Published online Sep 21, 2013. doi: 10.3748/wjg.v19.i35.5870
Revised: July 15, 2013
Accepted: August 4, 2013
Published online: September 21, 2013
Processing time: 169 Days and 20.1 Hours
AIM: To evaluate the association between the genetic polymorphisms and haplotypes of the ITGA1 gene and the risk of gastric cancer.
METHODS: The study subjects were 477 age- and sex-matched case-control pairs. Genotyping was performed for 15 single nucleotide polymorphisms (SNPs) in ITGA1. The associations between gastric cancer and these SNPs and haplotypes were analyzed with multivariate conditional logistic regression models. Multiple testing corrections were carried out following methodology for controlling the false discovery rate. Gene-based association tests were performed using the versatile gene-based association study (VEGAS) method.
RESULTS: In the codominant model, the ORs for SNPs rs2432143 (1.517; 95%CI: 1.144-2.011) and rs2447867 (1.258; 95%CI: 1.051-1.505) were statistically significant. In the dominant model, polymorphisms of rs1862610 and rs2447867 were found to be significant risk factors, with ORs of 1.337 (95%CI: 1.029-1.737) and 1.412 (95%CI: 1.061-1.881), respectively. In the recessive model, only the rs2432143 polymorphism was significant (OR = 1.559, 95%CI: 1.150-2.114). The C-C type of ITGA1 haplotype block 2 was a significant protective factor against gastric cancer in the both codominant model (OR = 0.602, 95%CI: 0.212-0.709, P = 0.021) and the dominant model (OR = 0.653, 95%CI: 0.483-0.884). The ITGA1 gene showed a significant gene-based association with gastric cancer in the VEGAS test. In the dominant model, the A-T type of ITGA1 haplotype block 2 was a significant risk factor (OR = 1.341, 95%CI: 1.034-1.741). SNP rs2447867 might be related to the severity of gastric epithelial injury due to inflammation and, thus, to the risk of developing gastric cancer.
CONCLUSION: ITGA1 gene SNPs rs1862610, rs24321
43, and rs2447867 and the ITGA1 haplotype block that includes SNPs rs1862610 and rs2432143 were significantly associated with gastric cancer.
Core tip: There are few studies addressing the role of the integrin α 1 subunit in the development of gastric cancer. To the best of our knowledge, this study is the first to show that ITGA1 gene single nucleotide polymorphisms and haplotypes are associated with gastric cancer risk.
-
Citation: Yim DH, Zhang YW, Eom SY, Moon SI, Yun HY, Song YJ, Youn SJ, Hyun T, Park JS, Kim BS, Lee JY, Kim YD, Kim H.
ITGA1 polymorphisms and haplotypes are associated with gastric cancer risk in a Korean population. World J Gastroenterol 2013; 19(35): 5870-5876 - URL: https://www.wjgnet.com/1007-9327/full/v19/i35/5870.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i35.5870
Gastric cancer is the second most common cancer in South Korea and represents the second leading cause of cancer death for both men and women worldwide[1,2]. Approximately one million new cases of stomach cancer are estimated to have occurred (989000 cases, 7.8% of the total), currently making it the fourth most common malignancy in the world, following cancers of the lung, breast and colo-rectum[2]. Epidemiological studies have provided evidence that a high intake of salt and nitrite-rich foods and Helicobacter pylori (H. pylori) infection are associated with a high incidence of gastric cancer in South Korea[3-7].
The risk of developing gastric cancer is estimated to be increased 2-6 fold in patients with H. pylori infection[8]. The risk of gastric cancer among individuals infected with H. pylori is influenced by bacterial virulence. The most widely studied H. pylori virulence factors are the cag (cytotoxin-associated gene) antigens[9]. Compared to individuals infected with cagA-negative H. pylori strains, those infected with cagA-positive H. pylori strains show a higher risk of developing gastric cancer[10]. To introduce cagA into host cells, the cagL protein of H. pylori binds to integrins on the basolateral surface of gastric epithelial cells[11,12].
Integrins are members of a family of heterodimeric cell-surface proteins that mediate cell-matrix and cell-cell interactions. The 18 integrin α-subunits and 8 β-subunits together form at least 25 different integrins[13]. Integrins mediate signaling events that are essential for stable cell adhesion, spreading, migration, survival, proliferation and differentiation. Several integrins, including α1β1, bind to extracellular matrix proteins present in the basal membrane of mature vessels[14,15]. The tumor progression and metastasis of various cancers are associated with integrins[16,17].
The ITGA1 gene, located on chromosome 5q11.2, encodes the integrin α1 subunit, which is involved in the adhesion of gastric cancer cells to the peritoneum. The adhesion of integrin α1-positive gastric cancer cells to the extracellular matrix is a critical process in peritoneal dissemination[18,19]. There are few studies addressing the roles of integrins in the development of gastric cancer. An association with an increased risk of gastric cancer has only been reported for the ITGA2 C807T polymorphism in a Chinese population[20]. As the level of integrin α1β1 is up-regulated in association with inflammation of the gastrointestinal tract mucosa, which is the first step in gastric carcinogenesis[21], it is possible that the integrin α1 subunit plays an important role in gastric cancer development.
The purpose of this study was to evaluate the association between the genetic polymorphisms and haplotypes of the ITGA1 gene and the risk of gastric cancer.
This subjects included in this study consisted of 477 newly diagnosed gastric cancer patients and an equal number of age- (within 3 years) and sex-matched controls. The diagnoses of the gastric cancer patients were all histologically confirmed at Chungbuk National University Hospital and Eulji University Hospital, which are located in a geographically central region of South Korea. Controls were selected from individuals receiving routine medical examinations in these hospitals, and individuals with a previous diagnosis of any type of cancer were excluded. Trained interviewers used a structured questionnaire including questions about demographic factors, smoking habits, alcohol consumption and dietary habits to interview all subjects who provided written informed consent. Peripheral blood samples were collected from all subjects. This study was approved by the institutional review boards of Chungbuk National University Hospital, South Korea (IRB No. 2011-09-071).
At the International HapMap Project website (http://hapmap.ncbi.nlm.nih.gov/), tag SNPs were selected using a cut-off minimum minor allele frequency in the JPT population of 0.05 and pairwise tagging (r2 = 1-0.8). SNPs that significantly deviated from Hardy-Weinberg equilibrium were discarded.
Genomic DNA was extracted from whole blood using the QuickGene-810 nucleic acid isolation system (Fujifilm, Tokyo, Japan) and the QuickGene DNA Whole Blood Kit (Kurabo, Osaka, Japan), in accordance with the manufacturer’s instructions. DNA was stored at 4 °C until use. SNP genotyping was performed using a GoldenGate Genotyping Assay with VeraCode technology (Illumina, San Diego, CA, United States). A custom GoldenGate assay was designed for the analysis of the selected SNPs in the ITGA1 gene. Those SNPs were then assessed for suitability for the GoldenGate genotyping platform, and the analysis was carried out on the validated SNPs. The average call rate was 99.2%. Genotyping was carried out by Macrogen (Seoul, South Korea).
The study power was calculated using the “case-control for discrete traits” mode in the Genetic Power Calculator[22]. The following parameters were applied: risk allele frequency -0.4, alpha error -0.01, and disease prevalence -0.1%. The power of a codominant model was 0.7768 when the heterozygous OR was set to 1.5. For a dominant model, when the OR for a genotype with one or 2 risk allele(s) was taken as 2, the power was 0.8821. When a value of 2 was input for the OR for a genotype with 2 risk allele(s), the power of a recessive model was 0.8182.
Testing for deviation from the HWP was performed for each SNP in both cases and in controls using Pearson’s χ2 test. D values were measured using Lewontin’s method for all combinations of biallelic loci[23,24], and linkage disequilibrium blocks were structured using Haploview version 4.2 (Daly Lab at the Broad Institute Cambridge, MA, United States). Haplotype blocks were constructed and statistically compared between cases and controls with SNP Analyzer version 2.0 (ISTEC Inc., Goyang, South Korea).
Student’s t test was used to compare continuous variables between patients and control subjects. Associations between gastric cancer and the investigated SNPs and haplotypes were estimated via the OR and their corresponding 95%CI derived from multivariate conditional logistic regression models, after adjusting for potential confounding factors such as age, sex, smoking history, and alcohol intake. The genotypes of major homozygotes, heterozygotes and minor homozygotes were coded as 0, 1, and 2 in the codominant model, 0, 1 and 1 in the dominant model, and 0, 0 and 1 in the recessive model, respectively. Multiple testing corrections were carried out using Benjaminin and Hochberg’s methods for controlling the false discovery rate (FDR)[25]. A two-sided adjusted P value of < 0.05 was considered statistically significant. FDR Q values were calculated separately for the SNPs and haplotypes based on these numbers. Gene-based association tests were performed using the versatile gene-based association study (VEGAS) method[26]. For these statistical analyses, SAS version 9.2 (SAS Institute, Cary, NC, United States) was employed.
Patient characteristics are summarized in Table 1. No significant difference was observed between the distributions of the age, sex, and smoking and drinking habits of the cases and controls.
| Variables | Controls (n = 477) | Cases (n = 477) | OR (95%CI) |
| Age (yr) mean ± SD | 57.8 ± 10.2 | 58.7 ± 9.9 | |
| Sex | |||
| Males | 301 (63.1) | 301 (63.1) | |
| Females | 176 (36.9) | 176 (36.9) | |
| Smoking status | |||
| Non-smokers | 225 (47.6) | 194 (41.0) | 1.00 (reference) |
| Smokers | 248 (52.4) | 279 (59.0) | 1.64 (0.95-2.84) |
| Alcohol intake status | |||
| Non-drinkers | 194 (40.7) | 189 (39.6) | 1.00 (reference) |
| Drinkers | 283 (59.3) | 288 (60.4) | 1.18 (0.71-1.76) |
Table 2 lists and provides the frequencies of the 15 selected SNPs in the study subjects. None of the polymorphisms were significantly deviated from Hardy-Weinberg equilibrium. All the minor allele frequencies of the cases and controls were greater than 10%.
| SNP | Chromosomal position | Amino acid change | Genotype case/control | Case | Control | |||||
| Frequency | HWE1 | Frequency | HWE1 | |||||||
| rs13188662 | 2686006 | - | AA | AG | GG | N | 0.280 | 0.573 | 0.276 | 0.597 |
| 249/253 | 186/186 | 40/38 | 475/477 | |||||||
| rs11740785 | 2707341 | - | AA | AC | CC | N | 0.241 | 0.866 | 0.229 | 0.259 |
| 279/290 | 166/156 | 32/31 | 477/477 | |||||||
| rs1820167 | 2713715 | - | AA | AG | GG | N | 0.435 | 0.806 | 0.420 | 0.904 |
| 151/162 | 237/229 | 89/83 | 477/477 | |||||||
| rs1862610 | 2722239 | - | CC | AC | AA | N | 0.369 | 0.861 | 0.387 | 0.484 |
| 172/205 | 223/192 | 82/80 | 477/477 | |||||||
| rs2432143 | 2725674 | - | TT | TC | CC | N | 0.104 | 0.671 | 0.146 | 0.658 |
| 382/346 | 87/121 | 8/10 | 477/477 | |||||||
| rs2447867 | 2751733 | C/C | CC | TC | TT | N | 0.490 | 0.742 | 0.430 | 0.769 |
| 123/155 | 241/229 | 113/89 | 477/473 | |||||||
| rs4865745 | 2770258 | - | TT | TC | CC | N | 0.270 | 0.892 | 0.268 | 0.124 |
| 253/247 | 186/198 | 35/28 | 474/473 | |||||||
| rs13163497 | 2773367 | - | GG | AG | AA | N | 0.110 | 0.409 | 0.108 | 0.515 |
| 375/381 | 97/89 | 4/7 | 476/477 | |||||||
| rs1904163 | 2780355 | - | CC | TC | TT | N | 0.298 | 0.196 | 0.272 | 0.698 |
| 238/245 | 184/187 | 48/33 | 470/465 | |||||||
| rs1466445 | 2789486 | - | CC | TC | TT | N | 0.460 | 0.783 | 0.455 | 0.696 |
| 139/142 | 233/229 | 101/100 | 473/471 | |||||||
| rs16880453 | 2789866 | - | GG | GC | CC | N | 0.466 | 0.914 | 0.465 | 0.424 |
| 133/130 | 235/243 | 100/98 | 468/471 | |||||||
| rs2452864 | 2796757 | - | TT | TC | CC | N | 0.367 | 0.874 | 0.369 | 0.368 |
| 190/183 | 224/230 | 63/59 | 477/472 | |||||||
| rs1275659 | 2828018 | - | AA | AG | GG | N | 0.257 | 0.185 | 0.278 | 0.864 |
| 256/247 | 192/189 | 26/37 | 474/473 | |||||||
| rs1871186 | 2828974 | - | TT | TC | CC | N | 0.221 | 0.723 | 0.213 | 0.674 |
| 287/296 | 166/157 | 22/23 | 475/476 | |||||||
| rs988574 | 2835169 | E/G | TT | TC | CC | N | 0.180 | 0.723 | 0.183 | 0.674 |
| 319/309 | 141/155 | 15/9 | 475/473 | |||||||
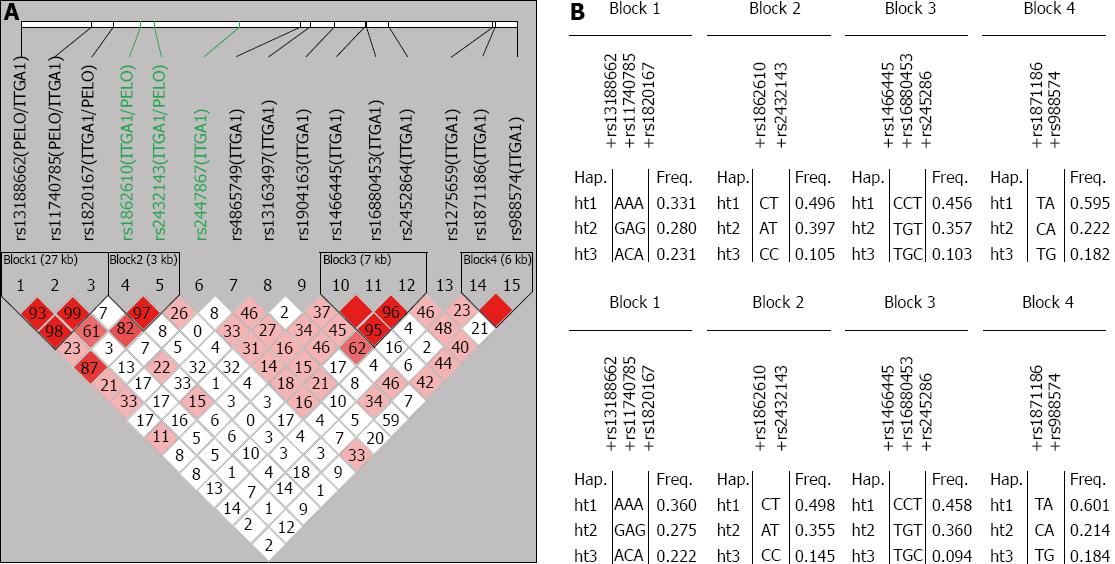
The haplotype linkage disequilibrium blocks and haplotype frequencies for ITGA1 are shown in Figure 1. D values were measured using Lewontin’s method. Four block haplotypes were constructed using Haploview version 4.2. The common haplotypes (frequency > 10%) in each block accounted for 84.2%, 99.8%, 91.6% and 99.9% for the cases and 85.7%, 99.8%, 91.2% and 99.9% for the controls.
The observed associations between the genetic polymorphisms in the ITGA1 gene and the risk of gastric cancer are shown in Table 3. In the codominant model, the OR of 1.517 obtained for SNP rs2432143 (95%CI: 1.144-2.011; P = 0.003; FDR Q = 0.045) was statistically significant, even after controlling the FDR, and that for rs2447867, of 1.258 (95%CI: 1.051-1.505; P = 0.012; FDR Q = 0.090), was marginally significant. In the dominant model, the rs1862610 and rs2447867 polymorphisms were not statistically significant risk factors for gastric cancer, displaying ORs of 1.337 (95%CI: 1.029-1.737; P = 0.029; FDR Q = 0.217) and 1.412 (95%CI: 1.061-1.881; P = 0.018; FDR Q = 0.217), respectively. Only the rs2432143 polymorphism was marginally significant in the recessive model, exhibiting an OR of 1.559 (95%CI: 1.150-2.114; P = 0.004; FDR Q = 0.060).
| SNP | Chromosomal position | Codominant | Dominant | Recessive | ||||||
| OR (95%CI) | P value1 | Q2 | OR (95%CI) | P value1 | Q2 | OR (95%CI) | P value1 | Q2 | ||
| rs13188662 | 2686006 | 1.040 (0.840-1.281) | 0.161 | 0.483 | 1.060 (0.811-1.379) | 0.689 | 0.866 | 1.060 (0.660-1.690) | 0.811 | 0.963 |
| rs11740785 | 2707341 | 1.069 (0.869-1.313) | 0.528 | 0.965 | 1.106 (0.848-1.442) | 0.457 | 0.866 | 1.032 (0.630-1.692) | 0.899 | 0.963 |
| rs1820167 | 2713715 | 1.066 (0.884-1.286) | 0.503 | 0.964 | 1.115 (0.846-1.468) | 0.440 | 0.866 | 1.043 (0.751-1.447) | 0.801 | 0.963 |
| rs1862610 | 2722239 | 1.151 (0.965-1.372) | 0.118 | 0.483 | 1.337 (1.029-1.737) | 0.029 | 0.217 | 1.029 (0.740-1.429) | 0.866 | 0.963 |
| rs2432143 | 2725674 | 1.517 (1.144-2.011) | 0.003 | 0.045 | 1.800 (0.603-5.371) | 0.292 | 0.883 | 1.559 (1.150-2.114) | 0.004 | 0.060 |
| rs2447867 | 2751733 | 1.258 (1.051-1.505) | 0.012 | 0.090 | 1.412 (1.061-1.881) | 0.018 | 0.217 | 1.303 (0.966-1.756) | 0.083 | 0.415 |
| rs4865745 | 2770258 | 1.016 (0.829-1.246) | 0.875 | 0.965 | 0.967 (0.750-1.247) | 0.795 | 0.863 | 1.269 (0.759-2.122) | 0.363 | 0.927 |
| rs13163497 | 2773367 | 1.021 (0.768-1.357) | 0.884 | 0.965 | 1.064 (0.781-1.449) | 0.693 | 0.866 | 0.571 (0.167-1.952) | 0.371 | 0.927 |
| rs1904163 | 2780355 | 1.157 (0.943-1.420) | 0.161 | 0.483 | 1.104 (0.849-1.436) | 0.461 | 0.866 | 1.593 (0.984-2.577) | 0.058 | 0.415 |
| rs1466445 | 2789486 | 1.013 (0.845-1.213) | 0.890 | 0.965 | 1.032 (0.778-1.368) | 0.829 | 0.883 | 1.000 (0.736-1.358) | 1.000 | 1.000 |
| rs16880453 | 2789866 | 1.000 (0.832-1.201) | 1.000 | 1.000 | 0.979 (0.734-1.305) | 0.883 | 0.883 | 1.025 (0.752-1.398) | 0.874 | 9.632 |
| rs2452864 | 2796757 | 0.986 (0.816-1.191) | 0.885 | 0.965 | 0.947 (0.728-1.233) | 0.687 | 0.883 | 1.056 (0.728-1.532) | 0.775 | 9.632 |
| rs1275659 | 2828018 | 1.136 (0.919-1.404) | 0.237 | 0.592 | 1.522 (0.899-2.575) | 0.117 | 0.585 | 1.095 (0.841-1.427) | 0.500 | 9.632 |
| rs1871186 | 2828974 | 1.043 (0.841-1.293) | 0.701 | 0.965 | 1.072 (0.828-1.388) | 0.597 | 0.866 | 0.957 (0.533-1.716) | 0.881 | 9.632 |
| rs988574 | 2835169 | 0.985 (0.772-1.256) | 0.901 | 0.965 | 0.927 (0.707-1.215) | 0.581 | 0.866 | 1.667 (0.729-3.808) | 0.225 | 0.843 |
| VEGAS statistics (P) | 23.986 (0.105) | 16.823 (0.364) | 18.732 (0.260) | |||||||
When the P values for the minor alleles of the codominant, dominant and recessive models were subjected to the VEGAS test, no significant gene-based associations were found. However, when the lower P value generated by the dominant and recessive models was input for every SNP, the value of the test statistic was 29.622, which was statistically significant (P = 0.037).
Four haplotype blocks were constructed using SNP Analyzer version 2.0. These blocks were evaluated for an association with the risk of gastric cancer (Table 4). The C-C type of ITGA1 haplotype block 2 was marginally significant in the codominant model (OR = 0.602, 95%CI: 0.212-0.709; P = 0.021; FDR Q = 0.063) and was a significant protective factor against gastric cancer in the dominant model (OR = 0.653, 95%CI: 0483-0.884; P = 0.006; FDR Q = 0.018). In the dominant model, the A-T type of ITGA1 haplotype block 2 was a significant risk factor (OR = 1.341, 95%CI: 1.034-1.741; P = 0.027; FDR Q = 0.045). No haplotype block was found to be significant in the recessive model.
| Haplotypes | Codominant | Dominant | Recessive | |||||||
| OR (95%CI) | P value1 | Q2 | OR (95%CI) | P value1 | Q2 | OR (95%CI) | P value1 | Q2 | ||
| ITGA1 | AAA | 0.771 (0.510-1.165) | 0.414 | 0.973 | 0.860 (0.666-1.112) | 0.250 | 0.750 | 0.819 (0.555-1.210) | 0.316 | 0.913 |
| Haplotype | GAG | 1.039 (0.643-1.678) | 0.973 | 0.973 | 1.030 (0.799-1.328) | 0.819 | 0.819 | 1.026 (0.644-1.636) | 0.913 | 0.913 |
| block 1 | ACA | 0.992 (0.559-1.760) | 0.768 | 0.973 | 1.088 (0.839-1.410) | 0.525 | 0.787 | 0.957 (0.544-1.683) | 0.879 | 0.913 |
| ITGA1 | CT | 0.982 (0.688-1.407) | 0.640 | 0.640 | 1.072 (0.800-1.437) | 0.641 | 0.641 | 0.911 (0.679-1.223) | 0.536 | 0.536 |
| Haplotype | AT | 1.316 (0.686-1.407) | 0.086 | 0.129 | 1.341 (1.034-1.741) | 0.027 | 0.045 | 1.121 (0.784-1.603) | 0.532 | 0.536 |
| block 2 | CC | 0.602 (0.212-0.709) | 0.021 | 0.063 | 0.653 (0.483-0.884) | 0.006 | 0.018 | 0.661 (0.233-1.872) | 0.433 | 0.536 |
| ITGA1 | CCT | 1.023 (0.707-1.480) | 0.677 | 0.794 | 0.934 (0.705-1.236) | 0.631 | 0.916 | 0.819 (0.555-1.210) | 0.316 | 0.913 |
| Haplotype | TGC | 0.973 (0.641-1.475) | 0.314 | 0.794 | 0.986 (0.761-1.278) | 0.916 | 0.916 | 1.026 (0.644-1.636) | 0.913 | 0.913 |
| block 3 | TGT | 1.418 (0.446-4.507) | 0.794 | 0.794 | 1.084 (0.782-1.505) | 0.627 | 0.916 | 0.957 (0.544-1.683) | 0.879 | 0.913 |
| ITGA1 | TA | 0.938 (0.641-1.370) | 0.907 | 0.907 | 0.928 (0.658-1.310) | 0.671 | 0.671 | 0.997 (0.765-1.299) | 0.981 | 0.981 |
| Haplotype | CA | 0.983 (0.536-1.803) | 0.803 | 0.907 | 1.079 (0.832-1.400) | 0.567 | 0.671 | 0.952 (0.523-1.733) | 0.873 | 0.981 |
| block 4 | TG | 1.619 (0.698-3.756) | 0.320 | 0.907 | 0.925 (0.708-1.209) | 0.569 | 0.671 | 1.685 (0.730-3.888) | 0.217 | 0.981 |
The present study focused on the association of genetic polymorphisms and haplotypes of the ITGA1 gene with gastric cancer risk. It has been suggested that the integrin α1 subunit could be involved in gastric cancer carcinogenesis. Integrins on gastric epithelial cells have been reported to serve as a portal for the entry of H. pylori cagA[11]. Additionally, the integrin α1 subunit is involved in the adhesion and dissemination of gastric cancer cells to the peritoneum[18], and an ITGA2 polymorphism has been reported to be associated with an increase in the risk of gastric cancer[20]. However, to our knowledge, no previous study has examined the association between ITGA1 polymorphisms and the risk of gastric cancer.
The SNPs rs1862610, rs2432143 and rs2447867 were significantly associated with an increase in the risk of gastric cancer. After controlling the FDR, only SNP rs2432143 in the codominant model was statistically significant. In a gene-based association test, the ITGA1 gene was found to be significantly associated with gastric cancer.
The C-C type of ITGA1 haplotype block 2, which includes rs1862610 and rs2432143 in intron 1 of the ITGA1 gene, was found to be a significant protective factor and the A-T type to be a risk factor for gastric cancer. This statistical significance was maintained after controlling the FDR. However, the precise molecular mechanism related to these SNPs is not clear. Based on SNP function prediction using computational methods, SNPs rs1862610 and rs2432143 are not predicted to be involved in any structural or functional changes in the integrin α1 subunit. However, we cannot rule out the possibility that these SNPs are either associated with the stability of ITGA1 mRNA, or in linkage disequilibrium with an as yet unknown functional polymorphism affecting the expression or function of the integrin α1 subunit.
We used public databases of SNPs related to gastric cancer and assessed the potential functions of selected SNPs with SNP function prediction software. Among the 15 selected SNPs, only two were located in exons, and one was non-synonymous. The potential function was not predicted for any of these SNPs, except for rs2447867, which was predicted to be an exonic splicing enhancer (ESE). ESEs are clinically significant because synonymous point mutations in ESEs that were previously thought to be silent mutations can lead to exon skipping and the production of a non-functional protein. As loss of integrin α1β1 has been observed in some other malignancies[27], non-functional integrin α1β1 could be associated with gastric cancer.
The increased expression of integrin molecules by epithelial cells during inflammation of the underlying lamina propria is probably an adaptive response to prevent extensive epithelial cell sloughing caused by inflammatory mediators. Loss of epithelial integrity due to a decrease in the function of integrin results in more severe injury of the epithelium[21]. At these sites of tissue injury, bone marrow-derived cells are recruited, and these cells can be a potential source of malignancy[28]. Because chronic infection with H. pylori also induces repopulation of the stomach with bone marrow-derived cells, there is a possibility that a non-functional integrin α1 subunit and H. pylori infection would have a synergistic effect in increasing the risk of gastric cancer. The major limitation of the present study is that we did not test for the presence of antibodies against H. pylori and the cagA antigen in the sera of the case and control subjects.
The OR obtained for SNPs rs1862610, rs2432143, and rs2447867 were all below 1.6, while the OR for the ITGA2 C807T polymorphism in relation to gastric cancer in a Chinese population is 1.57[20]. These relatively small values can be explained by the promiscuity and redundancy of integrins: one integrin can bind several different ligands, and many different integrins can bind to the same ligand[29]. Therefore, if an integrin is not functioning, other integrins can compensate for at least some of its function.
In conclusion, the ITGA1 gene SNPs rs2432143 and rs2447867 and the ITGA1 haplotype block that includes SNP rs2432143 are significantly associated with gastric cancer risk.
Integrins mediate signaling events that are essential for stable cell adhesion, cell spreading, migration, survival, proliferation and differentiation. Several integrins, including α1β1, bind to extracellular matrix proteins present in the basal membranes of mature vessels. Tumor progression and the metastasis of various cancers are associated with integrins. The ITGA1 gene, located on chromosome 5q11.2, encodes the integrin α1 subunit, which is involved in the adhesion of gastric cancer cells to the peritoneum. Adhesion of integrin α1-positive gastric cancer cells to the extracellular matrix is a critical process in peritoneal dissemination. As integrin α1β1 is up-regulated during inflammation in the gastrointestinal tract mucosa, which is the first step in gastric carcinogenesis, it is possible that the integrin α1 subunit plays an important role in the development of gastric cancer. It has been suggested that the integrin α1 subunit could be involved in gastric cancer carcinogenesis. Integrins on gastric epithelial cells have been reported to serve as a portal for the entry of Helicobacter pylori (H. pylori) cagA. As integrin α1β1 is up-regulated during inflammation in the gastrointestinal tract mucosa, which is the first step in the gastric carcinogenesis, it is possible that the integrin α1 subunit plays an important role in the development of gastric cancer.
There are few studies addressing the role of integrins in the development of gastric cancer. An association with an increased risk of gastric cancer has only been reported previously for the ITGA2 C807T polymorphism in a Chinese population. No earlier study has focused on the association of ITGA1 gene single nucleotide polymorphisms (SNPs) and haplotypes with gastric cancer risk.
To the best of the authors’ knowledge, this present study is the first to suggest a significant association of the genetic polymorphisms and haplotypes of ITGA1 gene with an increased gastric cancer risk.
Integrins on gastric epithelial cells have been reported to serve as a portal of entry for H. pylori cagA, and loss of epithelial integrity due to a decrease in the function of integrins results in more severe injury of the epithelium. Studies are needed addressing the interaction of non-functional integrin α1 subunit and H. pylori infection in increasing the risk of gastric cancer.
This paper is focused on the ITGA1 polymorphisms and haplotypes, and gastric cancer risk in a Korean population. The results showed the SNPs rs1862610, rs2432143, and rs2447867, and the ITGA1 haplotype block which includes SNPs rs1862610 and rs2432143 were significantly associated with gastric cancer. It is interesting.
P- Reviewers Du J, Lu JC S- Editor Gou SX L- Editor A E- Editor Ma S
| 1. | The Korea Central Cancer Registry, National Cancer Center. Annual report of cancer statistics in Korea in 2009. Seoul: Ministry of Health and Welfare 2011; . |
| 2. | Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893-2917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11128] [Cited by in RCA: 11837] [Article Influence: 845.5] [Reference Citation Analysis (4)] |
| 3. | Kim HJ, Chang WK, Kim MK, Lee SS, Choi BY. Dietary factors and gastric cancer in Korea: a case-control study. Int J Cancer. 2002;97:531-535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 157] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 4. | Lee JK, Park BJ, Yoo KY, Ahn YO. Dietary factors and stomach cancer: a case-control study in Korea. Int J Epidemiol. 1995;24:33-41. [PubMed] |
| 5. | Kim JH, Kim HY, Kim NY, Kim SW, Kim JG, Kim JJ, Roe IH, Seo JK, Sim JG, Ahn H. Seroepidemiological study of Helicobacter pylori infection in asymptomatic people in South Korea. J Gastroenterol Hepatol. 2001;16:969-975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 112] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 6. | Nan HM, Park JW, Song YJ, Yun HY, Park JS, Hyun T, Youn SJ, Kim YD, Kang JW, Kim H. Kimchi and soybean pastes are risk factors of gastric cancer. World J Gastroenterol. 2005;11:3175-3181. [PubMed] |
| 7. | Zhang YW, Eom SY, Kim YD, Song YJ, Yun HY, Park JS, Youn SJ, Kim BS, Kim H, Hein DW. Effects of dietary factors and the NAT2 acetylator status on gastric cancer in Koreans. Int J Cancer. 2009;125:139-145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 8. | Eslick GD, Lim LL, Byles JE, Xia HH, Talley NJ. Association of Helicobacter pylori infection with gastric carcinoma: a meta-analysis. Am J Gastroenterol. 1999;94:2373-2379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 295] [Cited by in RCA: 288] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 9. | Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci USA. 1993;90:5791-5795. [PubMed] |
| 10. | Arents NL, van Zwet AA, Thijs JC, Kooistra-Smid AM, van Slochteren KR, Degener JE, Kleibeuker JH, van Doorn LJ. The importance of vacA, cagA, and iceA genotypes of Helicobacter pylori infection in peptic ulcer disease and gastroesophageal reflux disease. Am J Gastroenterol. 2001;96:2603-2608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 44] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 11. | Kwok T, Zabler D, Urman S, Rohde M, Hartig R, Wessler S, Misselwitz R, Berger J, Sewald N, König W. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature. 2007;449:862-866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 473] [Cited by in RCA: 509] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 12. | Backert S, Meyer TF. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr Opin Microbiol. 2006;9:207-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 274] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 13. | Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nat Rev Cancer. 2002;2:91-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1292] [Cited by in RCA: 1282] [Article Influence: 55.7] [Reference Citation Analysis (0)] |
| 14. | Rüegg C, Dormond O, Mariotti A. Endothelial cell integrins and COX-2: mediators and therapeutic targets of tumor angiogenesis. Biochim Biophys Acta. 2004;1654:51-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 45] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 15. | Stupack DG, Cheresh DA. Get a ligand, get a life: integrins, signaling and cell survival. J Cell Sci. 2002;115:3729-3738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 448] [Cited by in RCA: 439] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 16. | Ura H, Denno R, Hirata K, Yamaguchi K, Yasoshima T. Separate functions of alpha2beta1 and alpha3beta1 integrins in the metastatic process of human gastric carcinoma. Surg Today. 1998;28:1001-1006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 52] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 17. | Matsuoka T, Yashiro M, Nishimura S, Inoue T, Fujihara T, Sawada T, Kato Y, Seki S, Hirakawa-Ys Chung K. Increased expression of alpha2beta1-integrin in the peritoneal dissemination of human gastric carcinoma. Int J Mol Med. 2000;5:21-25. [PubMed] |
| 18. | Fukuda K, Saikawa Y, Yagi H, Wada N, Takahashi T, Kitagawa Y. Role of integrin α1 subunits in gastric cancer patients with peritoneal dissemination. Mol Med Rep. 2012;5:336-340. [PubMed] |
| 19. | Orr FW, Wang HH, Lafrenie RM, Scherbarth S, Nance DM. Interactions between cancer cells and the endothelium in metastasis. J Pathol. 2000;190:310-329. [PubMed] |
| 20. | Chen J, Liu NN, Li JQ, Yang L, Zeng Y, Zhao XM, Xu LL, Luo X, Wang B, Wang XR. Association between ITGA2 C807T polymorphism and gastric cancer risk. World J Gastroenterol. 2011;17:2860-2866. [PubMed] |
| 21. | MacDonald TT, Horton MA, Choy MY, Richman PI. Increased expression of laminin/collagen receptor (VLA-1) on epithelium of inflamed human intestine. J Clin Pathol. 1990;43:313-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 22. | Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149-150. [PubMed] |
| 23. | Hedrick PW. Gametic disequilibrium measures: proceed with caution. Genetics. 1987;117:331-341. [PubMed] |
| 24. | Lee HJ, Kim SY, Koh JM, Bok J, Kim KJ, Kim KS, Park MH, Shin HD, Park BL, Kim TH. Polymorphisms and haplotypes of integrinalpha1 (ITGA1) are associated with bone mineral density and fracture risk in postmenopausal Koreans. Bone. 2007;41:979-986. [PubMed] |
| 25. | Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289-300. |
| 26. | Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, Hayward NK, Montgomery GW, Visscher PM, Martin NG. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;87:139-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 653] [Cited by in RCA: 655] [Article Influence: 43.7] [Reference Citation Analysis (0)] |
| 27. | Su AI, Welsh JB, Sapinoso LM, Kern SG, Dimitrov P, Lapp H, Schultz PG, Powell SM, Moskaluk CA, Frierson HF. Molecular classification of human carcinomas by use of gene expression signatures. Cancer Res. 2001;61:7388-7393. [PubMed] |
| 28. | Houghton J, Stoicov C, Nomura S, Rogers AB, Carlson J, Li H, Cai X, Fox JG, Goldenring JR, Wang TC. Gastric cancer originating from bone marrow-derived cells. Science. 2004;306:1568-1571. [PubMed] |
| 29. | Hynes RO. Integrins: versatility, modulation, and signaling in cell adhesion. Cell. 1992;69:11-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7430] [Cited by in RCA: 7404] [Article Influence: 224.4] [Reference Citation Analysis (0)] |