Published online Jun 7, 2014. doi: 10.3748/wjg.v20.i21.6547
Revised: December 13, 2013
Accepted: January 8, 2014
Published online: June 7, 2014
Processing time: 323 Days and 3.3 Hours
AIM: To investigate whether tissue samples processed by the rapid urease test (RUT) kit are suitable for dual-priming oligonucleotide-based multiplex polymerase chain reaction (DPO-PCR) to detect Helicobacter pylori (H. pylori).
METHODS: A total of 54 patients with specific gastrointestinal symptom were enrolled in this study. During endoscopy, gastric biopsy specimens were taken for histology, RUT, and DPO-PCR. DPO-PCR was performed on gastric biopsy samples and tissue samples that were analyzed by RUT at 2 separate institutes. In detecting H. pylori, the concordance rate of the DPO-PCR tests between the tissue samples that had been submitted to RUT and the gastric biopsy samples was investigated.
RESULTS: H. pylori co-occurred with 76.0% (19/25) of gastric ulcers, 64.3% (9/14) of duodenal ulcers, and 33.3% (4/12) of gastritis cases. H. pylori infection was found in 100% (3/3) of the patients with both gastric and duodenal ulcers. Overall, H. pylori was detected in 35 of 54 (64.8%) patients. The diagnostic sensitivities of histology, RUT, and DPO-PCR were 85.7% (30/35), 74.3% (26/35), and 97.1% (34/35), respectively (P = 0.02). The positive predictive value (PPV) of DPO-PCR was 94.4%, whereas the negative predictive value (NPV) was 94.7%. In the rapid urease test (CLOtest)-negative cases, the frequency of positive DPO-PCR and histologic results was 20.0% (7/35). The concordance rate of the DPO-PCR tests between the tissue samples from the RUT kit and the gastric biopsy samples was 94.4% (51/54). The rate of DPO-PCR and silver stain positivity in the RUT-negative cases was 20.0% (7/35).
CONCLUSION: In diagnosing H. pylori infection, DPO-PCR can be performed on tissue samples that have been processed by the RUT kit. Particularly, in patients with RUT-negative results, DPO-PCR on these tissue samples could be helpful in detecting of H. pylori infection.
Core tip: The rapid urease test (CLOtest) alone is unreliable in diagnosing Helicobacter pylori (H. pylori) infection and does not provide information about resistance to clarithromycin. Therefore, we investigated whether tissue samples that have been analyzed by the CLOtest kit are suitable for dual-priming oligonucleotide-based multiplex PCR (DPO-PCR) to detect H. pylori. Our results demonstrated that the DPO-based multiplex PCR test using tissue samples processed by the CLO kit is appropriate for detecting H. pylori and clarithromycin resistance. Particularly, in patients with CLO-negative results, this method is helpful for diagnosing H. pylori infection. Moreover, it would be beneficial in economical aspects.
-
Citation: Chung WC, Jung SH, Oh JH, Kim TH, Cheung DY, Kim BW, Kim SS, Kim JI, Sin EY. Dual-priming oligonucleotide-based multiplex PCR using tissue samples in rapid urease test in the detection of
Helicobacter pylori infection. World J Gastroenterol 2014; 20(21): 6547-6553 - URL: https://www.wjgnet.com/1007-9327/full/v20/i21/6547.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i21.6547
In patients with Helicobacter pylori (H. pylori) - related diseases, a reliable diagnosis of infection with this bacterium is crucial, but no single test can be considered the gold standard. The rapid urease test (RUT) is the most commonly used biopsy-based method to diagnose H. pylori infection because it is simple, rapid and accurate. However, it requires a high density of bacteria, and anything that reduces the bacterial load may produce false-negative tests[1]. Moreover, various medications may affect the presence of urease in the gastric mucosa. Within 2 wk of taking a proton pump inhibitor, bismuth or antibiotic, most of the H. pylori organisms have disappeared, making the RUT negative. Moreover, H. pylori does not distribute evenly in the stomach[2]. Several factors including gastric pH, inflammatory cells, atrophic gastritis, and intestinal metaplasia affect its distribution[3,4]. If the biopsy sample is taken from an area of intestinal metaplasia, the RUT will fail[2,3].
Molecular methods are widely used to diagnosis H. pylori infection, as are analyses of its virulence and resistance patterns[4-7]. Polymerase chain reaction (PCR) is the most sensitive and specific method for detecting of H. pylori in gastric biopsy specimens. It has great sensitivity with a detection limit of 0.02 pg H. pylori DNA, which corresponds to only 10 organisms[8]. However, in clinical practice, PCR is complicated and it is not always simple to achieve the desired result. It is a time-consuming and labor-intensive method. Recently, a commercial dual-priming oligonucleotide (DPO) primer has been developed to detect single-nucleotide polymorphisms (SNP) using a 1-step PCR assay[9]. Detection is accurate and rapid using the specific primers. Moreover, DPO-based multiplex PCR (DPO-PCR) can provide information about clarithromycin resistance because clarithromycin resistance is the main predictor of failure of eradication treatments; therefore, the detection of clarithromycin resistance is important.
Previously, tissue samples taken for rapid urease testing have also been analyzed by PCR, whitch can detect H. pylori DNA in gastric tissue samples obtained for the RUT kit[10]. When H. pylori infection is not detected in cases of peptic ulcer bleeding or peptic ulcer disease with chronic atrophy, an additional biopsy specimen and endoscopic procedure should be performed. In addition, in case repeated eradication therapy fails and the patient is clinically suspected of having an infection with a clarithromycin resistant strain, an additional biopsy specimen is necessary. Unfortunately, taking extra biopsy specimens is burdensome for clinicians and patients. In this study, we aimed to evaluate DPO-PCR in diagnosing of H. pylori infection, and to determine whether the tissue samples that already been submitted to the RUT kit are suitable for the DPO-PCR test compared with the result of DPO-PCR performed on gastric biopsy samples, RUT, and histologic results.
All patients with specific gastrointestinal symptoms were enrolled at a teaching hospital of the Catholic University of Medicine, St. Vincent’s Hospital, from November. 2011 to May 2012. Patients who were referred to the endoscopy unit were recruited for this prospective study.
Patients were eligible to enter the study if they were older than 18 year of age and had gastric H. pylori infection. None of the patients had a history of H. pylori eradication, had undergone previous gastric surgery or had taken antibiotics in the 2 mo preceding the study. Patients were also excluded if they had significant renal, hepatic, cardiovascular, metabolic or hematological disorders. Additionally, pregnant or lactating women were excluded from our investigation.
An estimated sample size of 50 subjects per group would give an 80% power to detect a difference of 15% in the H. pylori detection rate compared to other tests (assumed to have an detection rate of 85%), with a 2-sided alpha = 0.05. Thus, with a 10% drop out rate we needed to recruit at least 55 patients for each group.
n = [Zα/2(p0q0)2/1 + Zβ(p0q0)2/1]2/[(ε-|δ|)2]
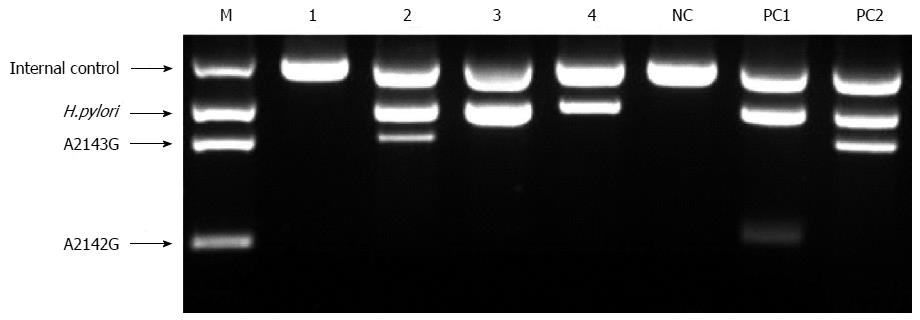
Genomic DNA from gastric biopsy and tissue samples analyzed by RUT (CLOtest; Kimberly- Clark, Utah, United States) were extracted using the QIAamp DNA Mini Kit (Qiagen, Valencia, CA, United States). DNAs was stored at -20 °C until it was required for analysis. A novel commercialized DPO-PCR (Seeplex® ClaR- H. pylori ACE Detection, Seoul, South Korea) was performed according to the manufacturer’s recommendations. This method uses 2 forward and 2 reverse DPO primers against the 23S rRNA gene. A 4-primer combination mixture (HP-F, HP-R, A2142G-F, and A2143G-R) that amplifies 3 fragments (i.e., a H. pylori common sequence, an A2142G mutant, and an A2143G mutant) is made for the multiplex PCR (Figure 1). The A2142G and A2143G mutations of the 23S rRNA gene in H. pylori are associated with resistance to clarithromycin[11,12]. DPO-PCR is a multiplex PCR that can be performed in any standard thermocycler. It is analyzed using a semi-automated system (i.e., Screen tape®), which allows ultra rapid migration and analysis of the PCR products in small polyacrylamide gels. 8-Methoxysporalen was added during the mix preparation to intercalate between the double-stranded nucleic acids generated during amplification, thereby limiting carry-over contamination after UV irradiation and before the PCR product analysis. The kit also includes a primer pair for internal control.
During endoscopy, gastric biopsy specimens were taken from the greater curvature of the mid -antrum and corpus for histology, CLOtest and DPO-PCR. The diagnosis of H. pylori infection was made based on (1) histologic evidence of H. pylori in any 2 specimens taken from the antrum or corpus by silver stain; or (2) positive CLOtest and serological test results. If only the CLOtest was positive, serological test was performed additionally. All patients with peptic ulcer disease were prescribed proton pump inhibitor (PPI) therapy for 2-4 wk, and the remaining patients were treated according to their symptoms for 2 wk. If the silver stain and CLOtest were H. pylori -negative, we obtained an additional biopsy specimen under endoscopy 4-8 wk after initial examination. Patients did not take PPIs for at least 2 wk before re-endoscopy.
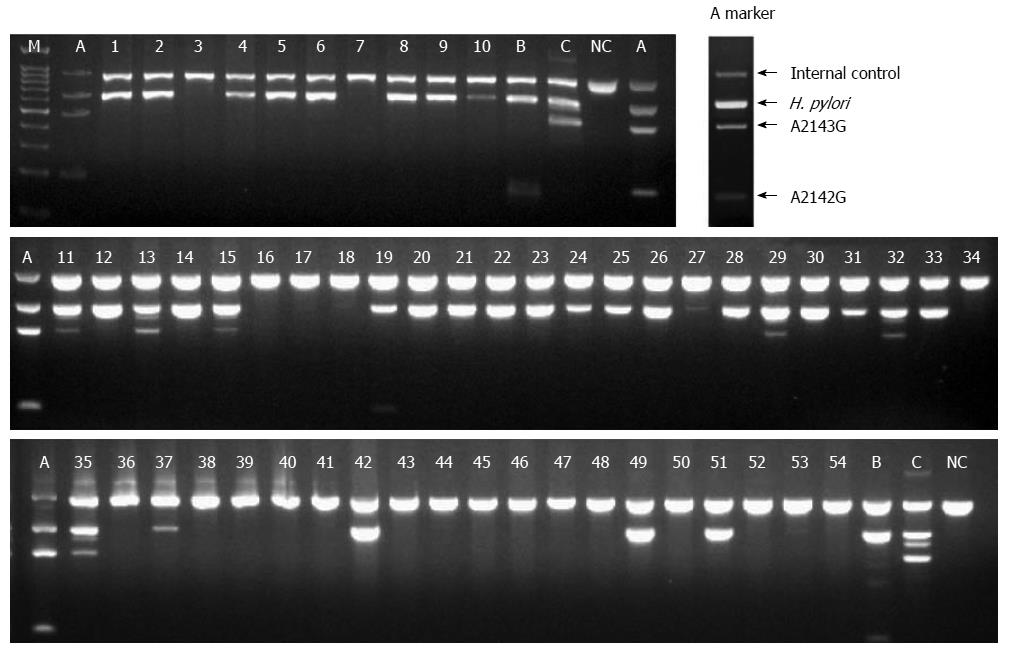
The specimens were fully immersed in the CLO reagent, and the test was interpreted at 1 and 9 h in the endoscopy room. If the CLOtest was positive, the specimens were placed at -15 to -20 °C. If the CLOtest was negative, it was re-interpreted 24 h later in ambient air. DPO-PCR tests were performed on the gastric biopsy samples (Seegene Institute of Life Science, Seoul, South Korea) (Figure 1) and on tissue samples obtained from the CLOtest kit at 2 separate institutes (Research Institute of St. Vincent Hospital, Suwon, South Korea) (Figure 2).
The study was approved by the institutional review board of the Catholic University of South Korea (VC11EISI0200). Each patient provided written informed consent to participate.
Gastric tissue samples were taken from 57 patients, but 3 patients were excluded because of inadequate DNA samples. A total of 54 patients (43 males, 11 females, mean age 58.7 ± 14.2 years) were enrolled in this study. Of these patients, 25 (46.3%) had gastric ulcers, while 14 (25.9%) had duodenal ulcers. Three (5.6%) patients had both gastric and duodenal ulcers, and 12 (22.2%) patients had chronic gastritis. H. pylori was detected in 76.0% (19/25) of the gastric ulcer patients, 64.3% (9/14) of the duodenal ulcer patients, and 33.3% (4/12) of the gastritis. H. pylori was detected in 100% (3/3) of the patients with both gastric and duodenal ulcers.
H. pylori was detected in 35 of 54 (64.8%) patients. The diagnostic sensitivities of histology, CLOtest, and DPO-PCR were 85.7% (30/35), 74.3% (26/35) and 97.1% (34/35), respectively (P = 0.02) (Table 1). The positive predictive value (PPV) is the proportion of patients with positive test results who are correctly diagnosed. The negative predictive value (NPV) is the proportion of patients with negative test results who are correctly diagnosed. The PPV of DPO-PCR was 94.4%, whereas its NPV was 94.7%. In CLOtest-negative cases, DPO-PCR and histology were both positive in 20.0% of the patients (7/35).
| Method of detection | Number of positivity | Number ofH.pylori infection |
| Silver stain - 1st session | 30 | 30 (85.7) |
| 2nd session | 2 | |
| CLOtest 1st session | 26 | 26 (74.3) |
| 2nd session | 3 | |
| DPO-PCR of gastric biopsy | 37 | 34 (97.1) |
| DPO-PCR using tissue sample of CLO kit | 34 | 33 (94.3) |
DPO-PCR was positive in 68.5% (37/54) of the gastric biopsy smaples, whereas the CLOtest kit was positive in 61.1% (33/54). The concordance rate of DPO-PCR tests between gastric biopsy samples and tissue samples analyzed by the CLOtest kit was 94.4% (51/54). There were only 2 false-positives in the gastric biopsy samples. Despite repeated histologic examinations, negative results were observed. In 1 case, both of DPO-PCR tests were all negative and there was positive histologic examination.
Among the 35 patients with H. pylori infection, 7 patients (20.0%) had 23S rRNA point mutations associated with clarithromycin resistance. The mutation subtypes included 6 patients with A2143G and 1 patient with A2142G.
A total of 28 patients with peptic ulcer disease were recommended to undergo eradication therapy of H. pylori, and follow-up was incomplete in 4 patients. Twenty-four patients completed the standard 7-d eradication therapy. In the absence of a 23S rRNA point mutation in H. pylori, the patients were treated with PPI-based triple therapy - twice daily with 1000 mg of amoxicillin, 500 mg of clarithromycin and 30 mg of lansoprazole. If a mutation was present, the patients took metronidazole containing triple therapy, which consisted of 1000 mg of amoxicillin and 30 mg of lansoprazole twice daily and 500 mg of metronidazole 3 times daily. Eradication was determined by the C13-urea breath test 6 wk after the eradication therapy. H. pylori eradication (intention to treat) was successful in 23/28 (90.3%) patients and the per- protocol analyses is showed a rate of 95.8% (23/ 24).
Despite the highly sensitive and specific nature of PCR, it can provide false-positive or false-negative results. To reduce the risk of false-positive results in PCR, a sterilization protocol to prevent the amplification of contaminants and highly specific primers should be applied. Compared to conventional PCR, DPO-PCR increases the specificity and sensitivity of detection by blocking non-specific binding sites; therefore, it eliminates imperfect primer annealing[13]. On the basis of the C13-urea breath test, H. pylori detection by DPO- PCR had a sensitivity of 87.5%, a specificity of 91.3%, a positive predictive value of 84.0%, a negative predictive value of 93.3%, and an accuracy of 90.0%[14].
However, in clinical practice, false-negative results can be a more significant problem. When the detection of H. pylori infection initially fails in patients with H. pylori-associated disease, additional biopsies and endoscopic procedures are required, which would be burdensome for clinicians and patients. Particularly in patients with recent upper gastrointestinal bleeding, the diagnosis of H. pylori infection can be discouraging, and its prevalence in bleeding peptic ulcers is usually underestimated[15]. Therefore, a diagnostic test at some point after the bleeding episode would be a good tool to diagnose of H. pylori infection[16,17]. From this point of view, our results are promising and the DPO-PCR test on the samples taken for RUT can reduce medical costs.
RUT is a convenient and inexpensive way to diagnosis H. pylori infection and is used worldwide clinically and for research. After interpretation, the biopsy specimen utilized for RUT is usually discarded. DPO- PCR is more expensive than RUT. It is highly dependent on the activity and equipment of the laboratory in which the test is performed. However, PCR tests using gastric biopsy specimens from the RUT kit can reduce the need for re-endoscopic examination with biopsy. Particularly when the RUT is negative and there is a suspicion of H. pylori, our method will greatly lighten the burdens of both clinicians and patients. In addition, when clarithromycin is the first-choice drug or in countries with high prevalence of primary clarithromycin resistance, our test will alleviate the social and economic costs of medical treatment.
Primary resistance to clarithromycin significantly affects the efficacy of eradication therapy and is considered to be a strong predictive factor for treatment failure[18,19]. The eradication rate could be increased to an ideal level by conducting a test for clarithromycin resistance. The A2142G and A2143G mutations of the 23S rRNA gene in H. pylori are associated with clarithromycin resistance[11,12]. Using the rapid and inexpensive DPO-based multiplex PCR test to detect clarithromycin resistance, clinicians can select the best regimen before eradication therapy. The DPO primer system differs from a conventional system by including a poly(I) linker between 2 unequal segments of primer sequences, which increases the specificity sufficiently to discriminate single-base changes by using 1-step PCR and allows accurate multiplex PCR. Therefore, there is no need for additional steps, expensive equipment, or specialized skills. In a previous clinical study, DPO-PCR was shown to be an alternative to culture and testing for clarithromycin resistance to H. pylori. The sensitivity of DPO-PCR was 97.7% and specificity was 83.1%, considering culture as the reference test[13]. Our results show that the frequency of clarithromycin resistance was 20%, but this result was not conclusive because of the small number of enrolled patients. A previous report from South Korea revealed that the antibiotic resistance rates for amoxicillin, metronidazole, and clarithromycin were 0%, 40.6%, and 5.9%, respectively, prior to 2000[20]. However, these rates increased to 18.5%, 66.2%, and 13.8%, respectively, in 2003[21]. Between 2003 and 2009, the resistance rates to amoxicillin and metronidazole decreased to 4.5% and 29.7%, respectively, but the resistance rate for clarithromycin increased drastically to 32.0%[22]. The recent Maastricht III consensus report recommended that the clarithromycin not be used or that a susceptibility test be performed when the resistance to this antibiotic is ≥ 20%[23]. Currently, DPO-based multiplex PCR can detect clarithromycin resistance before eradication therapy and help in the selection of the appropriate regimen. Hopefully, this process can prevent exposure to unnecessary antibiotics and increase the eradication rate.
Gastric biopsy specimens stored in a gel of the RUT kit can be used to confirm the diagnosis of H. pylori infection and to test clarithromycin susceptibility despite having been stored at room temperature for 30 d[24]. H. pylori DNA can be detected by PCR on gastric biopsy specimens processed by the RUT kit. The contents of RUT are bacterial agar containing urea, phenol red (phenolsulfonphthalein), and sodium phosphate. These materials do not damage DNA. We combined the rationales for DPO-PCR and RUT and designed this study to determine the diagnostic accuracy of a DPO-PCR test using tissue specimens previously processed by a RUT kit. DNA testing is becoming a popular method of clinical diagnosis. Furthermore, DNA profiling is being used more often and can provide individual medical information. However, DNA testing can result in ethical or legal issues if informed consent is not obtained. In clinical practice, an institutional device or method to prevent inadvertent disclosure of personal information should be established.
In conclusion, our results demonstrate that DPO-based multiplex PCR using tissue samples analyzed by RUT is appropriate for detecting of H. pylori and clarithromycin resistance. Particularly in patients with RUT-negative results, this test could be helpful for diagnosing H. pylori infection. Moreover, it would be beneficial in economical aspect. Further experience and large-scale studies are needed to compare the various diagnostic methods.
In diagnosing Helicobacter pylori (H. pylori) infection, the CLOtest alone is unreliable. PCR has the advantage of providing diagnostic results that are highly sensitive and specific. The authors aim to investigate whether tissue samples previously processed by the CLOtest kit are suitable for dual-priming oligonucleotide-based multiplex polymerase chain reaction (DPO-PCR) to detect H. pylori.
A reliable diagnosis of infection is crucial in patients with H. pylori-related diseases, but there is no single test that can be considered the gold standard. Recently, a commercial DPO primer has been developed to detect SNPs using a 1-step PCR assay. It achieves accurate and rapid detection using the specific primers. Moreover, DPO-PCR provides information about clarithromycin resistance.
Authors’ results demonstrate that DPO-PCR using a tissue sample previously analyzed by the CLOtest kit is appropriate for detecting H. pylori and clarithromycin resistance, particularly in patients with CLO-negative results.
In diagnosing H. pylori infection, the DPO-PCR test is accurate and economical. Further experience and large-scale studies are needed to compare the various diagnostic methods.
DPO-PCR is a novel, commercial dual-priming oligonucleotide-based multiplex PCR method used to detect H. pylori.
This manuscript investigates whether tissue samples from the CLOtest kit are suitable for DPO-PCR to detect H. pylori. Although it is possible that the small sample size could affect the results, the DPO-PCR test could be helpful in diagnosing H. pylori infection.
P- Reviewer: Yeung CY S- Editor: Wen LL L- Editor: A E- Editor: Liu XM
| 1. | Yakoob J, Jafri W, Abid S, Jafri N, Abbas Z, Hamid S, Islam M, Anis K, Shah HA, Shaikh H. Role of rapid urease test and histopathology in the diagnosis of Helicobacter pylori infection in a developing country. BMC Gastroenterol. 2005;5:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 23] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 2. | Lan HC, Chen TS, Li AF, Chang FY, Lin HC. Additional corpus biopsy enhances the detection of Helicobacter pylori infection in a background of gastritis with atrophy. BMC Gastroenterol. 2012;12:182. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 3. | Kim HD, Kim do H, Park H, Kim WJ, Ahn YS, Lee YJ, Park SM, Seo ES, Park C, Kim YH. Detection of Helicobacter pylori in Gastric Aspirates Using a Monoclonal Antibody-Based Test. Gut Liver. 2013;7:30-34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 4. | Mégraud F, Lehours P. Helicobacter pylori detection and antimicrobial susceptibility testing. Clin Microbiol Rev. 2007;20:280-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 476] [Cited by in RCA: 488] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 5. | Menoni SM, Bonon SH, Zeitune JM, Costa SC. PCR-Based Detection and Genotyping of Helicobacter pylori in Endoscopic Biopsy Samples from Brazilian Patients. Gastroenterol Res Pract. 2013;2013:951034. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 6. | Cerqueira L, Fernandes RM, Ferreira RM, Oleastro M, Carneiro F, Brandão C, Pimentel-Nunes P, Dinis-Ribeiro M, Figueiredo C, Keevil CW. Validation of a fluorescence in situ hybridization method using peptide nucleic acid probes for detection of Helicobacter pylori clarithromycin resistance in gastric biopsy specimens. J Clin Microbiol. 2013;51:1887-1893. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 7. | Fonseca TL, Moraes EP, Juliano CR, Silva AM, Scaini CJ, Mendoza-Sassi RA, Silva PE. Detection of Helicobacter pylori by phenotypic and genotypic methods. Dig Dis Sci. 2010;55:1643-1648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 8. | Linpisarn S, Koosirirat C, Prommuangyong K, Suwan W, Lertprasertsuke N, Phornphutkul K. Use of different PCR primers and gastric biopsy tissue from CLO test for the detection of Helicobacter pylori. Southeast Asian J Trop Med Public Health. 2005;36:135-140. [PubMed] |
| 9. | Chun JY, Kim KJ, Hwang IT, Kim YJ, Lee DH, Lee IK, Kim JK. Dual priming oligonucleotide system for the multiplex detection of respiratory viruses and SNP genotyping of CYP2C19 gene. Nucleic Acids Res. 2007;35:e40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 171] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 10. | Yula E, Nagiyev T, Kaya OA, Inci M, Celik MM, Köksal F. Detection of primary clarithromycin resistance of Helicobacter pylori and association between cagA (+) status and clinical outcome. Folia Microbiol (Praha). 2013;58:141-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 11. | Mégraud F. H pylori antibiotic resistance: prevalence, importance, and advances in testing. Gut. 2004;53:1374-1384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 607] [Cited by in RCA: 681] [Article Influence: 32.4] [Reference Citation Analysis (2)] |
| 12. | Versalovic J, Shortridge D, Kibler K, Griffy MV, Beyer J, Flamm RK, Tanaka SK, Graham DY, Go MF. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother. 1996;40:477-480. [PubMed] |
| 13. | Lehours P, Siffré E, Mégraud F. DPO multiplex PCR as an alternative to culture and susceptibility testing to detect Helicobacter pylori and its resistance to clarithromycin. BMC Gastroenterol. 2011;11:112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 66] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 14. | Lee HJ, Kim JI, Cheung DY, Kim TH, Jun EJ, Oh JH, Chung WC, Kim BW, Kim SS, Park SH. Eradication of Helicobacter pylori according to 23S ribosomal RNA point mutations associated with clarithromycin resistance. J Infect Dis. 2013;208:1123-1130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 15. | Ramírez-Lázaro MJ, Lario S, Casalots A, Sanfeliu E, Boix L, García-Iglesias P, Sánchez-Delgado J, Montserrat A, Bella-Cueto MR, Gallach M. Real-time PCR improves Helicobacter pylori detection in patients with peptic ulcer bleeding. PLoS One. 2011;6:e20009. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 52] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 16. | Gisbert JP, Abraira V. Accuracy of Helicobacter pylori diagnostic tests in patients with bleeding peptic ulcer: a systematic review and meta-analysis. Am J Gastroenterol. 2006;101:848-863. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 135] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 17. | Güell M, Artigau E, Esteve V, Sánchez-Delgado J, Junquera F, Calvet X. Usefulness of a delayed test for the diagnosis of Helicobacter pylori infection in bleeding peptic ulcer. Aliment Pharmacol Ther. 2006;23:53-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 18. | Vakil N, Megraud F. Eradication therapy for Helicobacter pylori. Gastroenterology. 2007;133:985-1001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 175] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 19. | Graham DY, Fischbach L. Helicobacter pylori treatment in the era of increasing antibiotic resistance. Gut. 2010;59:1143-1153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 652] [Cited by in RCA: 736] [Article Influence: 49.1] [Reference Citation Analysis (1)] |
| 20. | Kim JJ, Reddy R, Lee M, Kim JG, El-Zaatari FA, Osato MS, Graham DY, Kwon DH. Analysis of metronidazole, clarithromycin and tetracycline resistance of Helicobacter pylori isolates from Korea. J Antimicrob Chemother. 2001;47:459-461. [PubMed] |
| 21. | Kim JM, Kim JS, Jung HC, Kim N, Song IS. [Antibiotic resistance of Helicobacter pylori isolated from Korean patients in 2003]. Korean J Gastroenterol. 2004;44:126-135. [PubMed] |
| 22. | Hwang TJ, Kim N, Kim HB, Lee BH, Nam RH, Park JH, Lee MK, Park YS, Lee DH, Jung HC. Change in antibiotic resistance of Helicobacter pylori strains and the effect of A2143G point mutation of 23S rRNA on the eradication of H. pylori in a single center of Korea. J Clin Gastroenterol. 2010;44:536-543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 106] [Article Influence: 7.1] [Reference Citation Analysis (1)] |
| 23. | Malfertheiner P, Megraud F, O’Morain C, Bazzoli F, El-Omar E, Graham D, Hunt R, Rokkas T, Vakil N, Kuipers EJ. Current concepts in the management of Helicobacter pylori infection: the Maastricht III Consensus Report. Gut. 2007;56:772-781. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1396] [Cited by in RCA: 1352] [Article Influence: 75.1] [Reference Citation Analysis (1)] |
| 24. | Li Y, Rimbara E, Thirumurthi S, Trespalacios A, Reddy R, Sabounchi S, Attumi TA, Graham DY. Detection of clarithromycin resistance in Helicobacter pylori following noncryogenic storage of rapid urease tests for 30 days. J Dig Dis. 2012;13:54-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 24] [Article Influence: 1.8] [Reference Citation Analysis (0)] |