAGCGG | TTGGTATGCCTTTCCACCCAG | 151 | [23] |
| Normalising | ||||
| h-β-actin | TGCCATCCTAAAAGCCAC | TCAACTGGTCTCAAGTCAGTG | 289 | NM_001101 |
| m-β-actin | AAATCGTGCGTGACATCAAA | AAGGAAGGCTGGAAAAGAGC | 178 | [49] |
We utilized OPN specific siRNAs that blocked OPN (Stealth RNAiTM siRNA Duplex Oligoribonucleotides, Gibco, Invitrogen, United States), using reverse transfection process, achieving 70% transfection efficiency as per manufacturer’s instructions. Briefly, OPN RNAi oligonucleotides were incubated with LipofectamineTM RNAi MAX reagent for 30 min at room temperature in a 6-well plate. BLOCKiT Alexa Fluor 555 Red fluorescent (transfection control), Scrambled Stealth RNAi (high GC RNAi-scrambled control) and lipofectamine alone (negative control) were used as specific controls. For reverse transfection, cells (2 × 105 cells/mL) in Optimem media without antibiotics were added to the OPN RNAi lipofectamine mix and incubated at 37 °C for 24 h. Post-transfected cells were grown in serum free DMEM containing antibiotics for another 24 h for RNA extraction and 48 h for protein extraction and collection of CM. siOPN oligo that produced > 90% inhibition of OPN expression in LX2 was selected for further experiments.
Protein was isolated from frozen liver tissue using glass bead (Qiagen, Doncaster) homogenization in RIPA lysis buffer (Table 4). Cells ± alcohol ± 4-MP/antibodies/aptamer was lysed in RIPA lysis buffer for 20 min, centrifuged at 12000 g for 15 min at 4 °C and supernatant collected.
| Buffer | Composition |
| RIPA | 0.5% (w/v) sodium deoxycholate, 150 mmol/L NaCl, 50 mmol/L Tris pH 8.0, 20 mmol/L NaF, 10% (v/v) glycerol, 5.0 mmol/L EDTA pH = 8.0, 0.1% (w/v) SDS, 1.0% (v/v) Igepal CA-630 Electrophoresis reagent, 1 × phosSTOP, 1 × complete protease inhibitor |
| TBS | 50 mmol/L Tris, 150 mmol/L NaCl, pH = 7.5 |
| TBST | 0.1% (v/v) Tween 20 in TBS |
| Horseradish peroxidase inactivation | 0.05% (v/v) of sodium azide, 3% (w/v) skim milk and 0.1% (v/v) Tween 20 in TBS |
Western blots (WB) were performed with 40-60 μg of total protein per lane using the NuPAGE system (Invitrogen). To ensure expression of each protein was measured in the same sample, membranes were cut into strips according to the expected molecular weight of each protein and simultaneously probed with appropriate antibodies (Table 2). Each membrane was washed in buffered Tris-Tween20 (TBST) and incubated with horseradish peroxidase (HRP) conjugated secondary antibody. The primary and secondary antibodies to phospho-Erk (P-Erk) and P-Akt were removed by Restore PLUS Western Blot stripping Buffer (Theme Fisher Scientific Australia) and HRP inactivating buffer (Table 4) and re-probed with T-Erk and T-Akt antibodies. Enhanced ChemiLuminescence (Millipore) was used for visualization and bands quantitated with Image J software (National Institutes of Health, United States).
LX2 cells post-treatment were fixed in 100% ethanol, semi-permeabilised with 4% paraformaldehyde and stained with HSC-specific vimentin and OPN primary antibodies as previously described[1]. AlexaFluor 488 and 594 were used as secondary antibodies for visualisation.
Plasmin activity in serum was measured in the presence of plasminogen and substrate S2251 as previously described[23,30]. LX2 cells (5 × 104 cells/well) were seeded in 96-well plates overnight in DMEM which was replaced with serum-free media ± 10 mmol/L alcohol ± 1 mmol/L 4-MP. To block OPN-receptor binding, blocking antibodies (Table 2) were added and cells incubated for 16 h. Supernatants (50 μL) from wells were assayed for plasmin activation as previously described[23].
A novel migration assay was developed using prestained cells with 5 μmol/L Hoechst33342 (Molecular Probes) to monitor migration across membrane by confocal microscopy. LX2 cells (1 × 105 cells/well) were seeded in serum-free media into the top Boyden chamber with 8 μm transwell inserts (BD Sciences, NSW, Australia) in a 6-well plate format. For inhibition treatments, blocking antibodies (anti-OPN, anti-CD44v6, anti-ITGAV, Table 2) and OPN-R3 aptamer were added. Transforming growth factor (TGF) β (2 ng/mL) was added to the bottom chamber with serum-free media ± alcohol and incubated for 4 h. Membrane from each transwell was removed with scalpel and top-bottom orientation was noted. Membranes were placed on superfrost plus slides (Lomb Scientific Pty Ltd, NSW, Australia) with a drop of prolong gold antifade reagent (Invitrogen, Oregon, United States). Slides were analysed using Z-stacks by Leica SP5 Confocal Imaging. To distinguish between migrated (M) and non-migrated (NM) sides, cells were imaged at 405 nm on both sides of the membrane (imaged at 633nm). At least six fields per treatment for M and NM were captured using Volocity Version 5.3.0 software (Improvision Ltd). A program was designed in Axiovision 4.4 Software to calculate the number of nuclei (stained blue) per field on each side of the membrane to correlate with the number of cells to determine average M and NM cell count.
All experiments were conducted at least three times with internal replicates. Results are reported as average of all experiments tested for significance. Images represent typical experimental results. Data were analyzed using GraphPad Prism 5 with unpaired student’s t-test/non-parametric Mann-Whitney t-test for statistical inference. P-value < 0.05 was considered significant.
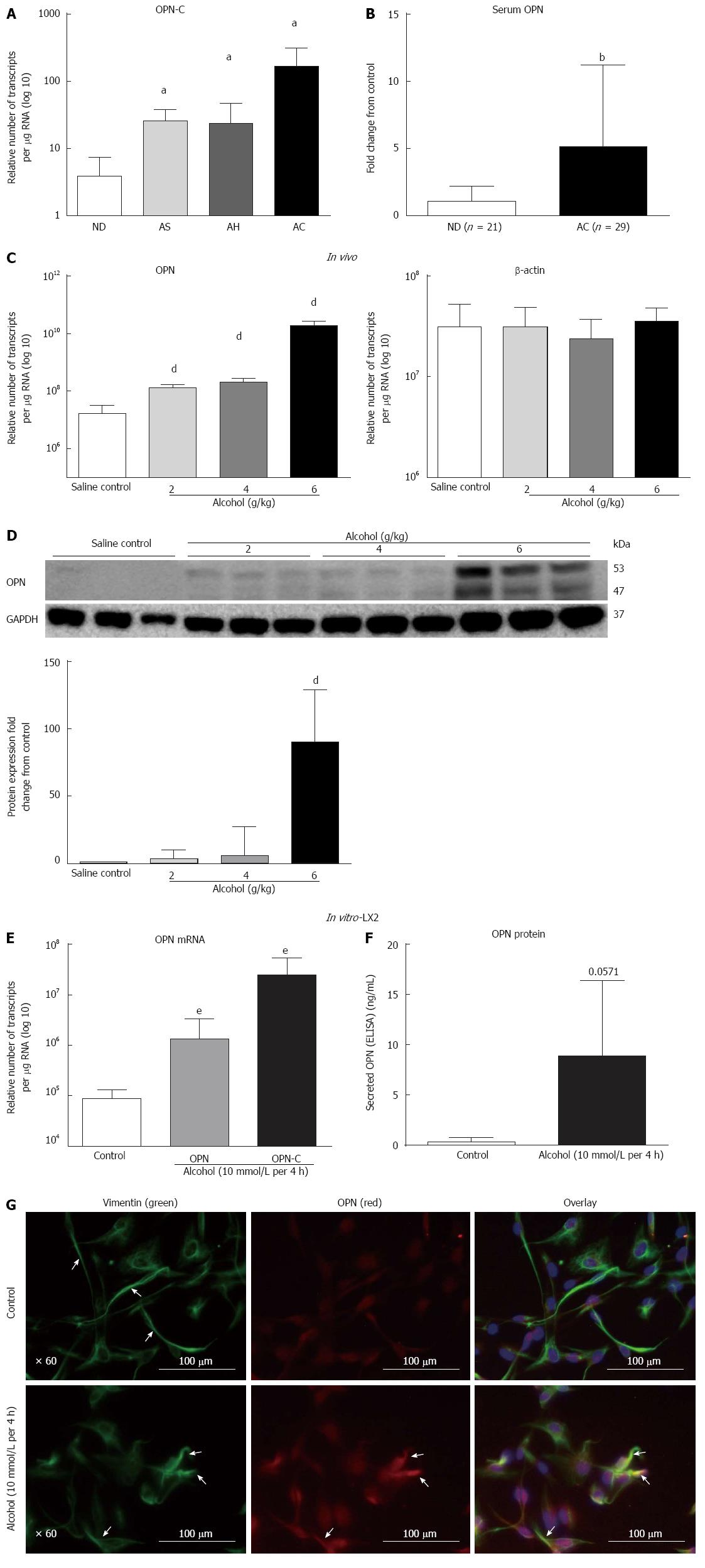
Human ALD: We tested the expression of OPN-C isoform mRNA in human livers. Similar to our previous report of total OPN up-regulation in human ALD[1], mRNA for the OPN-C isoform was increasingly expressed in patients with progressive stages of alcoholic steatosis (AS), alcoholic hepatitis (AH) and alcoholic cirrhosis (AC) (2.3-, 2.1-, 13-fold, respectively), compared to non-diseased (ND) control (Figure 1A). Significant increase in serum OPN (about 5-fold) in an independent cohort of alcoholic cirrhosis patients, compared to non-diseased alcoholics, confirmed mRNA data (Figure 1B).
In vitro mouse model of alcoholic steatosis: OPN mRNA expression increased (8.5-, 12.7-, 1171-fold) with increasing alcohol doses (2, 4, 6 g/kg, respectively) within 16 h in WT mice compared to saline control (Figure 1C). β-actin mRNA did not change. The increase in OPN expression was associated with increasing hepatic steatosis, increased liver enzymes and triglycerides reported earlier[23]. Protein expression (WB) confirmed mRNA increase. In addition, emergence of a cleaved form of OPN (about 47 kD) was evident with all doses of alcohol with maximum expression at 6 g/kg alcohol (Figure 1D). Quantitation showed significant up-regulation (3.2-, 5.9-, 90-fold) of full length (about 53 kD) OPN with 2, 4 and 6 g/kg alcohol doses, respectively (Figure 1D). Relative fold-increase for the cleaved 47 kD fragment could not be quantitated due to no visible expression in saline control.
In vitro LX2 cells exposed to acute alcohol: Expression of both full length OPN-A and isoform OPN-C mRNAs were tested in LX2 cells ± alcohol. OPN-A and OPN-C mRNAs significantly increased by 15-fold and 286-fold, respectively, within 4 h of 10 mmol/L alcohol exposure compared to untreated control (Figure 1E). An associated increase (about 15-fold) in secreted OPN (Figure 1F) and intracellular OPN (red, Figure 1G) protein confirmed mRNA result. Concomitant to increased OPN (red), alcohol induced LX2 stellate cell activation, seen as detracted processes distinct from control cells, was also evident on staining with vimentin (green) (Figure 1G).
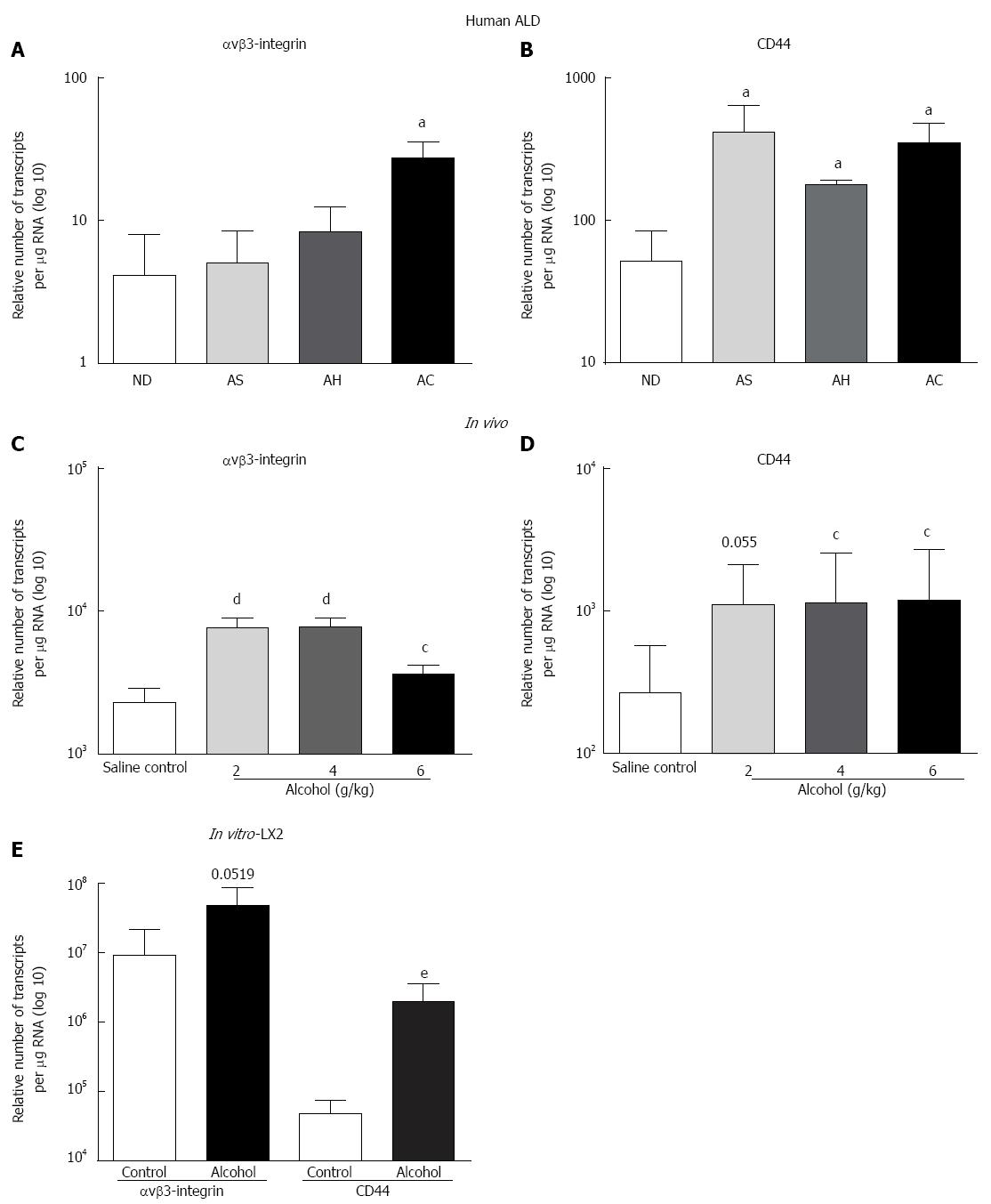
Significant increase in the expression of OPN receptor αvβ3-integrin (3-fold) was observed for AC compared to ND (Figure 2A), whereas CD44v6 mRNA increased (8.2-, 3.5-, 6.8-fold) in AS, AH and AC, respectively (Figure 2B). Similar to human data, all alcohol doses, except 2 g/kg (CD44), significantly induced both αvβ3-integrin (Figure 2C) and CD44 (Figure 2D) receptor mRNAs in WT mice compared to saline control. A single acute dose of alcohol also induced both receptor mRNAs in LX2 in vitro, but reached significance only in CD44 mRNA expression (Figure 2E).
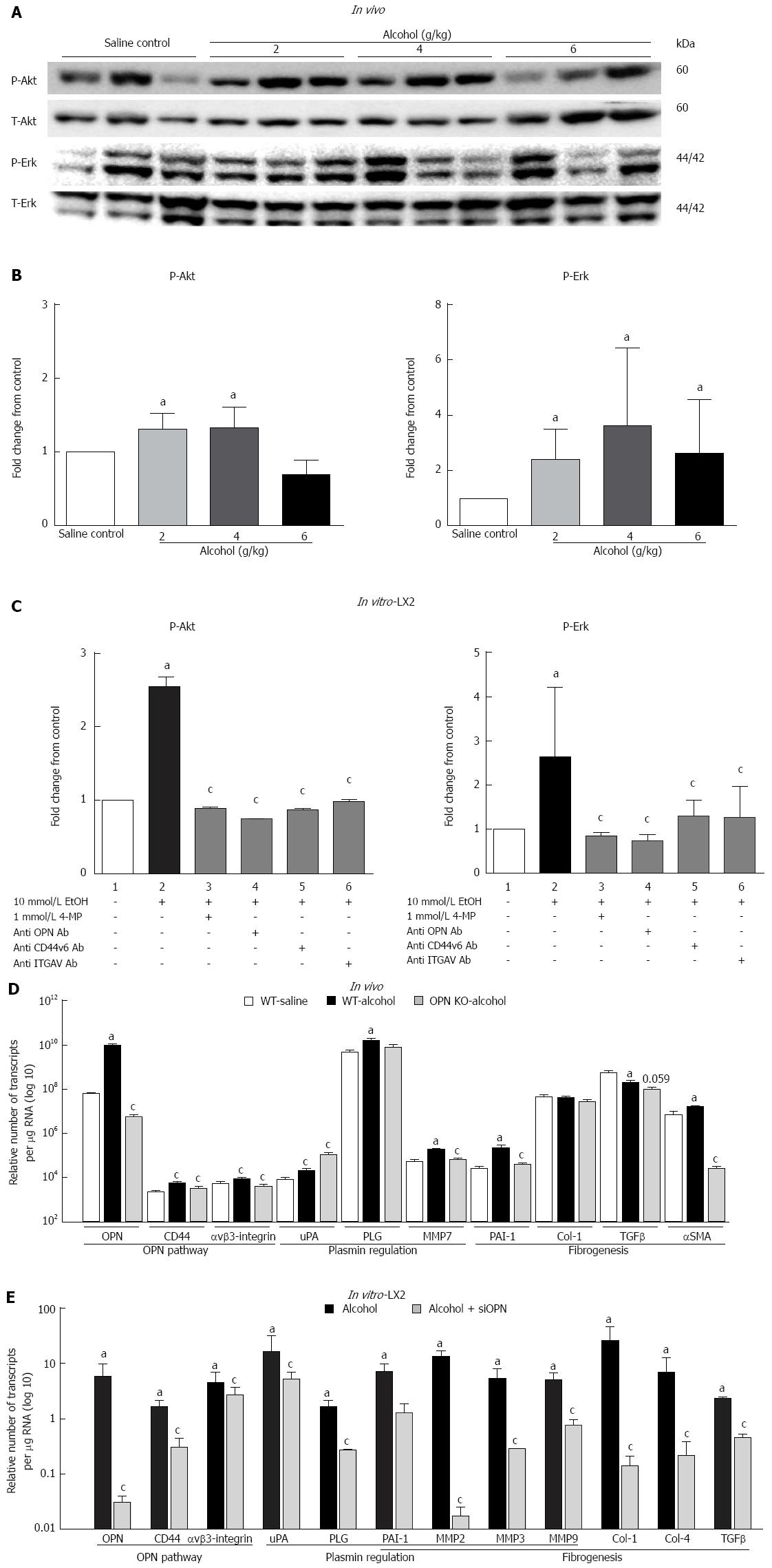
Parallel to increased OPN and its receptors, downstream phosphorylated Akt (P-Akt) and Erk (P-Erk) were also induced in mice administered a single dose of alcohol (Figure 3A). However, no dose specific response to alcohol was observed. It is noteworthy that individual mice showing high P-Erk had low P-Akt and vice versa as shown in the WB image. Densitometric quantitation normalized with total-Akt (T-Akt) and -Erk (T-Erk), showed moderate but significant activation of P-Akt with 2 and 4 g/kg and of P-Erk at all doses of alcohol (Figure 3B). In vitro, a single acute dose of alcohol also significantly induced P-Akt and P-Erk (about 2.5-fold) in LX2 cells (Figure 3C, lane 2) compared to untreated control (lane 1). This increase was significantly inhibited by alcohol dehydrogenase (ADH) inhibitor 4-MP (lane 3) and blocking antibodies to anti-OPN, -CD44v6 and -ITGAV (lanes 4, 5 and 6, respectively).
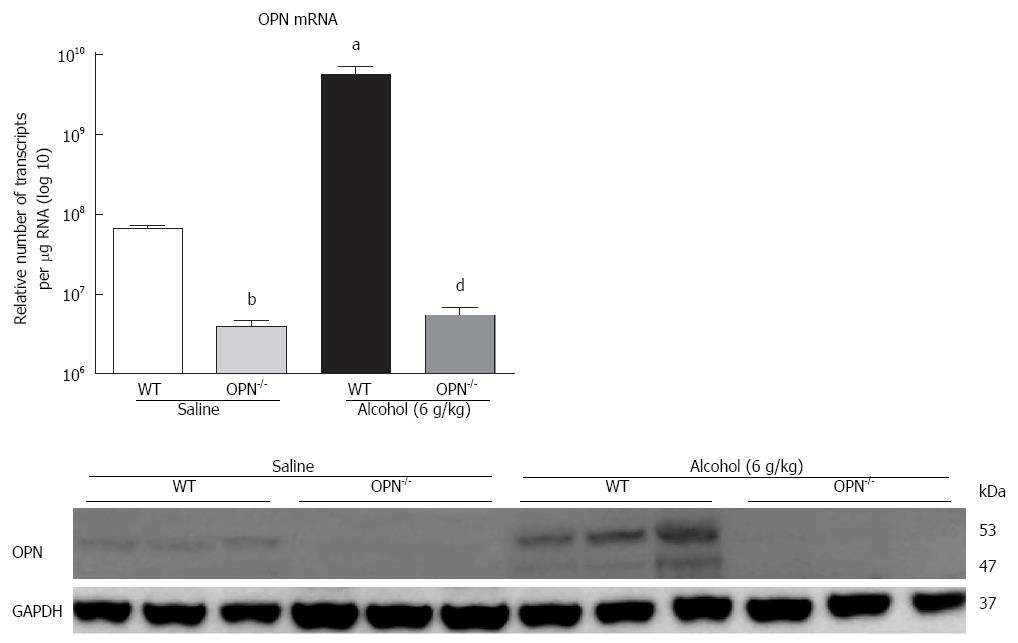
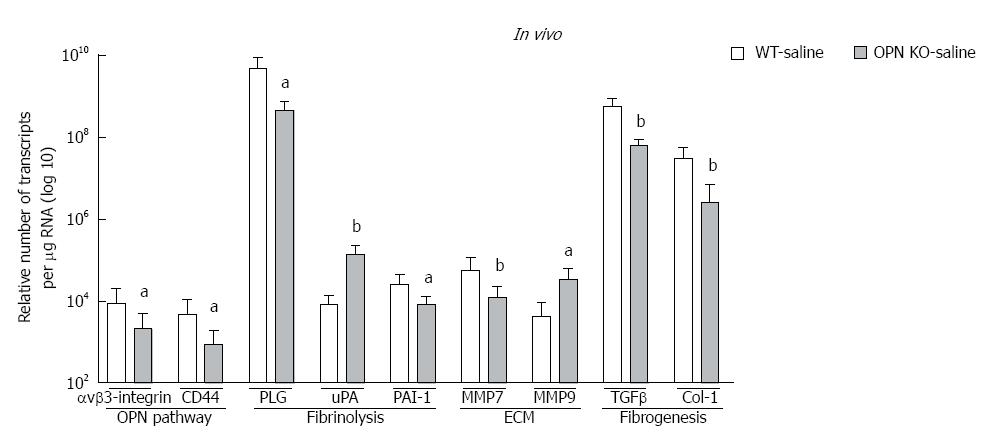
In vivo: mRNA expression of selected target genes related to plasmin, ECM remodelling and fibrogenesis pathways was tested by Q-PCR. In WT mice (dark bars), in addition to increased OPN and its receptors, alcohol (6 g/kg) also significantly increased uPA, PLG, MMP7, PAI-1 and αSMA, and down-regulated TGFβ, compared to saline control (white bars) (Figure 3D). Increasing expression was also observed with other doses of alcohol, with some of these genes reaching significance (data not shown). To identify OPN target genes, mRNA expression in OPN-/- mice was compared to WT, under 6 g/kg alcohol administration (grey bars). As expected, OPN mRNA was significantly inhibited (3 log fold) in knockout animals and OPN protein was not detected (Figure 4). Importantly, mRNA expression of CD44, αvβ3-integrin, MMP7, PAI-1 and αSMA remained significantly inhibited in OPN-/- animals compared to WT in the presence of alcohol (Figure 3D) and without any treatment (Figure 5). Conversely, uPA mRNA was significantly increased in the OPN-/- animals compared to WT, both in the presence of alcohol (Figure 3D) and with no treatment (Figure 5).
In vitro: Similar to in vivo data, a single dose of alcohol in LX2 cells induced OPN, receptors CD44 and αvβ3-integrin, shown as fold-change from control (Figure 3D, dark bars). In addition, μPA, PLG, PAI-1, MMP2, MMP3, MMP9, collagen-1 (Col-1), collagen-4, (Col-4) and TGFβ mRNAs were also up-regulated with alcohol in these cells. Furthermore, knocking down OPN in LX2 cells by siRNA in the presence of alcohol, most molecules, except uPA and αvβ3-integrin, were significantly inhibited compared to LX2-scrambled control (grey bars). However, in the absence of alcohol, expression of all mRNAs, except μPA, remained significantly inhibited by siRNA compared to scrambled control (data not shown).
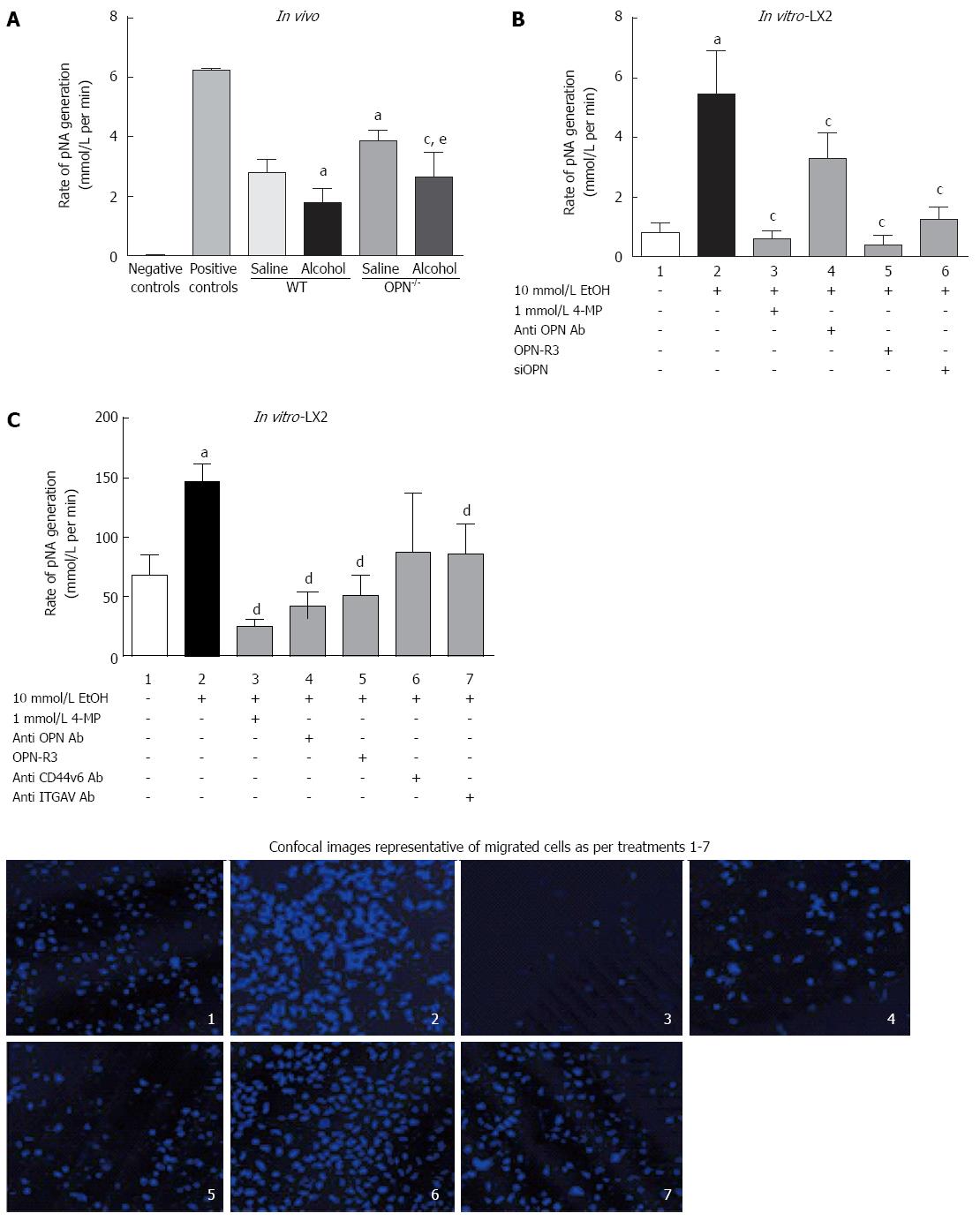
In vivo: Serum plasmin activity, measured as rate of pNA generation (pNA/mol/L), was significantly down-regulated with alcohol (6 g/kg) both in WT and OPN-/- mice, compared to their respective saline controls (Figure 6A). However, in untreated OPN-/- mice intrinsic plasmin activity was significantly higher (1.4-fold) compared to control WT.
In vitro: Plasmin activity significantly increased several fold in LX2 CM exposed to alcohol (Figure 6B, lane 2) compared to untreated cells (lane 1) and was significantly inhibited in the presence of 4-MP (0.2 pNA/mol/L, lane 3). Inhibiting OPN by anti-OPN antibody, OPN-R3 aptamer and siOPN also significantly inhibited alcohol-induced plasmin activation to 3.9 pNA/mol/L (lane 4), 0.38 pNA/mol/L (lane 5) and 1.8 pNA/mol/L (lane 6), respectively (Figure 6B).
LX2 cell migration, recorded as percent migrated over non-migrated cells, significantly increased (1.7-fold) on exposure to alcohol (Figure 6C, lane 2) compared to control (lane 1). Blocking alcohol metabolism by 4-MP (lane 3), and OPN by anti-OPN antibody (lane 4) and OPN-R3 (lane 5), significantly inhibited migration. Furthermore, blocking OPN receptors through anti-CD44v6 (lane 6) and -ITGAV (lane 7) antibodies also reduced LX2 migration but reached significance with anti-ITGAV (lane 7). Representative confocal images of cells stained with Hoechst dye (blue) shown in the panel below show typical numbers of migrated cells for treatments in lanes 1-7.
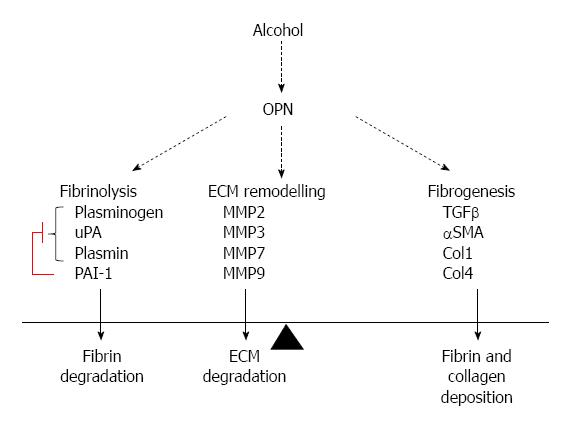
These are the first studies describing specific mechanisms by which OPN activation mediates the acute pro-fibrogenic effects of alcohol on HSC. Our novel in vivo and in vitro data provides causative evidence that OPN mediates alcohol-induced functions, such as cell signaling, plasmin activation and HSC migration. We also show that OPN is a key modulator of several alcohol-induced molecules related to fibrinolysis, ECM remodeling and fibrogenesis as downstream targets of OPN. Indeed, our data indicate that OPN negatively regulates μPA and plasmin and positively regulates PAI-1, α-SMA, TGFβ, collagens and MMPs, both in vivo and in vitro, suggesting a pro-fibrogenic role for this molecule. Human data provide further support for a clinical relevance of OPN pathway in ALD. Although human ALD is a chronic disorder, fatty liver or steatosis may be seen in humans after a single large dose of alcohol. Steatosis is in turn associated with early liver injury and when persistent, with an increased risk of fibrosis. Our single binge alcoholic steatosis model is therefore appropriate to study the role of this cytokine in early liver injury.
Extending our previous observation of elevated OPN mRNA in ALD[1], serum OPN levels also significantly increased in heavy drinkers with liver cirrhosis than those with no liver disease, confirming other recent observations[4,31] and underscoring the potential for OPN as a marker for liver injury. OPN exists as full length (OPN-A) and as alternatively spliced transcriptional isoforms (OPN-B, -C), but their specific functions are not clear. Our preliminary data comparing expression of all three OPN isoforms including hepatocyte cell lines (Huh7, HepG2) showed increased OPN-C primarily in human ALD and LX2 cells on alcohol exposure, but not in the hepatocyte cell lines[15,32]. Activated HSCs are known to produce OPN in animal models and human liver injury[33-35]. We now confirm that the greatest up-regulation of OPN-C was seen in patients with end stage cirrhosis and in activated LX2 stellate cells, the major fibrosis producing cell type. Increased OPN in alcoholic hepatitis patients, mainly in the areas of inflammation and fibrosis, and in HSCs was reported recently[4]. In our studies, the early activation of OPN-C isoform in LX2 and in advanced ALD, suggests a possible role for stellate cell derived OPN-C in a transient wound healing response, as well as in the process of fibrogenesis, perhaps even increasing the risk for malignant transformation in cirrhotic patients, as OPN-C is linked to HCC[13,14,28].
In addition to the human and in vitro data, our in vivo experiments provide clear evidence that a single dose of alcohol not only increased hepatic OPN in WT mice, but also up-regulated a cleaved form of OPN (cOPN) in a dose-dependent manner. Proteolytic cleavage via plasmin[36], MMPs 3 and 7[37] and thrombin[38] generate OPN forms that are known to enhance receptor binding, signalling and other cellular functions[11]. It is well known that amount of alcohol intake is an important factor in the severity of liver injury. Therefore, emergence of a potentially more “activated” cOPN at higher dose of alcohol was intriguing. As far as we know, this is the first report to suggest that OPN activation may be mediated through alcohol induced cleavage of OPN. It has been suggested that alcohol treatment alone does not cause OPN cleavage, particularly via thrombin, and required an additional hit with LPS in an experimental model of ASH[5]. Our model showing suppressed plasmin activity but increased MMP7 expression with concurrent increase in cleaved OPN, supports MMP7 as the key protease to cleave OPN with alcohol alone. A contribution from MMP3 cannot be ruled out, nonetheless, another recent report also suggests contribution of MMP7 in OPN cleavage in alcoholic hepatitis patients[4]. Moreover, transcription inhibition of MMP7 in OPN-/- mice and inhibition of several MMPs, including MMP7, in in vitro LX2 on silencing OPN, suggests that MMP7 is a downstream target of OPN. We propose that alcohol may directly activate OPN and further enhance its activation via increased MMP7 in a positive feedback loop. It will be important to tease out the exact mechanism/s of alcohol-induced OPN activation via proteolytic cleavage.
An associated increase in OPN receptors CD44 and αvβ3-integrin in human ALD, in vivo in acute alcoholic steatosis mouse model and in vitro in LX2 cells with a single dose of alcohol confirms that OPN pathway was indeed induced with alcohol. Alcohol treatment in vivo and in vitro, also activated downstream Akt and Erk phosphorylation, known to be the two major signaling pathways induced on OPN binding to CD44 and αvβ3-integrin. HSCs are known to metabolise alcohol via ADH[39], therefore, to confirm that the effect on OPN was mediated via alcohol, we utilized ADH inhibitor 4-MP to inhibit alcohol metabolism in vitro. Inhibition of alcohol-induced Erk- and Akt signaling, downstream plasmin activation and LX2 cell migration by 4-MP, signify that alcohol metabolism was required for OPN action on HSC functions.
We identified several downstream alcohol-induced genes in vivo and in vitro belonging to plasmin/ECM (uPA, PLG, PAI-1, MMPs 2, 3, 7, 9) and fibrogenesis (TGFβ, PAI-1, αSMA, collagens) pathways. Interestingly, inhibiting OPN in OPN-/- animals and silencing OPN in LX2 (LX2-siOPN), altered expression of several of these genes, identifying OPN-receptor signalling targets. As opposed to in vivo observations, alcohol increased expression of fibrogenic markers TGFβ, Col-1 and Col-4 (Figure 3E) and αSMA reported previously[23], confirming activation of stellate LX2 cells with alcohol. Downregulation of TGFβ in mice with alcohol exposure was unexpected, but more importantly, in the primary TGFβ producing LX2 stellate cells, TGFβ was significantly upregulated with alcohol.
The present study confirms a novel hypothesis that alcohol induced plasmin is mediated via OPN in hepatic stellate cells. In addition, we have also shown that OPN has a key role in alcohol-induced HSC functions such as signalling, cell migration, and activation of fibrogenic pathway. Alcohol effect on OPN activation and downstream target pathways was acute and required alcohol metabolism. Importantly, this study identified alcohol-induced transcriptional targets of OPN, in vivo and in vitro, related to fibrinolysis, ECM and fibrogenesis, significant pathways in disease progression, confirming a role for OPN in these processes.
Down-regulation of CD44 and αvβ3-integrin as a consequence of OPN inhibition both in vivo and in vitro, demonstrates that OPN may positively regulate (autoregulation) its cognate receptors and pathway. Similarly, inhibition of TGFβ, αSMA, PAI-1, MMPs, collagens 1 and 4 expression on silencing and knocking out OPN, suggests these fibrogenic molecules were also positively regulated by OPN. Most surprising and unlike other molecules, basal uPA expression was higher in OPN-/- mice (about 17-fold) and LX2-siOPN (50-fold) than their respective untreated controls (not shown). This points to a negative regulation of μPA by OPN, perhaps indirectly via regulating inhibitors of uPA, such as PAI-1. A markedly reduced basal expression of PAI-1 in OPN-/- mice and LX2-siOPN suggests that increased μPA may be a consequence of inhibited PAI-1, and that OPN may mediate, at least in part, the balance between μPA and PAI-1 expression in the liver.
It is intriguing that these effects are evident early and with only a single dose of alcohol. However, the limitation of this study is that it does not reproduce chronic injury. We are also exploring these pathways in a chronic alcoholic liver injury model (Figure 7).
Implications of the results: This study extends our knowledge of OPN biology in alcohol-induced liver injury. Contribution of OPN in modulating hepatic fibrinolysis and fibrogenesis in hepatic stellate cells within hours of alcohol exposure is significant.
Although alcohol-induced and OPN-mediated activation of uPA, PLG and MMPs may facilitate fibrinolysis and matrix proteolysis, our data suggest that the overall impact of OPN remains pro-fibrogenic in the context of stellate cells.
It is established that activated HSCs are an important hepatic source of plasmin[19]. We have identified OPN pathway as a novel mechanism that may contribute towards alcohol-induced plasmin activation in the HSCs. We show that in LX2 cells alcohol increased OPN and its cognate receptors resulting in Akt and Erk signalling, enhanced expression of downstream fibrinolytic targets uPA and PLG, and subsequent activation of plasmin. Inhibiting OPN action using three independent methods (neutralising OPN antibody, OPN-R3 aptamer, siOPN), clearly abrogated plasmin activation in these cells confirming a role for OPN in alcohol-induced function of the stellate LX2 cells. The anomalous observation of increased uPA mRNA but decreased plasmin in OPN inhibition experiments in LX2 cells may be explained by significantly reduced levels of plasminogen; so even though uPA was increased, reduced uPA substrate plasminogen may have limited the conversion to plasmin in these settings. In the knockout animals, however, increased plasmin was associated with increased uPA, signifying in vivo tissue- and in vitro cell type-specific differences in response to OPN inhibition.
In addition, inhibition of OPN pathway (via OPN-R3, neutralizing antibodies to OPN and OPN receptors) demonstrated that alcohol-induced stellate cell migration was clearly dependent on OPN. Blocking OPN by OPN neutralizing antibody and OPN-R3, had a stronger inhibitory effect on LX2 migration than blocking CD44v6 or αvβ3-integrin individually, implying a key role for OPN in HSC migration. Similar observations have been reported in highly invasive MDA-MB231 breast cancer cells where blocking OPN by OPN-R3 aptamer inhibited cell migration more than blocking either receptor[26]. We have identified a mechanism of alcohol-induced and OPN-mediated HSC migration similar to that in highly metastatic cancer cells.
In conclusion, we have demonstrated that alcohol activates the OPN pathway in human ALD and experimental in vitro and in vivo models of acute alcohol. In turn, activated OPN mediated several alcohol induced functions of stellate cells. The study identified OPN as a key mediator of acute alcohol-induced activation and initiation of pro-fibrogenic processes in hepatic stellate cells. Our functional data unequivocally shows that OPN mediated the alcohol-induced stellate cell functions such as signalling, plasmin activation and cell migration confirming a role for OPN and OPN pathway in alcoholic liver disease. Future research will explore the therapeutic role of interventions to target this pathway.
We acknowledge the work by Ms. Chiyoko Yagasaki for her thesis for which she received a first class Honours in 2011 from the University of Sydney. We acknowledge Dr. Alison Morgan for producing data on cell migration. The SPP1-/- mice originated from Dr. Denhardt’s laboratory (Rittling 1998), and breeding pairs were obtained from our collaborator Dr. Nilsson S, Australian Stem Cell Centre, Monash University, Melbourne.
The excessive use of alcohol is widely recognised as a major health and social problem worldwide. Alcohol is associated with over 60 medical diseases of which alcoholic liver disease (ALD) causes the most deaths and is consequently the greatest public health problem. Indeed, despite the increased incidence of other liver diseases (such as hepatitis C and fatty liver disease), alcohol abuse is still the most common cause of advanced liver disease. Alcohol abuse is also an important cause of liver cancer, the most rapidly rising form of cancer.
One key issue with ALD is the early identification of liver injury via an appropriate biomarker. Osteopontin, a multifunctional cytokine, is known to be associated with the severity of liver injury in ALD, HepC and non-alcoholic steatohepatitis (NASH), and indicates a poor prognosis in hepatocellular carcinoma. However, the mechanisms of action remain unclear. Existence of several, transcriptional and post-translational, forms of over-expression of Osteopontin (OPN) are suspected to account for its multiple roles in disease. In this study, the authors identify several interesting lines of further investigations: early induction of OPN-C isoform and cleaved OPN with a single dose of alcohol; alcohol-induced hepatic stellate cell (HSC) functions mediated via OPN; HSC derived OPN has a role in fibrogenesis via plasmin activation.
Authors were the first to identify increased levels of Osteopontin and fibrinolysis related molecules in human alcoholic liver disease. Since then, others have confirmed the findings in alcoholic hepatitis and others forms of liver injury. This study underscores the role of OPN and its forms in human, and in acute alcohol models in vivo and in vitro. This is the first report to show that acute alcohol induces potentially more active transcriptional and post-translational forms of OPN, specifically OPN-C induction in HSCs and patients with advanced ALD. Their in vitro functional data unequivocally shows a novel mechanism of OPN action in HSCs in the processes of wound healing and fibrogenesis.
By understanding the role and action of OPN and consequences of OPN inhibition, this study represents a potential for OPN as an early marker for ALD and a therapeutic agent in liver injury.
OPN-C and cOPN forms of OPN are reported as being more active forms due to increased binding capacities to cell surface receptors, integrin and CD44. In alcoholic liver injury, increased plasminogen activators are known to regulate liver matrix remodelling through activation of plasminogen to plasmin. Induction of OPN pathway early with acute alcohol and regulation of plasmin activation in HSCs via OPN defines a novel mechanism for fibrogenesis in ALD.
In this manuscript the expression levels and potential functions of OPN was studied in vitro (in a hepatic stellate cell line) and in vivo in patient samples as well as animal models. The effects on HSC are mainly convincing and therefore potentially relevant.
P- Reviewer: Buechler C, Breitkopf-Heinlein K S- Editor: Qi Y L- Editor: A E- Editor: Zhang DN
| 1. | Seth D, Gorrell MD, Cordoba S, McCaughan GW, Haber PS. Intrahepatic gene expression in human alcoholic hepatitis. J Hepatol. 2006;45:306-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 58] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 2. | Seth D, Leo MA, McGuinness PH, Lieber CS, Brennan Y, Williams R, Wang XM, McCaughan GW, Gorrell MD, Haber PS. Gene expression profiling of alcoholic liver disease in the baboon (Papio hamadryas) and human liver. Am J Pathol. 2003;163:2303-2317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 65] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 3. | Syn WK, Choi SS, Liaskou E, Karaca GF, Agboola KM, Oo YH, Mi Z, Pereira TA, Zdanowicz M, Malladi P. Osteopontin is induced by hedgehog pathway activation and promotes fibrosis progression in nonalcoholic steatohepatitis. Hepatology. 2011;53:106-115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 229] [Cited by in RCA: 216] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 4. | Morales-Ibanez O, Domínguez M, Ki SH, Marcos M, Chaves JF, Nguyen-Khac E, Houchi H, Affò S, Sancho-Bru P, Altamirano J. Human and experimental evidence supporting a role for osteopontin in alcoholic hepatitis. Hepatology. 2013;58:1742-1756. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 5. | Apte UM, Banerjee A, McRee R, Wellberg E, Ramaiah SK. Role of osteopontin in hepatic neutrophil infiltration during alcoholic steatohepatitis. Toxicol Appl Pharmacol. 2005;207:25-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 79] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 6. | Banerjee A, Apte UM, Smith R, Ramaiah SK. Higher neutrophil infiltration mediated by osteopontin is a likely contributing factor to the increased susceptibility of females to alcoholic liver disease. J Pathol. 2006;208:473-485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 76] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 7. | Sahai A, Pan X, Paul R, Malladi P, Kohli R, Whitington PF. Roles of phosphatidylinositol 3-kinase and osteopontin in steatosis and aminotransferase release by hepatocytes treated with methionine-choline-deficient medium. Am J Physiol Gastrointest Liver Physiol. 2006;291:G55-G62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 8. | Sahai A, Malladi P, Melin-Aldana H, Green RM, Whitington PF. Upregulation of osteopontin expression is involved in the development of nonalcoholic steatohepatitis in a dietary murine model. Am J Physiol Gastrointest Liver Physiol. 2004;287:G264-G273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 152] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 9. | Kawashima R, Mochida S, Matsui A, YouLuTuZ Y, Ishikawa K, Toshima K, Yamanobe F, Inao M, Ikeda H, Ohno A. Expression of osteopontin in Kupffer cells and hepatic macrophages and Stellate cells in rat liver after carbon tetrachloride intoxication: a possible factor for macrophage migration into hepatic necrotic areas. Biochem Biophys Res Commun. 1999;256:527-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 112] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 10. | Welch KD, Reilly TP, Bourdi M, Hays T, Pise-Masison CA, Radonovich MF, Brady JN, Dix DJ, Pohl LR. Genomic identification of potential risk factors during acetaminophen-induced liver disease in susceptible and resistant strains of mice. Chem Res Toxicol. 2006;19:223-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 33] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 11. | Bellahcène A, Castronovo V, Ogbureke KU, Fisher LW, Fedarko NS. Small integrin-binding ligand N-linked glycoproteins (SIBLINGs): multifunctional proteins in cancer. Nat Rev Cancer. 2008;8:212-226. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 360] [Cited by in RCA: 353] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 12. | Patani N, Jouhra F, Jiang W, Mokbel K. Osteopontin expression profiles predict pathological and clinical outcome in breast cancer. Anticancer Res. 2008;28:4105-4110. [PubMed] |
| 13. | Takafuji V, Forgues M, Unsworth E, Goldsmith P, Wang XW. An osteopontin fragment is essential for tumor cell invasion in hepatocellular carcinoma. Oncogene. 2007;26:6361-6371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 141] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 14. | Chae S, Jun HO, Lee EG, Yang SJ, Lee DC, Jung JK, Park KC, Yeom YI, Kim KW. Osteopontin splice variants differentially modulate the migratory activity of hepatocellular carcinoma cell lines. Int J Oncol. 2009;35:1409-1416. [PubMed] |
| 15. | Seth D, Beard M, Haber PS. Characterizing role of osteopontin in alcoholic liver injury. Clin Exp Res. 2007;31:79A-281. |
| 16. | Das R, Philip S, Mahabeleshwar GH, Bulbule A, Kundu GC. Osteopontin: it’s role in regulation of cell motility and nuclear factor kappa B-mediated urokinase type plasminogen activator expression. IUBMB Life. 2005;57:441-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 65] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 17. | Das R, Mahabeleshwar GH, Kundu GC. Osteopontin induces AP-1-mediated secretion of urokinase-type plasminogen activator through c-Src-dependent epidermal growth factor receptor transactivation in breast cancer cells. J Biol Chem. 2004;279:11051-11064. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 80] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 18. | Philip S, Kundu GC. Osteopontin induces nuclear factor kappa B-mediated promatrix metalloproteinase-2 activation through I kappa B alpha/IKK signaling pathways, and curcumin (diferulolylmethane) down-regulates these pathways. J Biol Chem. 2003;278:14487-14497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 196] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 19. | Leyland H, Gentry J, Arthur MJ, Benyon RC. The plasminogen-activating system in hepatic stellate cells. Hepatology. 1996;24:1172-1178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 71] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 20. | Neubauer K, Knittel T, Armbrust T, Ramadori G. Accumulation and cellular localization of fibrinogen/fibrin during short-term and long-term rat liver injury. Gastroenterology. 1995;108:1124-1135. [PubMed] |
| 21. | Leiper K, Croll A, Booth NA, Moore NR, Sinclair T, Bennett B. Tissue plasminogen activator, plasminogen activator inhibitors, and activator-inhibitor complex in liver disease. J Clin Pathol. 1994;47:214-217. [PubMed] |
| 22. | Shanmukhappa K, Sabla GE, Degen JL, Bezerra JA. Urokinase-type plasminogen activator supports liver repair independent of its cellular receptor. BMC Gastroenterol. 2006;6:40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 32] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 23. | Seth D, Hogg PJ, Gorrell MD, McCaughan GW, Haber PS. Direct effects of alcohol on hepatic fibrinolytic balance: implications for alcoholic liver disease. J Hepatol. 2008;48:614-627. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 38] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 24. | Rittling SR, Matsumoto HN, McKee MD, Nanci A, An XR, Novick KE, Kowalski AJ, Noda M, Denhardt DT. Mice lacking osteopontin show normal development and bone structure but display altered osteoclast formation in vitro. J Bone Miner Res. 1998;13:1101-1111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 324] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 25. | Wang XM, Yu DM, McCaughan GW, Gorrell MD. Fibroblast activation protein increases apoptosis, cell adhesion, and migration by the LX-2 human stellate cell line. Hepatology. 2005;42:935-945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 170] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 26. | Mi Z, Guo H, Russell MB, Liu Y, Sullenger BA, Kuo PC. RNA aptamer blockade of osteopontin inhibits growth and metastasis of MDA-MB231 breast cancer cells. Mol Ther. 2009;17:153-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 119] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 27. | Helbig KJ, Ruszkiewicz A, Semendric L, Harley HA, McColl SR, Beard MR. Expression of the CXCR3 ligand I-TAC by hepatocytes in chronic hepatitis C and its correlation with hepatic inflammation. Hepatology. 2004;39:1220-1229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 98] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 28. | Phillips RJ, Helbig KJ, Van der Hoek KH, Seth D, Beard MR. Osteopontin increases hepatocellular carcinoma cell growth in a CD44 dependant manner. World J Gastroenterol. 2012;18:3389-3399. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 22] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 29. | Seth D, Gorrell MD, McGuinness PH, Leo MA, Lieber CS, McCaughan GW, Haber PS. SMART amplification maintains representation of relative gene expression: quantitative validation by real time PCR and application to studies of alcoholic liver disease in primates. J Biochem Biophys Methods. 2003;55:53-66. [PubMed] |
| 30. | Couto LT, Donato JL, de Nucci G. Analysis of five streptokinase formulations using the euglobulin lysis test and the plasminogen activation assay. Braz J Med Biol Res. 2004;37:1889-1894. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 31. | Patouraux S, Bonnafous S, Voican CS, Anty R, Saint-Paul MC, Rosenthal-Allieri MA, Agostini H, Njike M, Barri-Ova N, Naveau S. The osteopontin level in liver, adipose tissue and serum is correlated with fibrosis in patients with alcoholic liver disease. PLoS One. 2012;7:e35612. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 66] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 32. | Seth D, Beard M, Gorrell M, McCaughan GW, Haber P. Osteopontin: an early marker for alcoholic liver injury? Alcohol Clin Exp Res. 2006;30:89A-336. |
| 33. | Lee SH, Seo GS, Park YN, Yoo TM, Sohn DH. Effects and regulation of osteopontin in rat hepatic stellate cells. Biochem Pharmacol. 2004;68:2367-2378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 68] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 34. | Syn WK, Agboola KM, Swiderska M, Michelotti GA, Liaskou E, Pang H, Xie G, Philips G, Chan IS, Karaca GF. NKT-associated hedgehog and osteopontin drive fibrogenesis in non-alcoholic fatty liver disease. Gut. 2012;61:1323-1329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 215] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 35. | Xiao X, Gang Y, Gu Y, Zhao L, Chu J, Zhou J, Cai X, Zhang H, Xu L, Nie Y. Osteopontin contributes to TGF-β1 mediated hepatic stellate cell activation. Dig Dis Sci. 2012;57:2883-2891. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 36. | Christensen B, Schack L, Kläning E, Sørensen ES. Osteopontin is cleaved at multiple sites close to its integrin-binding motifs in milk and is a novel substrate for plasmin and cathepsin D. J Biol Chem. 2010;285:7929-7937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 95] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 37. | Agnihotri R, Crawford HC, Haro H, Matrisian LM, Havrda MC, Liaw L. Osteopontin, a novel substrate for matrix metalloproteinase-3 (stromelysin-1) and matrix metalloproteinase-7 (matrilysin). J Biol Chem. 2001;276:28261-28267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 304] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 38. | Shio K, Kobayashi H, Asano T, Saito R, Iwadate H, Watanabe H, Sakuma H, Segawa T, Maeda M, Ohira H. Thrombin-cleaved osteopontin is increased in urine of patients with rheumatoid arthritis. J Rheumatol. 2010;37:704-710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 39. | Casini A, Pellegrini G, Ceni E, Salzano R, Parola M, Robino G, Milani S, Dianzani MU, Surrenti C. Human hepatic stellate cells express class I alcohol dehydrogenase and aldehyde dehydrogenase but not cytochrome P4502E1. J Hepatol. 1998;28:40-45. [PubMed] |
| 40. | He B, Mirza M, Weber GF. An osteopontin splice variant induces anchorage independence in human breast cancer cells. Oncogene. 2006;25:2192-2202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 146] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 41. | Celetti A, Testa D, Staibano S, Merolla F, Guarino V, Castellone MD, Iovine R, Mansueto G, Somma P, De Rosa G. Overexpression of the cytokine osteopontin identifies aggressive laryngeal squamous cell carcinomas and enhances carcinoma cell proliferation and invasiveness. Clin Cancer Res. 2005;11:8019-8027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 50] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 42. | Schordan S, Schordan E, Endlich K, Endlich N. AlphaV-integrins mediate the mechanoprotective action of osteopontin in podocytes. Am J Physiol Renal Physiol. 2011;300:F119-F132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 43] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 43. | Sretavan DW, Feng L, Puré E, Reichardt LF. Embryonic neurons of the developing optic chiasm express L1 and CD44, cell surface molecules with opposing effects on retinal axon growth. Neuron. 1994;12:957-975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 127] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 44. | Sahai A, Malladi P, Pan X, Paul R, Melin-Aldana H, Green RM, Whitington PF. Obese and diabetic db/db mice develop marked liver fibrosis in a model of nonalcoholic steatohepatitis: role of short-form leptin receptors and osteopontin. Am J Physiol Gastrointest Liver Physiol. 2004;287:G1035-G1043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 224] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 45. | Syn WK, Jung Y, Omenetti A, Abdelmalek M, Guy CD, Yang L, Wang J, Witek RP, Fearing CM, Pereira TA. Hedgehog-mediated epithelial-to-mesenchymal transition and fibrogenic repair in nonalcoholic fatty liver disease. Gastroenterology. 2009;137:1478-1488.e8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 216] [Cited by in RCA: 213] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 46. | Maquoi E, Munaut C, Colige A, Collen D, Lijnen HR. Modulation of adipose tissue expression of murine matrix metalloproteinases and their tissue inhibitors with obesity. Diabetes. 2002;51:1093-1101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 210] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 47. | Ochoa B, Syn WK, Delgado I, Karaca GF, Jung Y, Wang J, Zubiaga AM, Fresnedo O, Omenetti A, Zdanowicz M. Hedgehog signaling is critical for normal liver regeneration after partial hepatectomy in mice. Hepatology. 2010;51:1712-1723. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 144] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 48. | Ong K, Horsfall W, Conway EM, Schuh AC. Early embryonic expression of murine coagulation system components. Thromb Haemost. 2000;84:1023-1030. [PubMed] |
| 49. | Voronova A, Fischer A, Ryan T, Al Madhoun A, Skerjanc IS. Ascl1/Mash1 is a novel target of Gli2 during Gli2-induced neurogenesis in P19 EC cells. PLoS One. 2011;6:e19174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 40] [Article Influence: 2.9] [Reference Citation Analysis (0)] |