Published online Feb 21, 2013. doi: 10.3748/wjg.v19.i7.1030
Revised: January 7, 2013
Accepted: January 29, 2013
Published online: February 21, 2013
Processing time: 155 Days and 18.5 Hours
Current progress in epigenetic research supports the view that diet and dietary components are important in cancer etiology by enhancing or inhibiting carcinogenesis. Since diet and dietary factors may significantly contribute to the causation and progression of many cancers, it is important to find the molecular mechanisms of action of such dietary factors for cancer prevention and treatment. Recently, the role of epigenetic mechanisms in the cancer development and progression has attracted more attention as additional evidence along with traditional DNA sequence based mechanisms such as mutations and structural re-arrangements. Such an increasing interest in cancer epigenetics has also accelerated the development and application of molecular assays and tools for DNA methylation detection and histone modification enrichment analysis. In this paper, key assays and methods for epigenetic research are reviewed and discussed in terms of their utility and usability. In addition, more advanced methods for genome-wide analysis are introduced as part of upcoming research trends and directions.
- Citation: Jang H, Shin H. Current trends in the development and application of molecular technologies for cancer epigenetics. World J Gastroenterol 2013; 19(7): 1030-1039
- URL: https://www.wjgnet.com/1007-9327/full/v19/i7/1030.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i7.1030
Diet and dietary factors play an important role in many biological processes and are also involved in the regulation of pathological progressions including cancers. Several epidemiological and preclinical studies suggested that increased intake of bioactive dietary components may modulate cancer risk. Many studies provide compelling evidence that part of the anti-cancer properties contributed to several bioactive dietary components may relate to modulation of epigenetic process including DNA methylation and histone protein modifications. Here, we provide a brief overview of dietary modulation of DNA methylation and histone modifications and its potential role in cancer prevention. Also, we will discuss several new epigenetic methods to help understand the effect of dietary factors on epigenetic modifications.
The growing interest in the role of epigenetics in cancer came from the demonstration that epigenetic modifications are involved in tumor development and progression. Epigenetics can be defined as phenomena that alter the expression of the information in the genome at the transcriptional, translational, or posttranslational level without change in DNA sequence[1,2]. Epigenetic information is maintained to preserve cellular identity in normal cells, while cancer cells are characterized by profound alteration of epigenetic regulation[3-7]. The overall disruption of epigenetic phenomena is a common feature of all human tumors and includes alteration of DNA methylation and histone modification patterns[8]. DNA methylation patterns of neoplastic cells have been recognized as being substantially altered compared with normal cells[3,4]. Two types of changes in the DNA methylation pattern can occur in cancer: global DNA hypomethylation and hypermethylation of CpG islands which are associated with gene silencing[3,4,7]. DNA in eukaryotic cells is intimately associated with a family of small, basic histone proteins forming a highly ordered and condensed DNA-protein complex termed chromatin. Because of this chromatin structure, changes in DNA methylation in cancer cells are not isolated events; they occur in the context of more complex epigenetic deregulation[9]. Chromatin is the physiological template of the genetic information and is composed of DNA, histones, and other chromosomal proteins. The fundamental repeating unit of chromatin is the nucleosome octamer, which consists of 147 base pairs of DNA wrapped around 2 copies each of histones H2A, H2B, H3 and H4[10]. The amino-terminal tails of histones are subject to posttranslational modifications, including acetylation, methylation, phosphorylation, ubiquitination, SUMOylation, and ADP-ribosylation[11], and multiple histone modifications may occur on a given histone tail[12]. Histone modifications patterns distinguish the structure of chromatin status, in particular, acetylation of histone H3 and H4 is associated with active gene expression with open chromatin structure. Histone acetylation is regulated by several enzymes such as histone acetyltransferase and histone deacetylases activity. Aberrations in post-translational modifications of histones have been shown to occur in cancer cells. Although alterations in global histone modification patterns in cancer cells have remained unknown, recent studies on global histone modifications at specific amino acids have been suggested as predictive clinical outcomes for various cancers[13-15]. Additionally, a number of studies have been focused only on changes of a particular histone modification at individual gene promoters in cancer cells.
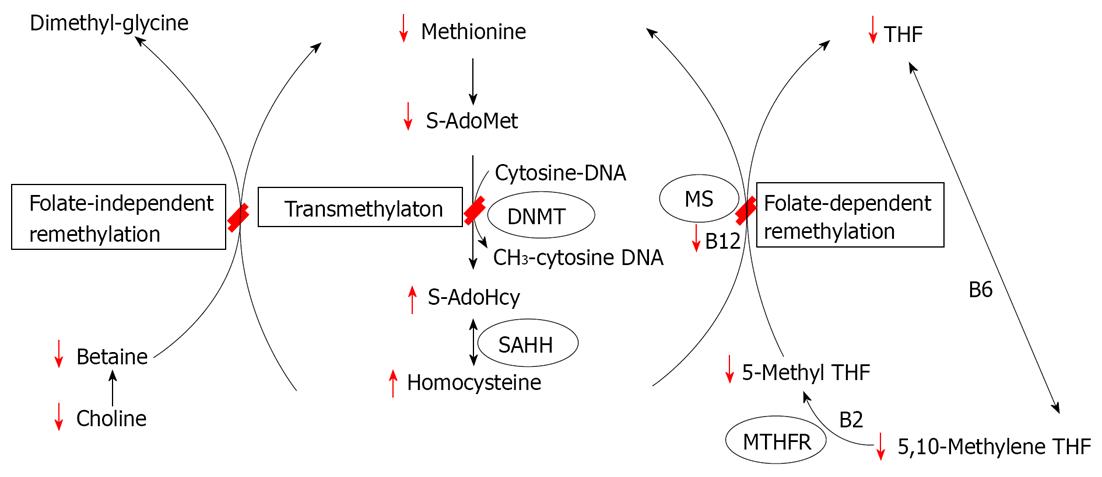
In last 4 decades, researchers have developed various tools for exploring DNA methylation, and started to apply those new technologies to the field of nutrition science. The methyl-deficient model of endogenous hepatocarcinogenesis is one of which in DNA methylation has been extensively studied. This animal model is unique in that dietary omission rather than chemical carcinogens addition can lead to tumor formation[16]. Specifically, deficiency of the major dietary sources of methyl groups - methionine, choline, folic acid and vitamin B12 - leads to the development of liver cancers in rats and certain mouse strains[17-19]. From early 1990, these animal models have shown that the methyl-deficiency is associated with several defects, including genome-wide DNA hypomethylation and gene-specific hypermethylation[20-22]. Importantly, the aberrant epigenetic alterations imposed by this diet have been hypothesized to be the primary mechanism responsible for malignant transformation of rat liver cells[15,20,22,23]. Figure 1 displays a simplified version of biological methylation pathway from one-carbon metabolism, emphasizing that various dietary methyl sources (methionine, choline, various co-enzymatic forms of folate and vitamin B2, B6 and B12) play an important roles in DNA methylation. Methyl source deficiency has marked effects on the flow of one-carbon units through this web of reactions as the effect of methyl source deficiency are highlighted in red. The major effect observed in methyl deficiency models is a rapid decrease in hepatic S-adenosylmethionine (SAdoMet) levels and genomic DNA hypomethylation. In other recent studies examining the early stages of hepatocarcinogenesis induced by methyl deficiency in rats, substantial alterations in other aspects of the epigenetic machinery have been observed, including aberrant expression of DNA methyltransferases and methyl-CpG binding proteins[24], defects in histone methyltransferase protein expression and histone posttranslational modifications[15]. In Table 1, various DNA methylation assays were summarized in methyl-deficient model of hepatocarcinogenesis in rodents.
| Dietary component | Model | Observations | Methylation assay | Ref. |
| Amino acid-defined diet lacking choline, methionine, folic acid and vitamin B12 | Rat | Depletion of SAdoMet and DNA hypomethylation | Liver DNA methyltransferase activity assay with labeled SAdoMet | [20] |
| Amino acid-defined diet lacking choline, methionine, folic acid and vitamin B12 | Rat | Hypomethylation of CCGG site of c-myc, c-fos and c-Ha-ras | Enzyme digestion by HpaII/MspI | [21] |
| Diet low in methionine lacking in choline and folic acid | Rat | Hypermethylation of p16INK4A | MS-PCR | [94] |
| Diet low in methionine lacking in choline and folic acid | Rat | Decrease in the total percent of methylated CCGG sites in DNA | HpaII/MspI-based cytosine extension assay | [22] |
| Diet low in methionine lacking in choline and folic acid | Rat | Depletion of S-AdoMet, decrease in S-AdoMet/S-AdoHcy and global DNA hypomethylation | HpaII/MspI-based cytosine extension assay | [23] |
| Diet low in methionine lacking in choline and folic acid | Rat | Hypomethylation of ID element and LINE-1 in preneoplastic livers and liver tumors; Decrease in histone H4-Lys20 trimethylation and increase in histone H3-Lys9 trimethylation; Decrease in histone H4-Lys20 trimethylation at the LINE-1 regulatory region | ID methylation by methylation-sensitive McrBC-PCR array; LINE-1 methylation by COBRA-assay; global histone methylation by Western blotting; LINE-1-associated histone methylation by ChIP | [15] |
| Diet deficient in methionine lacking in choline and folic acid | Rat | Changes in the DNA methylation machinery | Indirect methods by DNA methyltransferases and Methyl CpG binding proteins | [24] |
| Amino acid-defined diet lacking choline | Rat | Hypermethylation of upstream of E-cadherin and Cx26 | Bisulfite sequencing | [95] |
| Diet deficient in methionine lacking in choline and folic acid | Rat | global loss of DNA methylation; hypermethylation of CpG islands | Global DNA methylation by cytosine extension assay and [3H-methyl] incorporation; CpG island methylation by [32P]dGTP incorporation | [30] |
| Diet deficient in methionine lacking in choline and folic acid | Mouse | Global DNA hypomethylation; substantial loss of repetitive sequences (LINE-1, SINES, IAP elements) cytosine methylation. Increase in histone H3-Lys9 trimethylation and decrease in histone H4-Lys20 trimethylation | Global DNA methylation by cytosine extension assay; methylation-sensitive McrBC-qPCR assay; global histone modifications by Western blot | [96] |
| Diet deficient in methionine lacking in choline and folic acid | Mouse | Detection of CpG island methylation profiles | MeDIP | [97] |
One of widely used methods for global DNA methylation assay is a radioassay that utilizes the enzyme SssI DNA methyltransferase to catalyze the de novo methylation of the CpG sites with radiolabeled [3H]-SAdoMet, a universal methyl donor in vitro[20,25-28]. And another method was developed thanks to the discovery of methylation-sensitive restriction endonucleases. In 1999, Pogribny et al[29] developed a new method based on methylation-sensitive endonucleases followed by single nucleotide extension with radiolabeled [3H]-dCTP. This cytosine extension assay was used in various studies of methyl-deficient model of hepatocarcinogenesis for genomic DNA methylation[22,23,29,30]. These enzyme based methods have wide variations in precision as a result of inconsistency in the activity of methyl-sensitive endonucleases and the instability of methyltransferase activity[31]. In 2002, Friso et al[32] developed a method for quantitative determination of 5-methyl-2’deoxycytidine using liquid chromatography/electrospray ionization/mass spectrometry (LC/ESI/MS). This method allows accurate measurement of the absolute amount of 5-methyl-2’deoxycytidine relative to the total amount of cytosine residues, furthermore, it requires relatively lower amount of DNA and has a shorter run time for each sample than other high-performance liquid chromatography-based methods[33-35]. DNA methylation assay by LC/ESI/MS has been widely used for quantitative DNA methylation in animal studies and population-based studies in the light of its greater reproducibility and precision in large number of samples[32,36-40].
DNA methylation has long been recognized as an important factor on the silencing of genes, therefore it has become important to know the methylation status of individual CpG site. The first generation of DNA methylation detection assay is Southern blot or polymerase chain reaction (PCR) amplification that follows the enzyme digestion with methylation-sensitive restriction endonucleases[41-46]. Currently, the most commonly used methods for gene-specific DNA methylation can be categorized into three major methods.
Bisulfite DNA sequencing and methylation-specific PCR: Treatment of DNA with bisulfite converts cytosine residue to uracil, but leaves 5-methylcytosine residue unaffected. Bisulfite sequencing involves chemical conversion of cytosine to uracil, followed by PCR, and DNA sequencing[47]. While providing single-base resolution, the high cost and labor-intensive steps limit the use of this method for high-throughput analyses[48]. Methylation-specific-PCR also employs bisulfite conversion, but avoids the need to sequence the area of interest. Instead, methyl-specific and unmethyl-specific primer sets are designed, to distinguish methylated from unmethylated DNA in bisulfate-converted DNA[49]. This method is powerful to explore CpG islands with high methylation density, as increased numbers of CpG in the primer increase the specificity of the assay. However, these two methods using bisulfite conversion are not currently suitable for whole-genome analysis on multiple samples but commonly used for data validation from array-based methods.
Methods that focus specific single-CpG: These include Combined Bisulfite Restriction Analysis (COBRA)[50], MethyLight[51], and bisulfite pyrosequencing[52]. In COBRA, the combination of bisulfite conversion and PCR amplification is used, therefore it results in sequence conversion (unmethylated cytosine residue to thymidine and methylated cytosine to cytosine) which can lead to new methylation-dependent restriction enzyme sites. The following digestion of the PCR product with at least one CpG site in the recognition sequence only proceeds if the CpG site is protected from bisulfite conversion by methylation. For this reason, the signal ratio of restriction products indicating methylation to undigested PCR product representing unmethylated sequences can be used as a measure for the methylation level of this specific CpG. MethyLight is a bisulfite-dependent, fluorescence-based, quantitative real-time PCR method for DNA methylation. MethyLight relies on methylation-specific priming combined with methylation-specific fluorescent probing. This combination of methylation-specific detection principles results in a highly methylation-specific detection technology, with an accompanying ability to sensitively detect very low frequencies of hypermethylated alleles. Bisulfite pyrosequencing has been used to analyze bisulfite-converted DNA without using methylation-specific PCR. Following PCR amplification of the region of interest, pyrosequencing is used to determine the bisulfite-converted sequence of specific CpG sites in the region. The ratio of cytosine to thymidine at individual sites can be determined quantitatively based on the amount of cytosine and thymidine incorporation during the sequence extension. While the methods mentioned above are sensitive, specific, and relatively inexpensive, none of these methods is suitable for analysis of the whole genome, which includes about 28 million CpGs.
Microarray-based methods: These enable to interrogate larger numbers of CpG, there are three major types of microarray-based methylation analysis. Direct hybridization to CpG island arrays is the first high-throughput approach capable of detecting DNA methylaion in genes across several CpG sites. Based on the bisulfite modification of DNA, this method utilizes methylation-specific oligonucleotides arrayed on glass slides for detection of all possible methylation in target DNA[53]. Methylated DNA immunoprecipitation (MeDIP) is also a large-scale, genome-wide purification method that is used to enrich for methylated DNA sequence using antibody raised against 5-methylcytosine[54]. DNA from MeDIP can be used for either array-based hybridization (MeDIP-chip) or high-throughput sequencing (MeDIP-seq). Although MeDIP helps generate comprehensive DNA methylation profiles, both applications have their typical limitation of array-based technology, restricted resolution. The HELP assay (HpaII tiny fragment enrichment by ligation-mediated PCR) is comparative isoschizomer profiling of DNA methylation[55]. DNA is digested by HpaII in parallel with MspI (resistant to DNA methylation), and then the HpaII and MspI products are either amplified by ligation-mediated PCR and hybridized using separate fluorochromes to a customized array, or directly sequenced[56]. These high-throughput array-based approaches for DNA methylation are relatively inexpensive tool suitable for genome-wide analysis, therefore, help to target aberrant methylation patterns in various cancer models. Furthermore, methylation profiling achieved from high throughput methods will offer differentially methylated regions to understand the effect of dietary factors on epigenetic modifications in cancer, subsequently, provide insight in prevention strategies to reduce the burden of cancer.
In addition to the effects on DNA methylation, dietary components can affect posttranslational modifications of histones. The dietary agent best studied in histone modifications is the short chain fatty acid butyrate which is generated in the colon as a result of bacterial fermentation of dietary fiber. Higher intake of dietary fiber is associated with reduced risk of colorectal cancer[57,58]. The molecular mechanisms underlying this anti-cancer effect of dietary fiber are poorly understood, however, the strongest evidence is based on the anti-carcinogenic actions of butyrate. Butyrate can be found at millimolar concentrations in the lumen of the colon[59], and has inhibitory effects on types I and II histone deacetylase enzymes. Butyrate-induced alterations in histone marks, especially acetylation at histone H3 and/or H4, have been associated with several processes, including cellular differentiation[60,61], cell cycle arrest[62-64], apoptosis[65-67], and inhibition of invasion[68] in a number of cancer cell studies. Table 2 summarized some of evidence of the effects of butyrate on histone acetylation. Although butyrate has strong marks on histone acetylation, a small fraction of cellular genes is regulated in response to butyrate[69-71]. Therefore, it should be noted that site-specific approach by chromatin immunoprecipitation (ChIP) based experimental tools will provide a better understanding on the chemopreventive effects of butyrate, showing gene-specific histone acetylation and its associated gene expression.
| Dietary component | Cell culture model | Observations | Histone modification assay | Ref. |
| Sodium butyrate | SW620 human colon carcinoma cells | Increased global histone H4 acetylation | Western blot | [70] |
| Sodium butyrate | A375 human melanoma and S91 mouse melanoma | Increased global histone H4 acetylation | Western blot | [98] |
| Sodium butyrate | Colo-320 human colon cancer cells | Increased acetylation of histone H3 and H4 within CDKN1A promoter site | ChIP | [64] |
| Sodium butyrate | EBC-1 human lung epithelial cells | Increased histone H3 and H4 acetylation associated with promoter of cathelicidin | ChIP | [99] |
| Sodium butyrate | HepG2 human hepatocarcinoma | Increased global histone H3 and H4 acetylation; Genome-wide changes in acetylation of DNA-bound histones | Western blot; ChIP-chip (ChIP and microarray hybridization) | [100] |
The first estimates for the rate of acetylation turnover were measured by pulse, pulse-chase, and steady-state acetylation labeling in hepatoma tissue culture cells in 1975[72]. Boffa et al[73] showed that sodium butyrate suppressed histone deacetylation in vivo and in vitro by measuring the kinetics of [3H] acetate release from histone proteins. Since specific antibodies to modified histones were developed, Western blot has been used to detect histone modifications. As shown in Table 2, butyrate-induced histone acetylation was confirmed by Western blot in many studies.
ChIP: The antibodies to acetylated histone H3 and H4 have been used for ChIP to determine histone acetylation in specific regions of gene promoter and other regulatory regions. ChIP is a specialized immunoprecipitation used to detect the covalent interaction between the DNA sequence and DNA-binding proteins such as transcription factors or histone proteins. ChIP using histone antibodies is able to determine the specific location in the genome that various histone modifications are associated with, indicating the target of the histone modifiers[74]. For example, ChIP experiment unveiled that butyrate induced an increase in histone H3 and H4 acetylation within the CDKN1A promoter, which regulates the p21 protein, in Colo-320 human colon cancer cells[64]. Due to its ability to precisely detect the DNA binding of modified histones, transcription factors, and non-histone chromosomal proteins, ChIP has been widely used to generate and test numerous hypotheses regarding transcriptional and epigenetic regulations. However, it remains to be still challenging to conduct ChIP on an “epigenome” level, since ensuring an antibody of high specificity is often laborious and time-consuming. Another important concern with ChIP scalability is the maximum range of target regions that can be investigated by a single assay. For instance, a typical experiment of ChIP coupled with qPCR is designed to measure the enrichment levels of a DNA binding protein at a handful of sites (e.g., gene promoters). However, in general, even a single epigenetic event in the cell pervasively occurs over a wide range of genomic regions, often involving thousands of genes and their associated regulatory elements. Thus, it becomes more important to have an ability to run the assay on a genome-wide scale for having a more balanced and unbiased perspective on the underlying mechanisms. Coupled with genomic profiling technologies such as tiling arrays or next generation sequencing (NGS), ChIP can be extended over the whole genome. In the following sections, we will introduce two major methods coupled with ChIP that enable epigenome-scale research of histone marks and transcription factors.
ChIP-chip: ChIP-chip is based on the combination of ChIP and a genomic tiling array technology (i.e., chip), in which DNA sequences extracted after ChIP hybridize with probes that are designed to cover the whole genome or specific regions of interest such as promoter[75,76]. Due to bias in microarray hybridization, a control experiment using chromatin input or DNA from non-specific immunoprecipitation (IP) (e.g., IP against immunoglobulin G) is often recommended. Most algorithms for ChIP-chip are designed to compute the normalized ratio between the hybridizations of ChIP and control after removing random and/or systemic noise. Then, they call binding sites as those significantly enriched in ChIP over control[77-79]. Since its emergence in the early and mid 2000s, ChIP-chip has been widely adopted in many transcriptional and epigenetic regulation studies, assisting scientists to more understand the role of each histone mark in physiological and pathological processes[80-83]. However, the utility of ChIP-chip is heavily restricted by a tiling array probe design, which determines the resolution of the measurement (i.e., intervals between adjacent probes) and the regions that can be explored (e.g., omission of repetitive sequence areas). These weaknesses of ChIP-chip have accelerated the major platform shift to NGS.
ChIP-Seq: In ChIP-Seq, the extracted DNA sequences are directly sequenced using a NGS technology instead of being hybridized onto tiling arrays. NGS refers to sequencing technologies that newly emerged since the mid 2000s as an alternative to the traditional automated Sanger sequencing. NGS is characterized as massive parallel sequencing of template DNA or RNA (cDNA) molecules by a relatively short length ranging over 50-400 bp[84]. One advantage of ChIP-Seq over ChIP-chip is that ChIP-Seq does not require any predefined array design, which allows a more unbiased assay at a much higher resolution (100-1000 bp in ChIP-chip vs 10-100 bp in ChIP-Seq). Since NGS generally produces a notoriously large amount of data than array-based methods, more powerful bioinformatics support is essential for data processing and analysis[85,86]. Bioinformatics analysis for ChIP-Seq in epigenetic research includes the pre-processing for sequence data such as quality control and read mapping, the identification of candidate sites enriched by the target histone mark, and further down-stream analysis for revealing biological implications of the observations from the precedent steps[85,86].
In cancer studies, the down-stream analysis is focused on finding the most associated genes or regulatory elements (e.g., promoters or enhancers) with the histone mark of interest and investigating how these genes and regulatory elements can be understood in the context of biological pathways. Since Barski et al[87] and Wang et al[88] studies on 19 histone methylations and 18 histone acetylations using the human CD4+ T cell, many studies have been done to understand the biological implications of histone marks in normal conditions[80-82]. However, due to the plasticity of epigenome and heterogeneity of cancer, cancer epigenetics of examining histone modifications on a genome scale still remains in its beginning stage. For this reason, most of currently on-going efforts in cancer epigenetics still largely target DNA methylation (e.g., The Cancer Genome Atlas, http://cancergenome.nih.gov/)[89]. Therefore, it will be a long-term goal to accumulate the knowledge on cancer epigenetics from histone modifications and use it for cancer studies, which will require a great amount of public and private investments. Another interesting research direction is an attempt to comprehend how genetic variations lead to epigenetic changes in cancer. In 2011, several studies have been published about the possibility of multiple chromatin remodelers and histone enzymes as potential oncogenes or tumor suppressor genes[90-93]. These studies suggest that the disruption of chromatin remodelers and histone enzymes due to driving somatic mutations in their coding regions may cause aberrant epigenetic changes, which eventually lead to cancer development or evolution in at least several cancer indications. Such approach is particularly interesting because it may be able to provide a genuine perspective on the target histone mark by observing somatic mutations in several key chromatin remodelers and histone enzymes.
In conclusion, a number of aberrant epigenetic modifications have been found in cancer cells, and diet and dietary factors play an important role to prevent cancer as well as to stimulate carcinogenesis. The use of epigenetic technology offers significant advantages to study the epigenetic mechanisms of cancer development and progression. Also, the newly developed technologies for epigenetic study expand the scope of nutrition study in the field of cancer research by helping monitor and pin down specific epigenetic pathways in diet-related cancers.
P- Reviewer Prasad KK, Lakatos PL S- Editor Huang XZ L- Editor A E- Editor Xiong L
| 1. | Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2210] [Cited by in RCA: 2202] [Article Influence: 104.9] [Reference Citation Analysis (0)] |
| 2. | Trosko JE, Upham BL. The emperor wears no clothes in the field of carcinogen risk assessment: ignored concepts in cancer risk assessment. Mutagenesis. 2005;20:81-92. [PubMed] |
| 3. | Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3397] [Cited by in RCA: 3720] [Article Influence: 161.7] [Reference Citation Analysis (0)] |
| 4. | Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1674] [Cited by in RCA: 1532] [Article Influence: 73.0] [Reference Citation Analysis (0)] |
| 6. | Hake SB, Xiao A, Allis CD. Linking the epigenetic ‘language’ of covalent histone modifications to cancer. Br J Cancer. 2004;90:761-769. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 243] [Cited by in RCA: 240] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 7. | Esteller M. Aberrant DNA methylation as a cancer-inducing mechanism. Annu Rev Pharmacol Toxicol. 2005;45:629-656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 373] [Cited by in RCA: 378] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 8. | Esteller M. Epigenetics provides a new generation of oncogenes and tumour-suppressor genes. Br J Cancer. 2006;94:179-183. [PubMed] |
| 9. | Fraga MF, Esteller M. Towards the human cancer epigenome: a first draft of histone modifications. Cell Cycle. 2005;4:1377-1381. [PubMed] |
| 10. | Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6680] [Cited by in RCA: 6760] [Article Influence: 241.4] [Reference Citation Analysis (0)] |
| 11. | Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6156] [Cited by in RCA: 6150] [Article Influence: 246.0] [Reference Citation Analysis (0)] |
| 12. | Fischle W, Wang Y, Allis CD. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003;425:475-479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 485] [Cited by in RCA: 472] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 13. | Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, Kurdistani SK. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262-1266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 796] [Cited by in RCA: 773] [Article Influence: 38.7] [Reference Citation Analysis (0)] |
| 14. | Tzao C, Tung HJ, Jin JS, Sun GH, Hsu HS, Chen BH, Yu CP, Lee SC. Prognostic significance of global histone modifications in resected squamous cell carcinoma of the esophagus. Mod Pathol. 2009;22:252-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 88] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 15. | Pogribny IP, Ross SA, Tryndyak VP, Pogribna M, Poirier LA, Karpinets TV. Histone H3 lysine 9 and H4 lysine 20 trimethylation and the expression of Suv4-20h2 and Suv-39h1 histone methyltransferases in hepatocarcinogenesis induced by methyl deficiency in rats. Carcinogenesis. 2006;27:1180-1186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 102] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 16. | Ghoshal AK, Farber E. The induction of liver cancer by dietary deficiency of choline and methionine without added carcinogens. Carcinogenesis. 1984;5:1367-1370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 180] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 17. | Newberne PM. Lipotropic factors and oncogenesis. Adv Exp Med Biol. 1986;206:223-251. [PubMed] |
| 18. | Poirier LA. Methyl group deficiency in hepatocarcinogenesis. Drug Metab Rev. 1994;26:185-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 49] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 19. | Denda A, Kitayama W, Kishida H, Murata N, Tsutsumi M, Tsujiuchi T, Nakae D, Konishi Y. Development of hepatocellular adenomas and carcinomas associated with fibrosis in C57BL/6J male mice given a choline-deficient, L-amino acid-defined diet. Jpn J Cancer Res. 2002;93:125-132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 67] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 20. | Wainfan E, Poirier LA. Methyl groups in carcinogenesis: effects on DNA methylation and gene expression. Cancer Res. 1992;52:2071s-2077s. [PubMed] |
| 21. | Christman JK, Sheikhnejad G, Dizik M, Abileah S, Wainfan E. Reversibility of changes in nucleic acid methylation and gene expression induced in rat liver by severe dietary methyl deficiency. Carcinogenesis. 1993;14:551-557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 142] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 22. | Pogribny IP, James SJ, Jernigan S, Pogribna M. Genomic hypomethylation is specific for preneoplastic liver in folate/methyl deficient rats and does not occur in non-target tissues. Mutat Res. 2004;548:53-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 84] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 23. | Pogribny IP, Ross SA, Wise C, Pogribna M, Jones EA, Tryndyak VP, James SJ, Dragan YP, Poirier LA. Irreversible global DNA hypomethylation as a key step in hepatocarcinogenesis induced by dietary methyl deficiency. Mutat Res. 2006;593:80-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 112] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 24. | Ghoshal K, Li X, Datta J, Bai S, Pogribny I, Pogribny M, Huang Y, Young D, Jacob ST. A folate- and methyl-deficient diet alters the expression of DNA methyltransferases and methyl CpG binding proteins involved in epigenetic gene silencing in livers of F344 rats. J Nutr. 2006;136:1522-1527. [PubMed] |
| 25. | Christman JK, Weich N, Schoenbrun B, Schneiderman N, Acs G. Hypomethylation of DNA during differentiation of Friend erythroleukemia cells. J Cell Biol. 1980;86:366-370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 125] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 26. | Balaghi M, Wagner C. DNA methylation in folate deficiency: use of CpG methylase. Biochem Biophys Res Commun. 1993;193:1184-1190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 181] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 27. | Kim YI, Giuliano A, Hatch KD, Schneider A, Nour MA, Dallal GE, Selhub J, Mason JB. Global DNA hypomethylation increases progressively in cervical dysplasia and carcinoma. Cancer. 1994;74:893-899. [PubMed] |
| 28. | Kim YI, Pogribny IP, Basnakian AG, Miller JW, Selhub J, James SJ, Mason JB. Folate deficiency in rats induces DNA strand breaks and hypomethylation within the p53 tumor suppressor gene. Am J Clin Nutr. 1997;65:46-52. [PubMed] |
| 29. | Pogribny I, Yi P, James SJ. A sensitive new method for rapid detection of abnormal methylation patterns in global DNA and within CpG islands. Biochem Biophys Res Commun. 1999;262:624-628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 154] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 30. | Pogribny IP, Shpyleva SI, Muskhelishvili L, Bagnyukova TV, James SJ, Beland FA. Role of DNA damage and alterations in cytosine DNA methylation in rat liver carcinogenesis induced by a methyl-deficient diet. Mutat Res. 2009;669:56-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 31. | Oakeley EJ. DNA methylation analysis: a review of current methodologies. Pharmacol Ther. 1999;84:389-400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 65] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 32. | Friso S, Choi SW, Dolnikowski GG, Selhub J. A method to assess genomic DNA methylation using high-performance liquid chromatography/electrospray ionization mass spectrometry. Anal Chem. 2002;74:4526-4531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 186] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 33. | Kuo KC, McCune RA, Gehrke CW, Midgett R, Ehrlich M. Quantitative reversed-phase high performance liquid chromatographic determination of major and modified deoxyribonucleosides in DNA. Nucleic Acids Res. 1980;8:4763-4776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 216] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 34. | Christman JK. Separation of major and minor deoxyribonucleoside monophosphates by reverse-phase high-performance liquid chromatography: a simple method applicable to quantitation of methylated nucleotides in DNA. Anal Biochem. 1982;119:38-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 35. | Kaur H, Halliwell B. Measurement of oxidized and methylated DNA bases by HPLC with electrochemical detection. Biochem J. 1996;318:21-23. [PubMed] |
| 36. | Friso S, Choi SW, Girelli D, Mason JB, Dolnikowski GG, Bagley PJ, Olivieri O, Jacques PF, Rosenberg IH, Corrocher R. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci USA. 2002;99:5606-5611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 684] [Cited by in RCA: 675] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 37. | Choi SW, Friso S, Ghandour H, Bagley PJ, Selhub J, Mason JB. Vitamin B-12 deficiency induces anomalies of base substitution and methylation in the DNA of rat colonic epithelium. J Nutr. 2004;134:750-755. [PubMed] |
| 38. | Friso S, Girelli D, Trabetti E, Olivieri O, Guarini P, Pignatti PF, Corrocher R, Choi SW. The MTHFR 1298A& gt; C polymorphism and genomic DNA methylation in human lymphocytes. Cancer Epidemiol Biomarkers Prev. 2005;14:938-943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 54] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 39. | Keyes MK, Jang H, Mason JB, Liu Z, Crott JW, Smith DE, Friso S, Choi SW. Older age and dietary folate are determinants of genomic and p16-specific DNA methylation in mouse colon. J Nutr. 2007;137:1713-1717. [PubMed] |
| 40. | Lim U, Flood A, Choi SW, Albanes D, Cross AJ, Schatzkin A, Sinha R, Katki HA, Cash B, Schoenfeld P. Genomic methylation of leukocyte DNA in relation to colorectal adenoma among asymptomatic women. Gastroenterology. 2008;134:47-55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 85] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 41. | Bird AP. Use of restriction enzymes to study eukaryotic DNA methylation: II. The symmetry of methylated sites supports semi-conservative copying of the methylation pattern. J Mol Biol. 1978;118:49-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 212] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 42. | Waalwijk C, Flavell RA. MspI, an isoschizomer of hpaII which cleaves both unmethylated and methylated hpaII sites. Nucleic Acids Res. 1978;5:3231-3236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 314] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 43. | Razin A, Riggs AD. DNA methylation and gene function. Science. 1980;210:604-610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1473] [Cited by in RCA: 1462] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 44. | Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1699] [Cited by in RCA: 1629] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 45. | Goelz SE, Vogelstein B, Hamilton SR, Feinberg AP. Hypomethylation of DNA from benign and malignant human colon neoplasms. Science. 1985;228:187-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 580] [Cited by in RCA: 548] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 46. | Singer-Sam J, LeBon JM, Tanguay RL, Riggs AD. A quantitative HpaII-PCR assay to measure methylation of DNA from a small number of cells. Nucleic Acids Res. 1990;18:687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 116] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 47. | Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827-1831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2250] [Cited by in RCA: 2268] [Article Influence: 68.7] [Reference Citation Analysis (0)] |
| 48. | Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378-1385. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1103] [Cited by in RCA: 953] [Article Influence: 50.2] [Reference Citation Analysis (0)] |
| 49. | Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821-9826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4183] [Cited by in RCA: 4248] [Article Influence: 146.5] [Reference Citation Analysis (0)] |
| 50. | Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532-2534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 913] [Cited by in RCA: 884] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 51. | Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1017] [Cited by in RCA: 1057] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 52. | Dupont JM, Tost J, Jammes H, Gut IG. De novo quantitative bisulfite sequencing using the pyrosequencing technology. Anal Biochem. 2004;333:119-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 189] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 53. | Gitan RS, Shi H, Chen CM, Yan PS, Huang TH. Methylation-specific oligonucleotide microarray: a new potential for high-throughput methylation analysis. Genome Res. 2002;12:158-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 189] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 54. | Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schübeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853-862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1315] [Cited by in RCA: 1291] [Article Influence: 64.6] [Reference Citation Analysis (0)] |
| 55. | Khulan B, Thompson RF, Ye K, Fazzari MJ, Suzuki M, Stasiek E, Figueroa ME, Glass JL, Chen Q, Montagna C. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 2006;16:1046-1055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 320] [Cited by in RCA: 297] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 56. | Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang H, Porter D, Hu M, Chin L, Richardson A. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 970] [Cited by in RCA: 990] [Article Influence: 47.1] [Reference Citation Analysis (0)] |
| 57. | Bingham SA. Mechanisms and experimental and epidemiological evidence relating dietary fibre (non-starch polysaccharides) and starch to protection against large bowel cancer. Proc Nutr Soc. 1990;49:153-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 115] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 58. | Murphy N, Norat T, Ferrari P, Jenab M, Bueno-de-Mesquita B, Skeie G, Dahm CC, Overvad K, Olsen A, Tjønneland A. Dietary fibre intake and risks of cancers of the colon and rectum in the European prospective investigation into cancer and nutrition (EPIC). PLoS One. 2012;7:e39361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 193] [Cited by in RCA: 181] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 59. | Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev. 2001;81:1031-1064. [PubMed] |
| 60. | McIntyre A, Gibson PR, Young GP. Butyrate production from dietary fibre and protection against large bowel cancer in a rat model. Gut. 1993;34:386-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 397] [Cited by in RCA: 383] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 61. | Gobbi G, Di Marcantonio D, Micheloni C, Carubbi C, Galli D, Vaccarezza M, Bucci G, Vitale M, Mirandola P. TRAIL up-regulation must be accompanied by a reciprocal PKC epsilon; down-regulation during differentiation of colonic epithelial cell: implications for colorectal cancer cell differentiation. J Cell Physiol. 2012;227:630-638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 62. | Gope R, Gope ML. Effect of sodium butyrate on the expression of retinoblastoma (RB1) and P53 gene and phosphorylation of retinoblastoma protein in human colon tumor cell line HT29. Cell Mol Biol (Noisy-le-grand). 1993;39:589-597. [PubMed] |
| 63. | Nakano K, Mizuno T, Sowa Y, Orita T, Yoshino T, Okuyama Y, Fujita T, Ohtani-Fujita N, Matsukawa Y, Tokino T. Butyrate activates the WAF1/Cip1 gene promoter through Sp1 sites in a p53-negative human colon cancer cell line. J Biol Chem. 1997;272:22199-22206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 309] [Cited by in RCA: 305] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 64. | Fang JY, Chen YX, Lu J, Lu R, Yang L, Zhu HY, Gu WQ, Lu LG. Epigenetic modification regulates both expression of tumor-associated genes and cell cycle progressing in human colon cancer cell lines: Colo-320 and SW1116. Cell Res. 2004;14:217-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 37] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 65. | Janson W, Brandner G, Siegel J. Butyrate modulates DNA-damage-induced p53 response by induction of p53-independent differentiation and apoptosis. Oncogene. 1997;15:1395-1406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 52] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 66. | Terui T, Murakami K, Takimoto R, Takahashi M, Takada K, Murakami T, Minami S, Matsunaga T, Takayama T, Kato J. Induction of PIG3 and NOXA through acetylation of p53 at 320 and 373 lysine residues as a mechanism for apoptotic cell death by histone deacetylase inhibitors. Cancer Res. 2003;63:8948-8954. [PubMed] |
| 67. | Bernhard D, Ausserlechner MJ, Tonko M, Löffler M, Hartmann BL, Csordas A, Kofler R. Apoptosis induced by the histone deacetylase inhibitor sodium butyrate in human leukemic lymphoblasts. FASEB J. 1999;13:1991-2001. [PubMed] |
| 68. | Kuwajima A, Iwashita J, Murata J, Abe T. The histone deacetylase inhibitor butyrate inhibits melanoma cell invasion of Matrigel. Anticancer Res. 2007;27:4163-4169. [PubMed] |
| 69. | Van Lint C, Emiliani S, Verdin E. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr. 1996;5:245-253. [PubMed] |
| 70. | Mariadason JM, Corner GA, Augenlicht LH. Genetic reprogramming in pathways of colonic cell maturation induced by short chain fatty acids: comparison with trichostatin A, sulindac, and curcumin and implications for chemoprevention of colon cancer. Cancer Res. 2000;60:4561-4572. [PubMed] |
| 71. | Davie JR. Inhibition of histone deacetylase activity by butyrate. J Nutr. 2003;133:2485S-2493S. [PubMed] |
| 72. | Jackson V, Shires A, Chalkley R, Granner DK. Studies on highly metabolically active acetylation and phosphorylation of histones. J Biol Chem. 1975;250:4856-4863. [PubMed] |
| 73. | Boffa LC, Vidali G, Mann RS, Allfrey VG. Suppression of histone deacetylation in vivo and in vitro by sodium butyrate. J Biol Chem. 1978;253:3364-3366. [PubMed] |
| 74. | Collas P. The current state of chromatin immunoprecipitation. Mol Biotechnol. 2010;45:87-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 156] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 75. | de Magalhães JP, Finch CE, Janssens G. Next-generation sequencing in aging research: emerging applications, problems, pitfalls and possible solutions. Ageing Res Rev. 2010;9:315-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 69] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 76. | Ho JW, Bishop E, Karchenko PV, Nègre N, White KP, Park PJ. ChIP-chip versus ChIP-seq: lessons for experimental design and data analysis. BMC Genomics. 2011;12:134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 97] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 77. | Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci USA. 2006;103:12457-12462. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 340] [Cited by in RCA: 348] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 78. | Song JS, Johnson WE, Zhu X, Zhang X, Li W, Manrai AK, Liu JS, Chen R, Liu XS. Model-based analysis of two-color arrays (MA2C). Genome Biol. 2007;8:R178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 91] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 79. | Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26:1293-1300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 604] [Cited by in RCA: 564] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 80. | Consortium EP. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636-640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1646] [Cited by in RCA: 1711] [Article Influence: 81.5] [Reference Citation Analysis (0)] |
| 81. | Liu T, Rechtsteiner A, Egelhofer TA, Vielle A, Latorre I, Cheung MS, Ercan S, Ikegami K, Jensen M, Kolasinska-Zwierz P. Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res. 2011;21:227-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 224] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 82. | Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, Lin MF. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787-1797. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1047] [Cited by in RCA: 959] [Article Influence: 63.9] [Reference Citation Analysis (0)] |
| 83. | Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045-1048. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1375] [Cited by in RCA: 1367] [Article Influence: 97.6] [Reference Citation Analysis (0)] |
| 84. | Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4795] [Cited by in RCA: 4098] [Article Influence: 256.1] [Reference Citation Analysis (0)] |
| 85. | Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008;26:1351-1359. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 805] [Cited by in RCA: 675] [Article Influence: 39.7] [Reference Citation Analysis (0)] |
| 86. | Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1327] [Cited by in RCA: 1392] [Article Influence: 87.0] [Reference Citation Analysis (0)] |
| 87. | Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823-837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5009] [Cited by in RCA: 5090] [Article Influence: 282.8] [Reference Citation Analysis (0)] |
| 88. | Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897-903. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1944] [Cited by in RCA: 1750] [Article Influence: 102.9] [Reference Citation Analysis (0)] |
| 89. | Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061-1068. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6404] [Cited by in RCA: 5995] [Article Influence: 352.6] [Reference Citation Analysis (0)] |
| 90. | Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field M. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298-303. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1337] [Cited by in RCA: 1307] [Article Influence: 93.4] [Reference Citation Analysis (1)] |
| 91. | Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, Wells VA, Grunn A, Messina M, Elliot O. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43:830-837. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 804] [Cited by in RCA: 794] [Article Influence: 56.7] [Reference Citation Analysis (0)] |
| 92. | Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, Wu R, Chen C, Li X, Zhou L. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43:875-878. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 534] [Cited by in RCA: 590] [Article Influence: 42.1] [Reference Citation Analysis (0)] |
| 93. | Li M, Zhao H, Zhang X, Wood LD, Anders RA, Choti MA, Pawlik TM, Daniel HD, Kannangai R, Offerhaus GJ. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat Genet. 2011;43:828-829. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 320] [Cited by in RCA: 344] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 94. | Pogribny IP, James SJ. De novo methylation of the p16INK4A gene in early preneoplastic liver and tumors induced by folate/methyl deficiency in rats. Cancer Lett. 2002;187:69-75. [PubMed] |
| 95. | Tsujiuchi T, Shimizu K, Itsuzaki Y, Onishi M, Sugata E, Fujii H, Honoki K. CpG site hypermethylation of E-cadherin and Connexin26 genes in hepatocellular carcinomas induced by a choline-deficient L-Amino Acid-defined diet in rats. Mol Carcinog. 2007;46:269-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 96. | Pogribny IP, Tryndyak VP, Bagnyukova TV, Melnyk S, Montgomery B, Ross SA, Latendresse JR, Rusyn I, Beland FA. Hepatic epigenetic phenotype predetermines individual susceptibility to hepatic steatosis in mice fed a lipogenic methyl-deficient diet. J Hepatol. 2009;51:176-186. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 137] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 97. | Tryndyak VP, Han T, Muskhelishvili L, Fuscoe JC, Ross SA, Beland FA, Pogribny IP. Coupling global methylation and gene expression profiles reveal key pathophysiological events in liver injury induced by a methyl-deficient diet. Mol Nutr Food Res. 2011;55:411-418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 66] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 98. | Demary K, Wong L, Spanjaard RA. Effects of retinoic acid and sodium butyrate on gene expression, histone acetylation and inhibition of proliferation of melanoma cells. Cancer Lett. 2001;163:103-107. [PubMed] |
| 99. | Kida Y, Shimizu T, Kuwano K. Sodium butyrate up-regulates cathelicidin gene expression via activator protein-1 and histone acetylation at the promoter region in a human lung epithelial cell line, EBC-1. Mol Immunol. 2006;43:1972-1981. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 81] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 100. | Rada-Iglesias A, Enroth S, Ameur A, Koch CM, Clelland GK, Respuela-Alonso P, Wilcox S, Dovey OM, Ellis PD, Langford CF. Butyrate mediates decrease of histone acetylation centered on transcription start sites and down-regulation of associated genes. Genome Res. 2007;17:708-719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 111] [Article Influence: 6.2] [Reference Citation Analysis (0)] |