Published online Apr 7, 2014. doi: 10.3748/wjg.v20.i13.3443
Revised: January 16, 2014
Accepted: March 6, 2014
Published online: April 7, 2014
Processing time: 188 Days and 17.5 Hours
The mechanisms that regulate disease progression during hepatitis C virus (HCV) infection and the response to treatment are not clearly identified. Numerous studies have demonstrated that a strong host immune response against HCV favors HCV clearance. In addition, genetic factors and metabolic machinery, particularly cholesterol modulation, are involved in HCV infection. It is likely that the interplay between all of these factors contributes to the outcome of HCV infection. In recent years, the world has experienced its largest epidemic of obesity. Mexico and the United States are the leading sufferers from this epidemic at the global level. Obesity is associated with the development of numerous pathologies including hypercholesterolemia which is one of the eight most important risk factors for mortality in Mexico. This may be related to the course of HCV infection in this population. Here, we focus on the urgent need to study the progression of HCV infection in relation to ethnic characteristics. Discoveries are discussed that hold promise in identifying immune, metabolic and genetic factors that, in conjunction, could be therapeutic targets or predictors of the progression of HCV infection.
Core tip: Immunologic, metabolic and genetic factors are involved in the progression of hepatitis C virus (HCV) infection and the response to treatment. The significant increase in obesity worldwide, including in Mexico, imposes a new metabolic stress factor in patients with HCV infection. Given that the lipid components associated with HCV infection are finely modulated, it is possible that the progression of HCV infection may be regulated by the characteristics of the population’s lipid composition.
- Citation: Fierro NA, Gonzalez-Aldaco K, Torres-Valadez R, Martinez-Lopez E, Roman S, Panduro A. Immunologic, metabolic and genetic factors in hepatitis C virus infection. World J Gastroenterol 2014; 20(13): 3443-3456
- URL: https://www.wjgnet.com/1007-9327/full/v20/i13/3443.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i13.3443
Hepatitis C infection is caused by the hepatitis C virus (HCV), a positive-stranded ribonucleic acid (RNA) virus of the Flaviviridae family with a hepatotropic lifecycle. HCV infection is an important cause of chronic liver disease and the third-leading cause of all death from cirrhosis and hepatocellular carcinoma (HCC) worldwide. Approximately 3% of the world’s population (160 million people) are currently infected with HCV, which in most cases establishes a lifelong chronic infection[1]. However, 25%-30% of infected individuals spontaneously clear the virus during acute infection. Because HCV is a non-cytopathic virus, it is accepted that the interplay between the virus and the host immune response may influence the outcome of infection[2]. In addition, host genetic factors, such as polymorphisms in cytokine and chemokine receptor genes promoters[3,4], are thought to be important contributors to the modulation of HCV outcome.
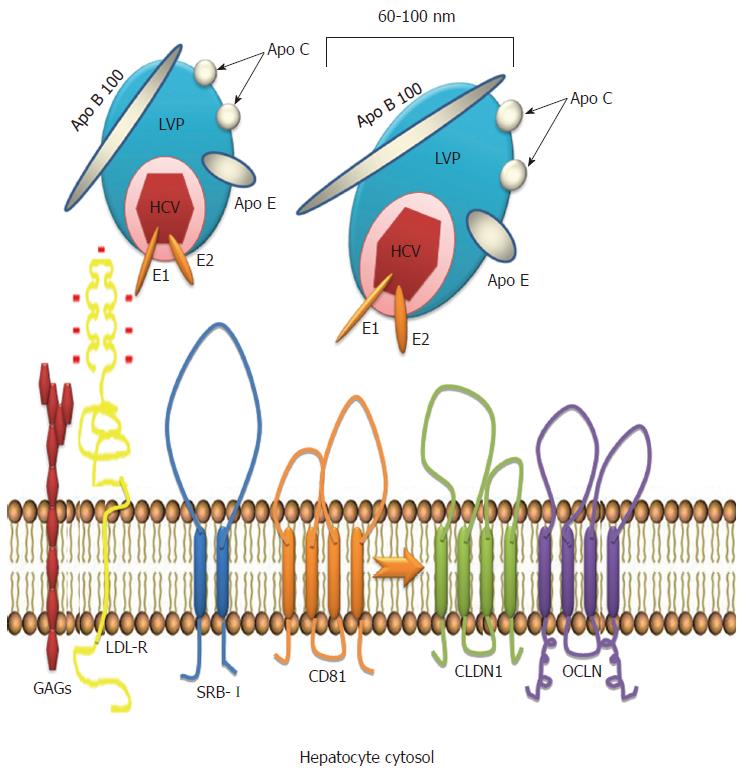
A key role for the low-density lipoprotein receptor (LDL-R) has been demonstrated during the first steps of HCV attachment to the cell surface[5]. LDL is responsible for transporting most of the cholesterol in plasma[6] and cholesterol levels are tightly modulated during HCV infection. Virus entry into the cell is also mediated by other lipoprotein receptors including scavenger receptor class B type I (SRB-I). In addition to providing a docking site for HCV particles, SRB-I facilitates entry of the virus into the hepatocyte[6,7]. The E2 envelope protein determines viral attachment to SRB-I.
Currently, Latinos represent the fastest growing ethnic group in the United States and are the most overweight. Mexico and the United States are experiencing the largest obesity epidemic in the world. In conjunction, genetic and cultural components, lifestyle and environmental factors are all associated with the development of obesity. In addition to representing a public health problem by itself, obesity is also associated with the development of numerous pathologies, including hypercholesterolemia. A recent analysis of the burden of disease revealed that hypercholesterolemia was among the eight most important risk factors for mortality in Mexico[8]. Thus, the specific characteristics of the lipid components already described suggest a unique mechanism of regulation of metabolic machinery among the Mexican people that could impact the progression of HCV infection.
Crosstalk between metabolic and immune components in conjunction with viral and genetic host factors may occur and predict HCV disease outcome. To date, the exact mechanisms responsible for HCV clearance and recovery are unknown. An understanding of these mechanisms will contribute to the development of novel, individualized, preventive strategies and an urgently needed vaccine. This review summarizes recent advances in understanding the crosstalk between the immune response, metabolism and genetics in HCV viral clearance and emphasizes characteristics of the Mexican population and their association with this process.
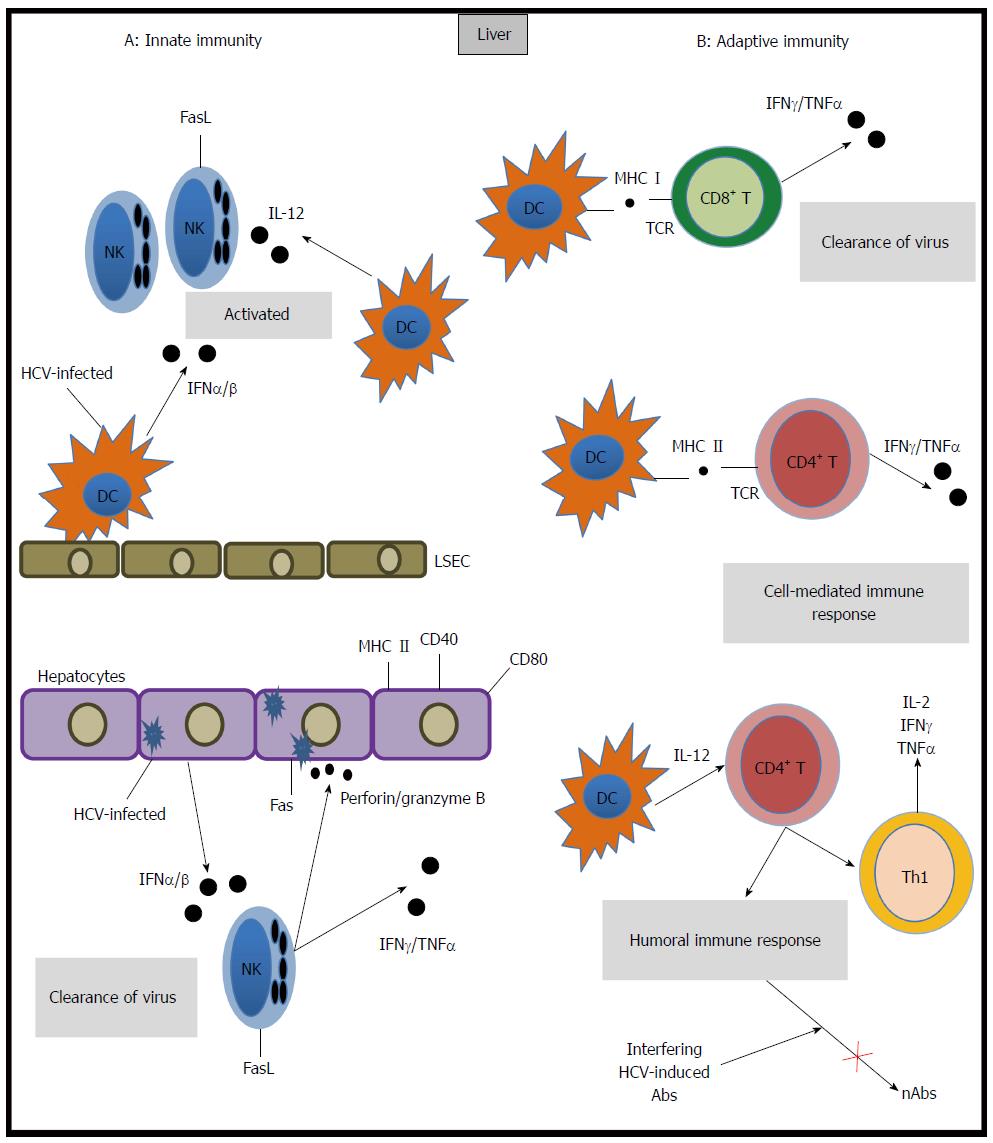
A robust innate immune response is activated when HCV infects the liver. This response includes the induction of several interferon-stimulated genes (ISGs)[9,10] and is mediated by the specific production of inflammatory and antiviral cytokines by macrophages, natural killer (NK) cells and neutrophils, which release perforin, granzyme B, interferon-gamma (IFN-γ) and tumor necrosis factor-beta. Immune cells also express Fas ligand which cause cell death in infected hepatocytes[11].
Animal models have enabled the precise description of the kinetics of viremia throughout infection. In particular, chimpanzee models have revealed that HCV RNA levels increase rapidly upon initial infection[12]. Thereafter, a slow decrease in viremia is observed. It is accepted that the innate immune response in hepatocytes contributes to the second phase of slowed viral replication[13]. This response includes type I interferon responses and the antiviral activity of plasmacytoid dendritic cells and NK cells. Thus, an ineffective innate immune response can result in disease progression.
The adaptive immune response to HCV is mediated by the humoral and cellular immune systems. HCV-specific T lymphocytes are detectable five to nine weeks after infection[14,15], which coincides with the onset of hepatitis. Both CD4+ and CD8+ T cells have been shown to play major roles in the outcome of HCV infection. CD8+ T cells inhibit HCV replication by cytolytic and non-cytolytic effector mechanisms[16] that are highly dependent on CD4+ T cell activation. CD4+ T cells are critical for the elimination of viruses through tightly regulated mechanisms. CD4+ T-helper cells are divided into four subsets (Th1, Th2, T regulatory, Th17) based on their expression profile of transcription factors and secreted cytokines. An effective Th1 cellular function is crucial for an adequate immune response against HCV, given the specific production of IFN-γ by this cellular subtype. Unbalanced Th1/Th2 T-cell responses in the liver are a characteristic of hepatic inflammation and subsequent liver fibrosis as a result of HCV infection[17]. Clinical data reveals increased interleukin-17 levels in HCV-infected patients who had increased alanine transaminase (ALT) values, suggesting that Th17 cells in chronic hepatitis C infection might be associated with control of liver injury. However, these observations are preliminary and not fully conclusive[18,19]. Indeed, vigorous peripheral and intrahepatic virus-specific T cell responses that target multiple epitopes have been described in patients who recover from HCV infection[13], while a weak and functionally impaired T cell response has been reported in patients who fail to clear the virus. Consistent with these findings, the role of T regulatory cells in HCV seems to range from suppressing T-cell responses directed against HCV to down-regulating the immune responses causing liver damage[20]. In addition, animal models have indicated that early expression of memory precursor markers on HCV-specific T cells or the expression of ligands for these memory markers in the liver predicts the outcome of acute infection[17] (Figure 1).
The generation of neutralizing antibodies represents a protective strategy in host immunity. Neutralizing anti-HCV antibodies (nAbs) were originally described in chimpanzees. These antibodies target epitopes within the hypervariable region 1 (HVR1) of envelope glycoprotein E2[21]. In humans, nAbs were identified in a cohort study of transplant recipients in which the incidence of HCV viremia was lower in patients receiving immunotherapy (anti-HBV immunoglobulins contaminated with anti-HCV immunoglobulins) compared to patients whose therapy did not include anti-HCV antibodies[22], indicating the potential contribution of neutralizing antibodies to viral control. Functional analysis and neutralization experiments using sera from chronically HCV-infected patients have demonstrated that host neutralizing responses target viral entry at a step after initial HCV binding. Initial HCV attachment to the cell surface is likely facilitated by interactions with attachment factors that include the LDL receptor[5]. Upon initial attachment, at least six host entry factors including scavenger SRB-I, CD81, the tight junction proteins claudin 1 and occludin[23], receptor tyrosine kinases[24] and the Niemann-Pick C1-like 1 cholesterol absorption receptor[25] are important for particle internalization. Indeed, several E2 domains have been shown to play pivotal roles in viral entry and neutralization. Two regions in the E2 viral envelope glycoprotein have increased genetic variability within quasispecies and among genotypes and have been identified as hypervariable regions. Antibodies that demonstrate broadly neutralizing activity tend to be directed against conserved and conformational epitopes within E2, which inhibit the interaction between CD81 and E2[26-30].
The strategies of HCV persistence and evasion of the immune response involve multiple mechanisms. The high genetic variability of HCV is a major contributor to the development of chronic HCV infection. It has been shown that HCV constantly circulates in the patient and rapidly evolves into genetically distinct but closely related variants within quasispecies. The simultaneous presence of different variants has been postulated to allow for the rapid selection of mutants that are best adapted to changes in the host environment. Genetic analysis reveals a positive correlation between distinct HCV quasispecies, viral clearance and a slowly adapting viral population, whereas distinct patterns of progression to chronicity are related to selective pressure on the HVR1 region of HCV E2. Moreover, chronic progression is associated with the rapid evolution of quasispecies[31], and single point mutations that result in glycosylation site modifications during viral genetic and conformational changes have been suggested to be responsible for masking the envelope glycoprotein binding sites[32-36]. The result is viral persistence in the body despite the presence of an immune response.
During the innate immune response, HCV interferes with innate signaling pathways using its structural and non-structural proteins to interact with factors that regulate ISGs, thus attenuating innate responses[37-39]. In addition, patients who fail to clear the virus show a weak and functionally impaired T cell response. This result strongly suggests that HCV also employs strategies to escape host adaptive immunity.
The action of nAbs may be blocked by the presence of interfering HCV-induced Abs[32,40,41]. There are two proposed mechanisms through which lipoproteins may provide HCV with protection from nAbs: masking viral epitopes by associating with LDL and very-low density lipoprotein (VLDL) or accelerating viral entry by interactions with high-density lipoprotein (HDL). In both cases, HCV lipoprotein-dependent protection results in limiting the exposure of viral epitopes to nAbs[42]. Recently, it has been suggested that in HCV genotype 2a, the viruses that combine with LDL and VLDL and are consequently distributed in low-density fractions are more capable of escaping neutralizing antibodies compared to viruses distributed in high-density fractions[43]. In addition, HCV is able to infect B-lymphocytes and induce hypermutations in the heavy chains of immunoglobulins. These hypermutations may decrease the affinity and specificity of anti-HCV antibodies and may allow the virus to escape from the immune system[44]. HCV dissemination by cell-to-cell transmission may also contribute to viral persistence by avoiding surveillance of nAbs present in the serum[45] (Figure 1).
After its discovery, HCV was found to be associated with lipoproteins. Thomssen et al[46] demonstrated the existence of distinct HCV particles categorized as high and low density particles. These particles, also recognized as lipo viro particles (LVP), are rich in triglycerides (TG) and can be almost completely precipitated by anti-Apo lipoprotein B and E. LVPs contain HCV RNA and structural viral proteins, mainly E1 and E2 that attach the virus to the hepatocyte surface through specific receptors[6,47].
Attachment and internalization of HCV into the cell represent the first steps in the viral replication cycle. The LDL receptor is likely the mediator of viral entry through Apolipoprotein E (ApoE), the major structural agent of infectious LVP and the natural ligand of LDL-R[48]. ApoE exists in three common isoforms (E2, E3 and E4) that affect LDL receptor binding[49]. Allele 4 is associated with clearance of HCV and protection against severe liver damage[50]. Allele 2 is associated with clearance and binds poorly to LDL-R; it is plausible that this defective binding could result in poor uptake of HCV LVP into hepatocytes, thus altering the balance between virus replication and the immune response by decreasing viral replication and favoring clearance. In contrast, allele 3 is associated with persistence[51]. Recently, an association between levels of ApoE and interferon sensitivity was found, and ApoE was shown to induce a higher LDL concentration, potentially inhibiting the binding of HCV to LDL-R. The association between ApoE and the peg-interferon treatment response suggests that lipid modulation is a potential target for modifying interferon sensitivity, favoring the clearance of HCV and avoiding progression to chronic infection[52] (Figure 2).
HCV RNA replication is strongly influenced by the intracellular levels and composition of fatty acids, including cholesterol. Recent studies have demonstrated that the expression of genes involved in biosynthesis, degradation and intracellular transport of lipids is altered during HCV infection[53,54]. The positive RNA strand of HCV induces remodeling of the intracellular membranes to generate compartments where RNA replication takes place. The NS5B viral protein is responsible for generating the membranous web required for RNA replication through diverse mechanisms[55]. Transcription of genes related to lipid metabolism is augmented during HCV infection. Sterol regulatory element-binding protein (SREBP) expression, which controls the transcription of specific genes required for cholesterol biosynthesis, is increased by the NS2 and NS4B HCV proteins. However, the exact mechanism responsible for disruption of host lipid metabolism by HCV infection is not yet clear. Understanding the components involved in this process will allow the possible design of specific therapeutic targets.
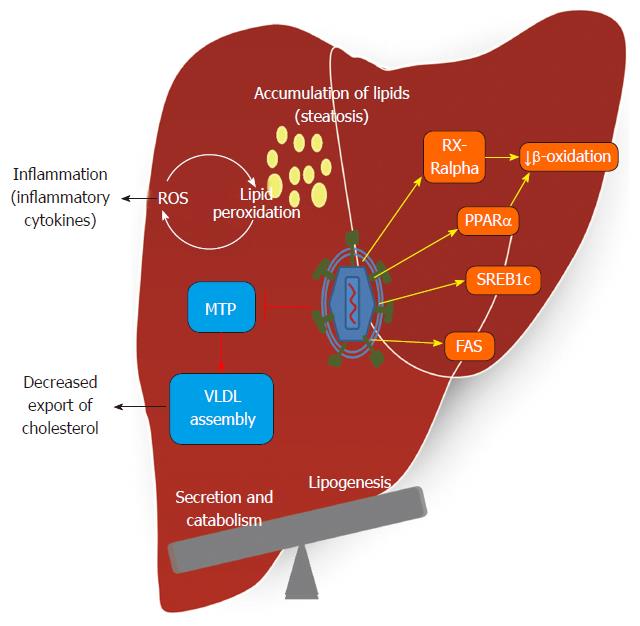
Some authors have reported that 30% of the total protein in complexes associated with HCV RNA is functionally involved in lipid metabolism[56]. Many lipids are crucial for the viral lifecycle, and inhibitors of cholesterol/fatty acid biosynthetic pathways inhibit viral replication, maturation and secretion[57]. In humans, apolipoprotein B (ApoB)-100 and ApoB-48 are obligatory proteins for the assembly of hepatic VLDL and intestinal chylomicron, respectively. VLDL assembly occurs via a two-step mechanism involving the formation of ApoB-containing VLDL precursor particles in the lumen of the endoplasmic reticulum, a step that requires microsomal triglyceride transfer protein (MTP)[58]. Hepatic VLDL assembly and secretion are also profoundly influenced by alterations in the de novo biosynthesis of phospholipids, such as phosphatidylethanolamine and phosphatidylcholine. Increased VLDL-TG concentration is a central feature of diabetic dyslipidemia and is largely caused by increased VLDL-TG secretion[59]. According to clinical data and experimental models, the HCV core protein has been shown to inhibit the MTP protein[60]. This enzyme plays a rate-limiting role in VLDL assembly and ApoB secretion[61].
Inhibition of VLDL assembly and secretion also affects virion morphogenesis and secretion, leading to the notion that HCV may hijack the VLDL secretion pathway for virion maturation and secretion. The reliance of HCV on host lipid metabolic pathways for its replication, morphogenesis and release requires the modulation of host lipid pathways by HCV to create a lipid-rich intracellular environment favorable for replication. HCV assembly and maturation in hepatocytes depend on MTP and ApoB in a manner that parallels the formation of VLDL[56,62]. Inhibitors of MTP and reduction of the expression of ApoB lowers virion production[56,63].
Steatosis, or the accumulation of hepatocellular lipid droplets, and altered serum lipid profiles are common consequences of HCV infection. The presence of steatosis in infected patients with HCV varies from 30% to 70% depending on ethnic and other factors, such as alcohol consumption, being overweight, obesity and factors that are also risk factors for non-alcoholic steatohepatitis[64]. In patients with chronic HCV, serum cholesterol is significantly reduced when compared to appropriately matched controls. This has been specifically analyzed in HCV genotype 3 infections in specific populations[65], and this metabolic effect can be reversed after successful HCV eradication. However, large-scale studies including samples from distinct populations and different genotypes are necessary to determine the roles of viral genotypes and host genetic factors in this process.
HCV modulates lipid metabolism to create an environment rich in lipids favorable for viral replication. Consistent with this lipid modification, each step of the viral replication cycle appears related to lipid metabolism[66]. The interaction of HCV proteins with hepatic cellular components contributes to the interference with lipid and carbohydrate metabolism, resulting in the release of cytokines, insulin resistance, inflammation, oxidative stress and steatosis[67,68]. The mechanism of triglyceride accumulation induced by HCV infection is multifactorial. de Gottardi et al[69] reported that host lipid metabolism may be modulated by HCV at three levels: impaired lipoprotein secretion, enhanced lipogenesis, and impaired fatty acid degradation. These detrimental alterations incurred during HCV infection then manifest as the pathological basis for some HCV-associated diseases, most notably steatosis and metabolic syndromes such as insulin resistance, obesity, and HCC[70-72]. MicroRNAs (miRNAs) exert regulatory control through modulation of many targets. In the liver, miRNA-122 is important for regulating lipid metabolism[73,74] and aberrant expression of miRNAs is linked to HCV infection[75]. It has recently been reported that HCV replication induces the expression of miR-27 in vitro and in vivo. This results in larger and more abundant lipid droplets and coincides with the repression of regulators of triglyceride homeostasis including peroxisome proliferator-activated receptor alpha (PPARα), identifying HCV’s up-regulation of miR-27 as a novel mechanism that may contribute to the development of steatosis[76].
In conjunction, the main lipid alterations in HCV are enhanced lipogenesis, reduced secretion and β-oxidation of lipids regulated by transcription factors, hormones and key enzymes that are briefly described below (Figure 3).
PPARs: PPARs are ligand-dependent transcription factors. The three subtypes of PPARs (PPARalpha, PPARdelta, and PPARgamma) are differentially expressed in tissues, and play pivotal roles in lipid, lipoprotein and glucose homeostasis[77]. PPARalpha, PPARdelta, and PPARgamma are differentially involved in HCV infection. A clear effect of PPARα in HCV RNA replication has been described and a recent report has shown that PPARα-selective antagonists inhibit HCV replication, while PPARα is not involved in this process[78]. PPARα is a transcription factor in the nuclear hormone receptor superfamily; it is involved in the differentiation of normal adipocytes, and its major function is to control fatty acid oxidation and activation. PPARα participates in enhancing the expression of adiponectin messenger RNA (mRNA) and serum adiponectin levels. PPARα deficiency causes defective hepatic fatty acid oxidation[79,80], while persistent activation of PPARα is essential for the pathogenesis of hepatic steatosis and HCC induced by HCV infection[81]. Dharancy et al[82] reported that PPARα mRNA expression was significantly reduced in the liver of chronic HCV patients infected with HCV genotype 3 when compared with HCV genotype 1 infections. A better understanding of the role of PPARα and its interaction with HCV proteins may help in developing novel therapies against HCV-induced steatosis and HCC.
Adiponectin and leptin: Adiponectin is an insulin-sensitizing protein that is abundantly expressed in white adipose tissue[83] and is a member of the adipocytokine family. Adiponectin improves hepatic insulin sensitivity, decreases lipid accumulation in macrophages and has anti-inflammatory properties[84-86]. Serum adiponectin levels in humans depend upon metabolic activity; it is present as a low molecular weight trimer and a high molecular weight (HMW) hexamer. HMW adiponectin is the biologically active form[87]. Adiponectin exerts its effect via two receptors, the adiponectin receptor 1 (AdipoR1) and adiponectin receptor 2 (AdipoR2). AdipoR1 is expressed in skeletal muscles, and AdipoR2 is expressed in the liver[88]. AdipoR1 is associated with AMP-activated protein kinase (AMPK) activation, while AdipoR2 is associated with PPARα activity. Some insights regarding the pathways of steatohepatitis have been identified by impaired lipid accumulation due to hepatic loss of adiponectin receptors, which play an important role in fatty acid accumulation by up-regulating the expression of enzymes during HCV infection, including AMPK, fatty acid synthase (FAS), acetyl-CoA carboxylase, liver gluconeogenic enzyme and phosphoenolpyruvate-carboxykinase[79,80]. Adiponectin is assumed to protect hepatocytes from triglyceride accumulation by increasing the β-oxidation of free fatty acids or decreasing de novo production of free fatty acid in hepatocytes[89]. In contrast to the anti-inflammatory role of adiponectin, leptin is a pro-inflammatory adipocytokine identified as one of the best markers of total body fat; its elevated expression can result in stimulation of cellular lipolysis and fatty oxidation, promoting a negative energy balance. Both adiponectin and leptin are crucial for hepatic steatosis[90]. However, data concerning the roles of adiponectin and leptin on HCV-related steatosis remain divergent. This is mainly due to the multiple functions involving adipocytokines. By using a HCV core-transgenic mouse model, a recent report reveals that HCV core-induced, non-obese hepatic steatosis is associated with down regulation of the leptin gene in visceral fat and hypoadiponectinemia[91]. A better understanding of this process may be valuable in the design of new therapeutic interventions, particularly in the cases of non-obese hepatic steatosis involving HCV infection.
AMPK: AMPK is a heterotrimeric protein with serine/threonine protein kinase activity that works as a sensor of cellular energy status and enables metabolic adaptation to the environment, such as in nutritional stress[92-94]. AMPK plays a key role in the regulation of both lipid and glucose metabolism. Activated AMPK inhibits energy-consuming biosynthetic pathways such as lipogenesis and activates ATP-producing catabolic pathways such as β-oxidation. A study demonstrated that phosphorylation of AMPK at threonine 172 causes dramatically reduced AMPK activity in cells infected with HCV or harboring an HCV subgenomic replicon. Thus, inhibition of AMPK is required for HCV replication and restoration of AMPK activity may represent a target for anti-HCV therapies[95,96]. The anti-HCV activity of 2-octynoic acid (2-OA), a compound used in perfumes, lipstick, and many food flavorings, has recently been revealed in vitro. This activity seems to be associated with the activation of AMPK by 2-OA, which regulates ISGs and suppresses miRNA-122 expression, inhibiting HCV infection. This represents a novel mechanism to explain inhibition of infection by AMPK[97].
SREBP: SREBP plays an important role in the regulation of lipid synthesis and cholesterol metabolism. The SREBP gene encodes three isoforms (SREBP-1a, SREBP-1c and SREBP-2[98] that regulate genes involved in cholesterol synthesis. SERBP-1a is an activator of cholesterol and fatty acid synthesis, while SERBP-1c regulates genes involved in lipid synthesis[99-102]. SREBP-1c activates the lipogenesis pathway in response to insulin. HCV infection and the HCV core protein up-regulate the expression of SREBP-1c and cause the development of fatty liver[103]. In non-alcoholic fatty liver disease, SERBP-1c is expressed at a level up to five-fold higher than observed in controls[104].
It has been demonstrated that HCV infection enhances the proteolytic cleavage of SREBP precursors in hepatic cells. The HCV NS2 and NS4B proteins can up-regulate SERBP-1c and consequently promote enhanced transcriptional activity of fatty acid synthase[105,106].
Animal models have been used to investigate the effect of the HCV core and NS proteins on SREBP gene regulation, and these models have indicated that HCV proteins that interfere with SREBP lead to steatosis[105,106]. The host subtilisin/kexin/isozyme/1 (SKI-1) or site 1 (S1P) plays a crucial role in the proteolytic activation of SREBP. The use of a SKI-1/S1P-specific protein-based inhibitor recently demonstrated that SKI-1/S1P inhibition blocks HCV infection in hepatoma cells by a mechanism that is associated with dramatic reduction in the number of lipid droplets and adipose differentiation-related protein/perilipin 2. The inhibition of virus assembly from infected cells identifies SKI-1/S1P as both a regulator of the HCV lifecycle and a potential host-directed therapeutic target against HCV infection[107].
Retinoid X receptor alpha: The retinoid X receptor (RXR) family is a family of nuclear receptors that target and regulate multiple signaling pathways. RXRs control gene expression by binding cooperatively as a dimer to hormone responsive elements[108,109]. RXRs form homodimers and are involved in 9-cis retinoic acid-mediated gene activation. RXRs exist as three isoforms, namely RXRα, RXRβ and RXRγ; the RXRα isoform is abundantly expressed in the liver and regulates cell proliferation and differentiation.
Some reports have suggested an interaction between HCV core proteins with RXRα and shown that the HCV core protein binds to the DNA-binding domain of RXRα, leading to an increase in binding of RXRα to its DNA response element[110,111]. In addition, RXRα is activated in cells expressing the HCV core protein and in the livers of HCV core-transgenic mice that develop hepatic steatosis and HCC. The HCV core protein may compete with p50- and p65-nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) subunits for direct interaction with RXRα. This competition may influence signaling via NF-κB, a regulator of many cellular functions and lipid synthesis, and up-regulate the enzymes acyl CoA oxidase and retinol binding protein II, exacerbating steatosis by increasing oxidative stress and decreasing β-oxidation[111,112]. These reports indicate that modulation of RXRα controls gene expression by interacting with the core protein and contributes to pathogenesis of HCV infection[112].
FAS: FAS is a protein that is directly linked to intracellular lipid synthesis and plays a central role in triglyceride accumulation in the liver. FAS catalyzes the conversion of acetyl CoA and malonyl CoA to saturated fatty acids, which are then converted to TG after etherification[106,113]. Yang et al[114] have demonstrated that HCV infection directly induces FAS expression, while Jackel-Cram et al[115] have reported that HCV-3a is a stronger teratogenic factor than HCV-1b. These authors reported that a single amino acid substitution at position 164 (phenylalanine) of the HCV-3a core protein can up-regulate FAS activity. Therefore, the short sequence YATG in HCV-1b and FATG in HCV-3a is responsible for FAS up-regulation at the transcriptional level[114]. However, further studies are required to understand the molecular mechanism of FAS activation.
Recent progress in the field of molecular genetics has revealed that the clinical outcome and response to treatment for HCV may be predetermined by genetic polymorphisms. Genes related to lipids, metabolism and the immune response have been studied (Table 1). Large-scale genetic epidemiological studies are required to attain adequate statistical power. Identifying the relationship between specific genes and the progression of HCV infection is a crucial step toward understanding the pathogenesis of infection.
| Lipid metabolism genes altered by HCV | ||
| Gene | Protein | Effect |
| SREBP | Sterol regulatory element-binding protein | Core protein increases the gene expression[110] |
| PPARα | Peroxisome proliferators-activated receptor alpha | HCV infection led to reduction in gene expression[126] |
| MTP | Microsomal triacylglycerol transfer protein | Core protein led to reduction in MTP activity and lower gene expression[127,128] |
| PEPCK | Phosphoenolpyruvate carboxykinase | NS5A increases gene expression and development of steatosis[129] |
| FAS | Fatty acid synthase | Core protein induces FAS promoter activity and severity of steatosis[115] |
| RXRα | Retinoid X receptor-alpha | Core protein enhances the transcriptional activity and contributes to the pathogenesis of infection[112] |
| APOE | Apolipoprotein E | HCV forms lipoviroparticles and hijacks ApoE for entry into hepatocyte[48] |
| Genes associated with spontaneous clearance of HCV | ||
| Gene | Protein | Allele |
| IL28B | Interleukin 28B | rs12979860 CC[130] |
| IL28B | Interleukin 28B | rs8099917 TT[131] |
| IL28B | Interleukin 28B | ss469415590DG[132] |
| KIR | Natural killer cell immunoglobulin-like receptor | KIR3DS1[133] |
| TNFα | Tumor necrosis factor-α | -863CC[134] |
| APOB | Apolipoprotein B | rs934197 TT[135] |
The lipid profile in chronic HCV-infected patients presents low levels of total cholesterol, HDL and LDL regarding non-infected individuals. In addition, these levels are significantly lower in patients who present with fatty liver. HCV-genotype 3-infected patients show a higher incidence of fatty liver development. The relationship of these lipid alterations with HCV infection is important for the lifecycle of the virus, and it is positively correlated with the response to antiviral treatment, especially in the setting of genotype 3 infections[57].
Studies have demonstrated the presence of hypobetalipoproteinemia in patients infected with HCV[116]. The pathogenesis of hypobetalipoproteinemia in HCV patients is unclear. Recently, hepatic steatosis has been correlated with this type of dyslipidemia. However, the influence of viral infection on these parameters has not been thoroughly investigated. It has been demonstrated that hepatic fibrosis is increased as serum levels of ApoB decrease. Thus, there is a strong negative correlation between fibrosis and ApoB levels[117].
Studies report that serum triglyceride levels are decreased in patients with HCV; however, in these studies, more than 50% of patients had liver cirrhosis and HCC[118]. Thus, the decrease in TG cannot be attributed to the presence of virus alone. In more recent studies, where HCV patients without cirrhosis or HCC were analyzed, the prevalence of TG was similar to control patients without HCV. However, TGs in the fasting state, transported only by VLDL, are found at significantly lower levels in HCV-infected patients compared to individuals without HCV infection. The levels of TGs in different stages of fibrosis in patients with HCV have been compared and no differences were found. Differences were only found in patients with cirrhosis. In contrast, the levels of TGs transported by VLDL declined with progression of liver damage until cirrhosis was reached. These data indicate that hypertriglyceridemia in HCV patients is not of viral origin. In contrast, the decreased serum levels of TG can be explained by the interference in maturation and excretion of VLDL caused by the viral lifecycle[119].
Chronic diseases associated with diet and lifestyles, including obesity and dyslipidemias, have genetic components. Currently, Mexico and the United States are experiencing the largest obesity epidemic in the world. The significant increase in prevalence of these diseases in recent years strongly suggests the influence of environmental factors in their development. High levels of cholesterol and lipid disorders are important risk factors for developing diseases including hepatic and cardiovascular diseases. Pathologies associated with high levels of cholesterol represent one of the eight main risk factors for mortality in Mexico[8,120]. Hypercholesterolemia has been described as a public health problem in Mexico since 1988 when the Secretariat of Health conducted the first national survey related to serum cholesterol levels among the Mexican population. In 1993, a new study was performed (National Survey of Chronic Diseases) revealing that the prevalence of hypercholesterolemia had risen to 35.5%, a 10% increase compared to the previous study. This trend continued during the coming years, resulting in an increase in prevalence of hypercholesterolemia from 35.3% to 42.6%[121]. Additionally, serum TGs are higher and HDL is lower among Mexicans when compared to other populations worldwide. In 2000, data from the Secretariat of Health revealed that fasting serum samples from 2351 adults had a mean total cholesterol of 197.5 mg/dL, HDL cholesterol of 38.4 mg/dL and TGs of 181.7 mg/dL. Only 6.1% of the study population was diagnosed with dyslipidemia and 85.88% did not know that they were ill[121]. Detailed analysis of the data revealed that the most frequent dyslipidemia was hypobetalipoproteinemia (low HDL), followed by hypertriglyceridemia and hypercholesterolemia. The most common combinations were high levels of TGs and low HDL cholesterol, and high levels of TGs with high total cholesterol (mixed dyslipidemia).
It is currently accepted that there is an important association between being overweight, obesity and various types of dyslipidemia[121]. As in the United States, the obesity epidemic in Mexico has dramatically increased in children and adults. In 2000, the Secretariat of Health in Mexico revealed an association between obesity and hypertriglyceridemia. Obese adults compared with individuals of normal weight were four times more at risk for a diagnosis of high cholesterol, low cholesterol HDL, high TGs or any combination of these conditions[121]. Currently, even children have been detected with high blood levels of cholesterol and triglycerides, most likely due to the marketing of processed foods with high sugar or fat content, changes in dietary patterns and the abuse of food rich in animal fat. It has been described that 100 mg of dietary cholesterol for each 1000 kcal results in 12 mg/dL increase in the concentration of blood cholesterol, and that children and adolescents with high levels of cholesterol are more likely to continue having high levels into adult stage[122].
Mexico has been reported to be a low endemic area for HCV[123,124]. Moreover, a low association between HCV infection and HCC has been reported in the country[125]. Given that lipid components associated with HCV infection are finely modulated in the Mexican population, it is plausible to hypothesize that the progression of infection may be regulated by the characteristics of this population’s lipid composition.
Ineffective viral escape from the immune response and effective T cell activity, together with the absence of interfering HCV-induced Abs, are factors that may contribute in combination to the spontaneous clearance of HCV. In contrast, exhausted T cell responses resulting in impaired T cell activity and viral adaptation to the host immune response may result in chronicity. In addition to the recognized role of lipoproteins during the initial steps of HCV infection, lipoproteins can also provide HCV with protection from nAbs, either by masking viral epitopes by associating with LDL and VLDL or by accelerating viral entry via HDL and thus limiting the exposure of viral epitopes to nAbs. ApoE alleles 2 and 4 are associated with HCV clearance and protection against severe liver damage. Furthermore, association between ApoE levels and IFN sensitivity suggest that lipid modulation is a potential target for preventing HCV disease progression.
When comparing serum lipid concentrations with those of other populations of the world, it is notable that Mexicans have higher concentrations of TGs and lower HDL cholesterol. Hypercholesterolemia is among the eight most important risk factors for mortality in the country. Moreover, in Mexico, as in the United States and other developed countries, obesity has significantly increased in recent years. Analysis of the 2000 Mexican National Health Survey significantly associated obesity with hypertriglyceridemia, followed by hypercholesterolemia. This reveals a unique lipid profile in the Mexican population that, in conjunction with genetics and lifestyle, might be responsible for the immune response during chronic diseases, including HCV infection.
Advances in understanding HCV disease outcome suggest that individualized therapies that account for factors, such as lipid modulation, lifestyle and genetics, in conjunction with immune-based therapies are required to establish better strategies for controlling infection with HCV.
P- Reviewers: Jin B, Sener A S- Editor: Gou SX L- Editor: A E- Editor: Ma S
| 1. | Lavanchy D. Evolving epidemiology of hepatitis C virus. Clin Microbiol Infect. 2011;17:107-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 919] [Cited by in RCA: 945] [Article Influence: 67.5] [Reference Citation Analysis (2)] |
| 2. | Fierro N, Castro-Garcia F, Panduro A. Rethinking cytokine function during hepatitis A and hepatitis C infections. Adv Biosci Biotechnol. 2013;4:13-18. [RCA] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 3. | Hellier S, Frodsham AJ, Hennig BJ, Klenerman P, Knapp S, Ramaley P, Satsangi J, Wright M, Zhang L, Thomas HC. Association of genetic variants of the chemokine receptor CCR5 and its ligands, RANTES and MCP-2, with outcome of HCV infection. Hepatology. 2003;38:1468-1476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 77] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 4. | Hsu CS, Hsu SJ, Chen HC, Liu CH, Jeng J, Liu CJ, Chen PJ, Chen DS, Kao JH. Association of IL28B genotypes with metabolic profiles and viral clearance rate in chronic hepatitis C patients. Hepatol Int. 2013;7:171-179. [RCA] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 5. | Molina S, Castet V, Fournier-Wirth C, Pichard-Garcia L, Avner R, Harats D, Roitelman J, Barbaras R, Graber P, Ghersa P. The low-density lipoprotein receptor plays a role in the infection of primary human hepatocytes by hepatitis C virus. J Hepatol. 2007;46:411-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 219] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 6. | Negro F. Abnormalities of lipid metabolism in hepatitis C virus infection. Gut. 2010;59:1279-1287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 131] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 7. | Popescu CI, Dubuisson J. Role of lipid metabolism in hepatitis C virus assembly and entry. Biol Cell. 2010;102:63-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 85] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 8. | Stevens G, Dias RH, Thomas KJ, Rivera JA, Carvalho N, Barquera S, Hill K, Ezzati M. Characterizing the epidemiological transition in Mexico: national and subnational burden of diseases, injuries, and risk factors. PLoS Med. 2008;5:e125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 139] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 9. | Marukian S, Andrus L, Sheahan TP, Jones CT, Charles ED, Ploss A, Rice CM, Dustin LB. Hepatitis C virus induces interferon-λ and interferon-stimulated genes in primary liver cultures. Hepatology. 2011;54:1913-1923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 147] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 10. | Thomas E, Gonzalez VD, Li Q, Modi AA, Chen W, Noureddin M, Rotman Y, Liang TJ. HCV infection induces a unique hepatic innate immune response associated with robust production of type III interferons. Gastroenterology. 2012;142:978-988. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 222] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 11. | Buonaguro L, Petrizzo A, Tornesello ML, Buonaguro FM. Innate immunity and hepatitis C virus infection: a microarray’s view. Infect Agent Cancer. 2012;7:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 12. | Dahari H, Major M, Zhang X, Mihalik K, Rice CM, Perelson AS, Feinstone SM, Neumann AU. Mathematical modeling of primary hepatitis C infection: noncytolytic clearance and early blockage of virion production. Gastroenterology. 2005;128:1056-1066. [PubMed] |
| 13. | Rehermann B. Hepatitis C virus versus innate and adaptive immune responses: a tale of coevolution and coexistence. J Clin Invest. 2009;119:1745-1754. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 419] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 14. | Thimme R, Bukh J, Spangenberg HC, Wieland S, Pemberton J, Steiger C, Govindarajan S, Purcell RH, Chisari FV. Viral and immunological determinants of hepatitis C virus clearance, persistence, and disease. Proc Natl Acad Sci USA. 2002;99:15661-15668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 479] [Cited by in RCA: 478] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 15. | Cox AL, Mosbruger T, Lauer GM, Pardoll D, Thomas DL, Ray SC. Comprehensive analyses of CD8+ T cell responses during longitudinal study of acute human hepatitis C. Hepatology. 2005;42:104-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 183] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 16. | Thimme R, Binder M, Bartenschlager R. Failure of innate and adaptive immune responses in controlling hepatitis C virus infection. FEMS Microbiol Rev. 2012;36:663-683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 97] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 17. | Rehermann B. Pathogenesis of chronic viral hepatitis: differential roles of T cells and NK cells. Nat Med. 2013;19:859-868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 327] [Cited by in RCA: 378] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 18. | Hammerich L, Heymann F, Tacke F. Role of IL-17 and Th17 cells in liver diseases. Clin Dev Immunol. 2011;2011:345803. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 164] [Cited by in RCA: 199] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 19. | Sousa GM, Oliveira IS, Andrade LJ, Sousa-Atta ML, Paraná R, Atta AM. Serum levels of Th17 associated cytokines in chronic hepatitis C virus infection. Cytokine. 2012;60:138-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 20. | Alatrakchi N, Koziel M. Regulatory T cells and viral liver disease. J Viral Hepat. 2009;16:223-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 78] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 21. | Farci P, Alter HJ, Wong DC, Miller RH, Govindarajan S, Engle R, Shapiro M, Purcell RH. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci USA. 1994;91:7792-7796. [PubMed] |
| 22. | Féray C, Gigou M, Samuel D, Ducot B, Maisonneuve P, Reynès M, Bismuth A, Bismuth H. Incidence of hepatitis C in patients receiving different preparations of hepatitis B immunoglobulins after liver transplantation. Ann Intern Med. 1998;128:810-816. [PubMed] |
| 23. | Zeisel MB, Fofana I, Fafi-Kremer S, Baumert TF. Hepatitis C virus entry into hepatocytes: molecular mechanisms and targets for antiviral therapies. J Hepatol. 2011;54:566-576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 140] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 24. | Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, Davis C, Mee CJ, Turek M, Gorke S. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med. 2011;17:589-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 603] [Cited by in RCA: 569] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 25. | Sainz B, Barretto N, Martin DN, Hiraga N, Imamura M, Hussain S, Marsh KA, Yu X, Chayama K, Alrefai WA. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat Med. 2012;18:281-285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 349] [Cited by in RCA: 353] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 26. | Hsu M, Zhang J, Flint M, Logvinoff C, Cheng-Mayer C, Rice CM, McKeating JA. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc Natl Acad Sci USA. 2003;100:7271-7276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 655] [Cited by in RCA: 644] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 27. | Triyatni M, Vergalla J, Davis AR, Hadlock KG, Foung SK, Liang TJ. Structural features of envelope proteins on hepatitis C virus-like particles as determined by anti-envelope monoclonal antibodies and CD81 binding. Virology. 2002;298:124-132. [PubMed] |
| 28. | Keck ZY, Li TK, Xia J, Gal-Tanamy M, Olson O, Li SH, Patel AH, Ball JK, Lemon SM, Foung SK. Definition of a conserved immunodominant domain on hepatitis C virus E2 glycoprotein by neutralizing human monoclonal antibodies. J Virol. 2008;82:6061-6066. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 104] [Cited by in RCA: 106] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 29. | Law M, Maruyama T, Lewis J, Giang E, Tarr AW, Stamataki Z, Gastaminza P, Chisari FV, Jones IM, Fox RI. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat Med. 2008;14:25-27. [PubMed] |
| 30. | Perotti M, Mancini N, Diotti RA, Tarr AW, Ball JK, Owsianka A, Adair R, Patel AH, Clementi M, Burioni R. Identification of a broadly cross-reacting and neutralizing human monoclonal antibody directed against the hepatitis C virus E2 protein. J Virol. 2008;82:1047-1052. [PubMed] |
| 31. | Farci P, Shimoda A, Coiana A, Diaz G, Peddis G, Melpolder JC, Strazzera A, Chien DY, Munoz SJ, Balestrieri A. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000;288:339-344. [PubMed] |
| 32. | Helle F, Duverlie G, Dubuisson J. The hepatitis C virus glycan shield and evasion of the humoral immune response. Viruses. 2011;3:1909-1932. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 77] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 33. | Falkowska E, Kajumo F, Garcia E, Reinus J, Dragic T. Hepatitis C virus envelope glycoprotein E2 glycans modulate entry, CD81 binding, and neutralization. J Virol. 2007;81:8072-8079. [PubMed] |
| 34. | Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1944] [Cited by in RCA: 1933] [Article Influence: 87.9] [Reference Citation Analysis (0)] |
| 35. | Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678-682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 719] [Cited by in RCA: 743] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 36. | Srivastava IK, Ulmer JB, Barnett SW. Role of neutralizing antibodies in protective immunity against HIV. Hum Vaccin. 2005;1:45-60. [PubMed] |
| 37. | Ait-Goughoulte M, Banerjee A, Meyer K, Mazumdar B, Saito K, Ray RB, Ray R. Hepatitis C virus core protein interacts with fibrinogen-beta and attenuates cytokine stimulated acute-phase response. Hepatology. 2010;51:1505-1513. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 38. | Arnaud N, Dabo S, Maillard P, Budkowska A, Kalliampakou KI, Mavromara P, Garcin D, Hugon J, Gatignol A, Akazawa D. Hepatitis C virus controls interferon production through PKR activation. PLoS One. 2010;5:e10575. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 97] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 39. | Polyak SJ, Khabar KS, Paschal DM, Ezelle HJ, Duverlie G, Barber GN, Levy DE, Mukaida N, Gretch DR. Hepatitis C virus nonstructural 5A protein induces interleukin-8, leading to partial inhibition of the interferon-induced antiviral response. J Virol. 2001;75:6095-6106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 234] [Cited by in RCA: 239] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 40. | Zhang P, Wu CG, Mihalik K, Virata-Theimer ML, Yu MY, Alter HJ, Feinstone SM. Hepatitis C virus epitope-specific neutralizing antibodies in Igs prepared from human plasma. Proc Natl Acad Sci USA. 2007;104:8449-8454. [PubMed] |
| 41. | Zhang P, Zhong L, Struble EB, Watanabe H, Kachko A, Mihalik K, Virata-Theimer ML, Alter HJ, Feinstone S, Major M. Depletion of interfering antibodies in chronic hepatitis C patients and vaccinated chimpanzees reveals broad cross-genotype neutralizing activity. Proc Natl Acad Sci USA. 2009;106:7537-7541. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 117] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 42. | Di Lorenzo C, Angus AG, Patel AH. Hepatitis C virus evasion mechanisms from neutralizing antibodies. Viruses. 2011;3:2280-2300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 50] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 43. | Bartosch B, Verney G, Dreux M, Donot P, Morice Y, Penin F, Pawlotsky JM, Lavillette D, Cosset FL. An interplay between hypervariable region 1 of the hepatitis C virus E2 glycoprotein, the scavenger receptor BI, and high-density lipoprotein promotes both enhancement of infection and protection against neutralizing antibodies. J Virol. 2005;79:8217-8229. [PubMed] |
| 44. | Machida K, Kondo Y, Huang JY, Chen YC, Cheng KT, Keck Z, Foung S, Dubuisson J, Sung VM, Lai MM. Hepatitis C virus (HCV)-induced immunoglobulin hypermutation reduces the affinity and neutralizing activities of antibodies against HCV envelope protein. J Virol. 2008;82:6711-6720. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 36] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 45. | Brimacombe CL, Grove J, Meredith LW, Hu K, Syder AJ, Flores MV, Timpe JM, Krieger SE, Baumert TF, Tellinghuisen TL. Neutralizing antibody-resistant hepatitis C virus cell-to-cell transmission. J Virol. 2011;85:596-605. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 210] [Cited by in RCA: 198] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 46. | Thomssen R, Bonk S, Thiele A. Density heterogeneities of hepatitis C virus in human sera due to the binding of beta-lipoproteins and immunoglobulins. Med Microbiol Immunol. 1993;182:329-334. [PubMed] |
| 47. | Dueñas-Carrera S. Hepatitis C virus and lipid metabolism: their implications in vaccine development and treatment. Biotecnol Apl. 2011;28:1-5 Available from: http://scielo.sld.cu/pdf/bta/v28n1/bta01111.pdf. |
| 48. | Owen DM, Huang H, Ye J, Gale M. Apolipoprotein E on hepatitis C virion facilitates infection through interaction with low-density lipoprotein receptor. Virology. 2009;394:99-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 174] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 49. | Mahley RW, Rall SC. Apolipoprotein E: far more than a lipid transport protein. Annu Rev Genomics Hum Genet. 2000;1:507-537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1164] [Cited by in RCA: 1269] [Article Influence: 55.2] [Reference Citation Analysis (0)] |
| 50. | Toniutto P, Fabris C, Fumo E, Apollonio L, Caldato M, Mariuzzi L, Avellini C, Minisini R, Pirisi M. Carriage of the apolipoprotein E-epsilon4 allele and histologic outcome of recurrent hepatitis C after antiviral treatment. Am J Clin Pathol. 2004;122:428-433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 9] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 51. | Price DA, Bassendine MF, Norris SM, Golding C, Toms GL, Schmid ML, Morris CM, Burt AD, Donaldson PT. Apolipoprotein epsilon3 allele is associated with persistent hepatitis C virus infection. Gut. 2006;55:715-718. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 72] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 52. | Sheridan DA, Bridge SH, Felmlee DJ, Crossey MM, Thomas HC, Taylor-Robinson SD, Toms GL, Neely RD, Bassendine MF. Apolipoprotein-E and hepatitis C lipoviral particles in genotype 1 infection: evidence for an association with interferon sensitivity. J Hepatol. 2012;57:32-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 53. | Dubuisson J, Penin F, Moradpour D. Interaction of hepatitis C virus proteins with host cell membranes and lipids. Trends Cell Biol. 2002;12:517-523. [PubMed] |
| 54. | Alvisi G, Madan V, Bartenschlager R. Hepatitis C virus and host cell lipids: an intimate connection. RNA Biol. 2011;8:258-269. [PubMed] |
| 55. | Liu HM, Aizaki H, Machida K, Ou JH, Lai MM. Hepatitis C virus translation preferentially depends on active RNA replication. PLoS One. 2012;7:e43600. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 56. | Huang H, Sun F, Owen DM, Li W, Chen Y, Gale M, Ye J. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc Natl Acad Sci USA. 2007;104:5848-5853. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 414] [Cited by in RCA: 429] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 57. | Syed GH, Amako Y, Siddiqui A. Hepatitis C virus hijacks host lipid metabolism. Trends Endocrinol Metab. 2010;21:33-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 258] [Cited by in RCA: 284] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 58. | Hussain MM, Shi J, Dreizen P. Microsomal triglyceride transfer protein and its role in apoB-lipoprotein assembly. J Lipid Res. 2003;44:22-32. [PubMed] |
| 59. | Sørensen LP, Andersen IR, Søndergaard E, Gormsen LC, Schmitz O, Christiansen JS, Nielsen S. Basal and insulin mediated VLDL-triglyceride kinetics in type 2 diabetic men. Diabetes. 2011;60:88-96. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 41] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 60. | Harris C, Herker E, Farese RV, Ott M. Hepatitis C virus core protein decreases lipid droplet turnover: a mechanism for core-induced steatosis. J Biol Chem. 2011;286:42615-42625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 66] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 61. | Hussain MM, Rava P, Walsh M, Rana M, Iqbal J. Multiple functions of microsomal triglyceride transfer protein. Nutr Metab (Lond). 2012;9:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 190] [Cited by in RCA: 194] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 62. | Gastaminza P, Cheng G, Wieland S, Zhong J, Liao W, Chisari FV. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J Virol. 2008;82:2120-2129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 347] [Cited by in RCA: 353] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 63. | Nahmias Y, Goldwasser J, Casali M, van Poll D, Wakita T, Chung RT, Yarmush ML. Apolipoprotein B-dependent hepatitis C virus secretion is inhibited by the grapefruit flavonoid naringenin. Hepatology. 2008;47:1437-1445. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 219] [Cited by in RCA: 190] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 64. | Clément S, Negro F. Hepatitis C virus: the viral way to fatty liver. J Hepatol. 2007;46:985-987. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 65. | Bassendine MF, Sheridan DA, Bridge SH, Felmlee DJ, Neely RD. Lipids and HCV. Semin Immunopathol. 2013;35:87-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 90] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 66. | Schaefer EA, Chung RT. HCV and host lipids: an intimate connection. Semin Liver Dis. 2013;33:358-368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 67. | Paracha UZ, Fatima K, Alqahtani M, Chaudhary A, Abuzenadah A, Damanhouri G, Qadri I. Oxidative stress and hepatitis C virus. Virol J. 2013;10:251. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 110] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 68. | Eslam M, Khattab MA, Harrison SA. Insulin resistance and hepatitis C: an evolving story. Gut. 2011;60:1139-1151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 50] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 69. | de Gottardi A, Pazienza V, Pugnale P, Bruttin F, Rubbia-Brandt L, Juge-Aubry CE, Meier CA, Hadengue A, Negro F. Peroxisome proliferator-activated receptor-alpha and -gamma mRNA levels are reduced in chronic hepatitis C with steatosis and genotype 3 infection. Aliment Pharmacol Ther. 2006;23:107-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 93] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 70. | Alter MJ. Epidemiology of hepatitis C virus infection. World J Gastroenterol. 2007;13:2436-2441. [PubMed] |
| 71. | Bartenschlager R, Lohmann V. Novel cell culture systems for the hepatitis C virus. Antiviral Res. 2001;52:1-17. [PubMed] |
| 72. | Sheehy P, Mullan B, Moreau I, Kenny-Walsh E, Shanahan F, Scallan M, Fanning LJ. In vitro replication models for the hepatitis C virus. J Viral Hepat. 2007;14:2-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 73. | Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, Berger U. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896-899. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1268] [Cited by in RCA: 1280] [Article Influence: 75.3] [Reference Citation Analysis (0)] |
| 74. | Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Ørum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1352] [Cited by in RCA: 1291] [Article Influence: 86.1] [Reference Citation Analysis (0)] |
| 75. | Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14460] [Cited by in RCA: 16076] [Article Influence: 1004.8] [Reference Citation Analysis (2)] |
| 76. | Singaravelu R, Chen R, Lyn RK, Jones DM, O’Hara S, Rouleau Y, Cheng J, Srinivasan P, Nasheri N, Russell RS. Hepatitis C virus induced up-regulation of microRNA-27: a novel mechanism for hepatic steatosis. Hepatology. 2014;59:98-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 88] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 77. | Lemberger T, Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: a nuclear receptor signaling pathway in lipid physiology. Annu Rev Cell Dev Biol. 1996;12:335-363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 541] [Cited by in RCA: 520] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 78. | Ban S, Ueda Y, Ohashi M, Matsuno K, Ikeda M, Kato N, Miyachi H. Peroxisome proliferator-activated receptor delta antagonists inhibit hepatitis C virus RNA replication. Bioorg Med Chem Lett. 2013;23:4774-4778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 79. | Leone TC, Weinheimer CJ, Kelly DP. A critical role for the peroxisome proliferator-activated receptor alpha (PPARalpha) in the cellular fasting response: the PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proc Natl Acad Sci USA. 1999;96:7473-7478. [PubMed] |
| 80. | Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest. 1999;103:1489-1498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1261] [Cited by in RCA: 1294] [Article Influence: 49.8] [Reference Citation Analysis (0)] |
| 81. | Tanaka N, Moriya K, Kiyosawa K, Koike K, Gonzalez FJ, Aoyama T. PPARalpha activation is essential for HCV core protein-induced hepatic steatosis and hepatocellular carcinoma in mice. J Clin Invest. 2008;118:683-694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 121] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 82. | Dharancy S, Lemoine M, Mathurin P, Serfaty L, Dubuquoy L. Peroxisome proliferator-activated receptors in HCV-related infection. PPAR Res. 2009;2009:357204. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 83. | Maeda K, Okubo K, Shimomura I, Funahashi T, Matsuzawa Y, Matsubara K. cDNA cloning and expression of a novel adipose specific collagen-like factor, apM1 (AdiPose Most abundant Gene transcript 1). Biochem Biophys Res Commun. 1996;221:286-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1483] [Cited by in RCA: 1444] [Article Influence: 49.8] [Reference Citation Analysis (0)] |
| 84. | Wolf AM, Wolf D, Rumpold H, Enrich B, Tilg H. Adiponectin induces the anti-inflammatory cytokines IL-10 and IL-1RA in human leukocytes. Biochem Biophys Res Commun. 2004;323:630-635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 546] [Cited by in RCA: 601] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 85. | Taliaferro-Smith L, Nagalingam A, Zhong D, Zhou W, Saxena NK, Sharma D. LKB1 is required for adiponectin-mediated modulation of AMPK-S6K axis and inhibition of migration and invasion of breast cancer cells. Oncogene. 2009;28:2621-2633. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 135] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 86. | AlSaleh A, Sanders TA, O’Dell SD. Effect of interaction between PPARG, PPARA and ADIPOQ gene variants and dietary fatty acids on plasma lipid profile and adiponectin concentration in a large intervention study. Proc Nutr Soc. 2012;71:141-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 87. | Tilg H, Moschen AR. Adipocytokines: mediators linking adipose tissue, inflammation and immunity. Nat Rev Immunol. 2006;6:772-783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2065] [Cited by in RCA: 2251] [Article Influence: 118.5] [Reference Citation Analysis (0)] |
| 88. | Yamauchi T, Kamon J, Ito Y, Tsuchida A, Yokomizo T, Kita S, Sugiyama T, Miyagishi M, Hara K, Tsunoda M. Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature. 2003;423:762-769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2258] [Cited by in RCA: 2314] [Article Influence: 105.2] [Reference Citation Analysis (0)] |
| 89. | You M, Matsumoto M, Pacold CM, Cho WK, Crabb DW. The role of AMP-activated protein kinase in the action of ethanol in the liver. Gastroenterology. 2004;127:1798-1808. [PubMed] |
| 90. | Janeckova R. The role of leptin in human physiology and pathophysiology. Physiol Res. 2001;50:443-459. [PubMed] |
| 91. | Chang ML, Yeh HC, Tsou YK, Wang CJ, Cheng HY, Sung CM, Ho YP, Chen TH, Yeh CT. HCV core-induced nonobese hepatic steatosis is associated with hypoadiponectinemia and is ameliorated by adiponectin administration. Obesity (Silver Spring). 2012;20:1474-1480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 92. | Hardie DG. Minireview: the AMP-activated protein kinase cascade: the key sensor of cellular energy status. Endocrinology. 2003;144:5179-5183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 742] [Cited by in RCA: 747] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 93. | Hardie DG. The AMP-activated protein kinase pathway--new players upstream and downstream. J Cell Sci. 2004;117:5479-5487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 872] [Cited by in RCA: 908] [Article Influence: 45.4] [Reference Citation Analysis (0)] |
| 94. | Khan M, Jahan S, Khaliq S, Ijaz B, Ahmad W, Samreen B, Hassan S. Interaction of the hepatitis C virus (HCV) core with cellular genes in the development of HCV-induced steatosis. Arch Virol. 2010;155:1735-1753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 95. | Nakashima K, Takeuchi K, Chihara K, Hotta H, Sada K. Inhibition of hepatitis C virus replication through adenosine monophosphate-activated protein kinase-dependent and -independent pathways. Microbiol Immunol. 2011;55:774-782. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 96. | Mankouri J, Tedbury PR, Gretton S, Hughes ME, Griffin SD, Dallas ML, Green KA, Hardie DG, Peers C, Harris M. Enhanced hepatitis C virus genome replication and lipid accumulation mediated by inhibition of AMP-activated protein kinase. Proc Natl Acad Sci USA. 2010;107:11549-11554. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 121] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 97. | Yang D, Xue B, Wang X, Yu X, Liu N, Gao Y, Liu C, Zhu H. 2-octynoic acid inhibits hepatitis C virus infection through activation of AMP-activated protein kinase. PLoS One. 2013;8:e64932. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 98. | Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331-340. [PubMed] |
| 99. | Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125-1131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 1746] [Article Influence: 75.9] [Reference Citation Analysis (0)] |
| 100. | Shimano H. Sterol regulatory element-binding proteins (SREBPs): transcriptional regulators of lipid synthetic genes. Prog Lipid Res. 2001;40:439-452. [PubMed] |
| 101. | Shimomura I, Bashmakov Y, Horton JD. Increased levels of nuclear SREBP-1c associated with fatty livers in two mouse models of diabetes mellitus. J Biol Chem. 1999;274:30028-30032. [PubMed] |
| 102. | Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA. 2003;100:12027-12032. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1021] [Cited by in RCA: 1123] [Article Influence: 51.0] [Reference Citation Analysis (0)] |
| 103. | Roingeard P. Hepatitis C virus diversity and hepatic steatosis. J Viral Hepat. 2013;20:77-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 104. | Kohjima M, Higuchi N, Kato M, Kotoh K, Yoshimoto T, Fujino T, Yada M, Yada R, Harada N, Enjoji M. SREBP-1c, regulated by the insulin and AMPK signaling pathways, plays a role in nonalcoholic fatty liver disease. Int J Mol Med. 2008;21:507-511. [PubMed] |
| 105. | Oem JK, Jackel-Cram C, Li YP, Zhou Y, Zhong J, Shimano H, Babiuk LA, Liu Q. Activation of sterol regulatory element-binding protein 1c and fatty acid synthase transcription by hepatitis C virus non-structural protein 2. J Gen Virol. 2008;89:1225-1230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 91] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 106. | Kapadia SB, Chisari FV. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc Natl Acad Sci USA. 2005;102:2561-2566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 386] [Cited by in RCA: 406] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 107. | Olmstead AD, Knecht W, Lazarov I, Dixit SB, Jean F. Human subtilase SKI-1/S1P is a master regulator of the HCV Lifecycle and a potential host cell target for developing indirect-acting antiviral agents. PLoS Pathog. 2012;8:e1002468. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 67] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 108. | Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841-850. [PubMed] |
| 109. | Ahuja HS, Szanto A, Nagy L, Davies PJ. The retinoid X receptor and its ligands: versatile regulators of metabolic function, cell differentiation and cell death. J Biol Regul Homeost Agents. 2003;17:29-45. [PubMed] |
| 110. | Kim KH, Hong SP, Kim K, Park MJ, Kim KJ, Cheong J. HCV core protein induces hepatic lipid accumulation by activating SREBP1 and PPARgamma. Biochem Biophys Res Commun. 2007;355:883-888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 108] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 111. | Koike K. Hepatitis C virus contributes to hepatocarcinogenesis by modulating metabolic and intracellular signaling pathways. J Gastroenterol Hepatol. 2007;22 Suppl 1:S108-S111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 93] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 112. | Tsutsumi T, Suzuki T, Shimoike T, Suzuki R, Moriya K, Shintani Y, Fujie H, Matsuura Y, Koike K, Miyamura T. Interaction of hepatitis C virus core protein with retinoid X receptor alpha modulates its transcriptional activity. Hepatology. 2002;35:937-946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 125] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 113. | Semenkovich CF. Regulation of fatty acid synthase (FAS). Prog Lipid Res. 1997;36:43-53. [PubMed] |
| 114. | Yang W, Huang M. Studying HCV RNA synthesis in vitro with replication complexes. Methods Mol Biol. 2009;510:177-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 115. | Jackel-Cram C, Babiuk LA, Liu Q. Up-regulation of fatty acid synthase promoter by hepatitis C virus core protein: genotype-3a core has a stronger effect than genotype-1b core. J Hepatol. 2007;46:999-1008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 119] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 116. | Bima AI, Hooper AJ, van Bockxmeer FM, Burnett JR. Hypobetalipoproteinaemia secondary to chronic hepatitis C virus infection in a patient with familial hypercholesterolaemia. Ann Clin Biochem. 2009;46:420-422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 117. | Petit JM, Benichou M, Duvillard L, Jooste V, Bour JB, Minello A, Verges B, Brun JM, Gambert P, Hillon P. Hepatitis C virus-associated hypobetalipoproteinemia is correlated with plasma viral load, steatosis, and liver fibrosis. Am J Gastroenterol. 2003;98:1150-1154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 91] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 118. | Dai CY, Yeh ML, Huang CF, Hou CH, Hsieh MY, Huang JF, Lin IL, Lin ZY, Chen SC, Wang LY. Chronic hepatitis C infection is associated with insulin resistance and lipid profiles. J Gastroenterol Hepatol. 2013;Jun 28; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 51] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 119. | Nishimura M, Yamamoto H, Yoshida T, Seimiya M, Sawabe Y, Matsushita K, Umemura H, Sogawa K, Takizawa H, Yokosuka O. Decreases in the serum VLDL-TG/non-VLDL-TG ratio from early stages of chronic hepatitis C: alterations in TG-rich lipoprotein levels. PLoS One. 2011;6:e17309. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 120. | Barrera-Cruz A, Rodríguez-González A, Molina-Ayala MA. [The current state of obesity in Mexico]. Rev Med Inst Mex Seguro Soc. 2013;51:292-299. [PubMed] |
| 121. | Aguilar-Salinas CA, Olaiz G, Valles V, Torres JM, Gómez Pérez FJ, Rull JA, Rojas R, Franco A, Sepulveda J. High prevalence of low HDL cholesterol concentrations and mixed hyperlipidemia in a Mexican nationwide survey. J Lipid Res. 2001;42:1298-1307. [PubMed] |
| 122. | Barquera S, Flores M, Olaiz-Fernández G, Monterrubio E, Villalpando S, González C, Rivera JÁ, Sepúlveda J. Dyslipidemias and obesity in Mexico. Salud Publica Mex. 2007;49 Suppl 3:S338-S347 Available from: http://www.scielo.org.mx/pdf/spm/v49s3/04.pdf. |
| 123. | Panduro A, Roman S, Khan A, Tanaka Y, Kurbanov F, Martinez-Lopez E, Campollo O, Hernandez-Nazara Z, Mizokami M. Molecular epidemiology of hepatitis C virus genotypes in west Mexico. Virus Res. 2010;151:19-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 124. | Panduro A, Escobedo Meléndez G, Fierro NA, Ruiz Madrigal B, Zepeda-Carrillo EA, Román S. [Epidemiology of viral hepatitis in Mexico]. Salud Publica Mex. 2011;53 Suppl 1:S37-S45. [PubMed] |
| 125. | Roman S, Fierro N, Moreno-Luna LE, Panduro A. Hepatitis B virus genotype H and environmental factors associated to the low prevalence of hepatocellular carcinoma in Mexico. J Cancer Ther. 2013;4:367-376. [RCA] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 126. | Cheng Y, Dharancy S, Malapel M, Desreumaux P. Hepatitis C virus infection down-regulates the expression of peroxisome proliferator-activated receptor alpha and carnitine palmitoyl acyl-CoA transferase 1A. World J Gastroenterol. 2005;11:7591-7596. [PubMed] |
| 127. | Yasui K, Harano Y, Mitsuyoshi H, Tsuji K, Endo M, Nakajima T, Minami M, Itoh Y, Zen Y, Nakanuma Y. Steatosis and hepatic expression of genes regulating lipid metabolism in Japanese patients infected with hepatitis C virus. J Gastroenterol. 2010;45:95-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 128. | Perlemuter G, Sabile A, Letteron P, Vona G, Topilco A, Chrétien Y, Koike K, Pessayre D, Chapman J, Barba G. Hepatitis C virus core protein inhibits microsomal triglyceride transfer protein activity and very low density lipoprotein secretion: a model of viral-related steatosis. FASEB J. 2002;16:185-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 419] [Cited by in RCA: 428] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 129. | Qadri I, Choudhury M, Rahman SM, Knotts TA, Janssen RC, Schaack J, Iwahashi M, Puljak L, Simon FR, Kilic G. Increased phosphoenolpyruvate carboxykinase gene expression and steatosis during hepatitis C virus subgenome replication: role of nonstructural component 5A and CCAAT/enhancer-binding protein β. J Biol Chem. 2012;287:37340-37351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 130. | Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798-801. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1706] [Cited by in RCA: 1687] [Article Influence: 105.4] [Reference Citation Analysis (0)] |
| 131. | Grebely J, Petoumenos K, Hellard M, Matthews GV, Suppiah V, Applegate T, Yeung B, Marks P, Rawlinson W, Lloyd AR. Potential role for interleukin-28B genotype in treatment decision-making in recent hepatitis C virus infection. Hepatology. 2010;52:1216-1224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 136] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 132. | Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, Hergott D, Porter-Gill P, Mumy A, Kohaar I. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet. 2013;45:164-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 701] [Cited by in RCA: 759] [Article Influence: 63.3] [Reference Citation Analysis (0)] |
| 133. | Rivero-Juarez A, Gonzalez R, Camacho A, Manzanares-Martin B, Caruz A, Martinez-Peinado A, Torre-Cisneros J, Pineda JA, Peña J, Rivero A. Natural killer KIR3DS1 is closely associated with HCV viral clearance and sustained virological response in HIV/HCV patients. PLoS One. 2013;8:e61992. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 134. | Lio D, Caruso C, Di Stefano R, Colonna Romano G, Ferraro D, Scola L, Crivello A, Licata A, Valenza LM, Candore G. IL-10 and TNF-alpha polymorphisms and the recovery from HCV infection. Hum Immunol. 2003;64:674-680. [PubMed] |
| 135. | Zhu C, Zhang R, Liu D, Mukhtar MM, Liu W, Peng G, Wang K, Hao Q, Xu Y, Liu F. Association of functional polymorphism of ApoB promoter with hepatitis C virus infection. Clin Chim Acta. 2009;401:124-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 0.8] [Reference Citation Analysis (0)] |