Published online Jun 14, 2013. doi: 10.3748/wjg.v19.i22.3385
Revised: December 6, 2011
Accepted: December 15, 2011
Published online: June 14, 2013
The discovery of induced pluripotent stem cells (iPSCs) unraveled a mystery in stem cell research, after identification of four re-programming factors for generating pluripotent stem cells without the need of embryos. This breakthrough in generating iPSCs from somatic cells has overcome the ethical issues and immune rejection involved in the use of human embryonic stem cells. Hence, iPSCs form a great potential source for developing disease models, drug toxicity screening and cell-based therapies. These cells have the potential to differentiate into desired cell types, including hepatocytes, under in vitro as well as under in vivo conditions given the proper microenvironment. iPSC-derived hepatocytes could be useful as an unlimited source, which can be utilized in disease modeling, drug toxicity testing and producing autologous cell therapies that would avoid immune rejection and enable correction of gene defects prior to cell transplantation. In this review, we discuss the induction methods, role of reprogramming factors, and characterization of iPSCs, along with hepatocyte differentiation from iPSCs and potential applications. Further, we discuss the location and detection of liver stem cells and their role in liver regeneration. Although tumor formation and genetic mutations are a cause of concern, iPSCs still form a promising source for clinical applications.
- Citation: Rao MS, Sasikala M, Reddy DN. Thinking outside the liver: Induced pluripotent stem cells for hepatic applications. World J Gastroenterol 2013; 19(22): 3385-3396
- URL: https://www.wjgnet.com/1007-9327/full/v19/i22/3385.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i22.3385
Patients suffering from chronic end-stage liver disease are currently receiving inadequate treatment due to the lack of organ donors for transplantation[1]. Alternatively, cell-based therapies are gaining importance as supportive therapy. Hepatocytes (adult, fetal) and liver stem cells form promising sources for cellular therapies in the treatment of liver diseases. However, inadequate proliferation, ethical issues and scanty numbers limit their applicability[2-5]. Therefore, it is essential to think outside the liver in favor of generating hepatocytes for drug screening, disease modeling and cell therapy applications. Identification of four reprogramming transcription factors revolutionized stem cell research in generating induced pluripotent stem cells (iPSCs). iPSCs generated from somatic cells can be utilized not only for cell-based therapies, but also for disease modeling and drug toxicity screening. Patient-specific iPSCs can be generated by reprogramming and differentiating somatic cells from the patient into the desired cell type. Key advantages of iPSCs over current transplantation approaches are that they form an unlimited potential source and are patient-specific. In addition, the possibility of correcting genetic defects in liver diseases is currently under investigation[6].
The identification of patient-specific pluripotent stem cells has long been an important goal for scientists working in the field of stem cells. In 2006, Takahashi et al[7] first reported that forced expression of four transcription factors [octamer-binding transcription factor (Oct) 3/4, SRY box-containing gene 2 (Sox2), Kruppel-like factor 4 (Klf4) and c-Myc] reprogrammed mouse somatic fibroblasts into embryonic stem cell (ESC)-like colonies, which were termed iPSCs. Later, human induced pluripotent stem cells (hiPSCs) were generated from embryonic, neonatal and adult fibroblasts[8-10]. In addition, derivation of patient-specific iPSCs for various diseases/disorders has also been reported[11-15]. Recently, several groups have investigated the possibilities of disease modeling using patient-derived iPSCs[6,16-23]. Apart from all these applications, hepatocytes derived from iPSCs will have larger implications in drug toxicity studies. Before iPSCs, several approaches were used to reprogram the differentiated cells to a pluripotent state. In the beginning, patient-specific human embryonic stem cells (hESCs) were derived using somatic cell nuclear transfer or therapeutic cloning. This technique requires the introduction of a nucleus from an adult donor cell into an enucleated oocyte to generate a nuclear transfer embryo. The objective of this technique is to produce pluripotent hESCs that carry the nuclear genome of the patient and then induce them to differentiate into cells which may be transplanted back into the patient[24-28]. Another method is the fusion of fibroblasts with ESCs[29,30]. However, the therapeutic application of either approach has been experiencing both ethical and technical difficulties, summarized in Table 1.
| Method | Results of reprogramming | Drawbacks | Ref. |
| Transfer of the nucleus from a somatic cell to an enucleated oocyte | The somatic cell nucleus is reprogrammed in the oocyte, and a whole organism develops as a result. Patient-specific hESCs can be derived | Low efficiency. Developmental abnormalities in cloned animals. Ethical and legal restrictions | [24-28] |
| Fusion of ESCs with differentiated cells | Hybrids of differentiated cells and ESCs display all properties of pluripotent cells | Cell hybrids lack a normal diploid chromosome set | [29,30] |
| Reprogramming of somatic cells to a pluripotent state can be generated by the ectopic expression of 4 transcription factors, Oct4, Klf4, Sox2 and c-Myc | Somatic cells regain a pluripotent state and become similar in properties to ESCs | Low efficiency of iPSC derivation. Viral integration. Tumor formation | [7] |
It was demonstrated that somatic cells can be re-programmed into pluripotent stem cells by ectopic expression of four transcription factors, namely Oct4, Klf4, Sox2, and c-Myc, using four independent retroviral vectors[7]. This achievement revolutionized stem cell research. Initially, iPSCs were derived from somatic cells by the retroviral or lentiviral transduction of transcription factors in which transgenes are randomly inserted into the genome of the hosts. Such integration of transgenes has the risk of tumorigenicity[31]. Later, trials to omit transgenic insertion of c-Myc resulted in low reprogramming efficiency and did not eliminate the risk of tumor formation[32], as overexpression of Oct3/4 and Klf4 can also cause tumor formation[33]. In Table 2, we have summarized the advantages and disadvantages of various strategies used for inducing iPSCs generation[7,8,9,32,34-49]. Furthermore, combining all four factors (Oct4, Klf4, Sox2, and c-Myc) into a single vector allowed derivation of iPSCs with a single lentiviral stem cell cassette containing a loxP sequence in the long terminal repeat (LTR)[43]. Following this, transgenes were removed using Cre-mediated excision. Although it left an incomplete LTR in the iPS genome, this method minimized the genomic alteration[44]. A transposon system encoding a reprogramming cassette has also been used for iPSC induction. The transduction of a plasmid-based transposon vector can integrate into the host genome with the help of transposase, and induces iPSC colony formation. The re-expression of the transposase after the establishment of iPSCs recognizes the terminal repeat of the integrated transposon vector, and excises it from the genome. The excision of the transposon does not leave a footprint in most cases, so it maintains the original endogenous sequences[45,46,50,51]. Several techniques have been used for obtaining transgene-free iPSCs.
| Methods | Advantages | Disadvantages | Ref. |
| Retroviral vectors | High efficiency | Genome integration, dividing target cells needed | [7-9,32,41,42] |
| Lentiviral vectors | High efficiency, target cells need not be dividing | Genome integration | [47-49] |
| Lentiviral vectors with Cre/Lox | High efficiency | Minimize genomic integration | [43,44] |
| Piggyback transposon | Precise deletion is possible | Minimize genomic integration, laborious | [45,46] |
| Viral vectors | No genome integration | Low efficiency | [34-37] |
| Adenoviral vectors | |||
| Sendai vectors | |||
| DNA vectors | |||
| Plasmid vectors | |||
| Episomal vectors | |||
| Minicircle vectors | |||
| Protein transduction | No genome integration | Low efficiency | [38] |
| Small molecules | No genetic modification | Low efficiency | [39] |
| Synthetic mRNA | No genetic modification, high efficiency | Multiple rounds of transfection are needed | [40] |
The first integration-free iPSCs were generated from adult mouse hepatocytes using non integrating adenoviral vectors. However, this required repeated transduction to maintain transgene expression[34,38]. Another technique used is transduction with the Sendai virus, an RNA virus, to deliver the reprogramming factors[35]. The Sendai virus does not integrate into the genome, but working with this system requires more than 15 passages to eliminate viral transgene expression. This complexity limits the general use of this method[48]. Transient transfection of plasmids, episome-based DNA vectors and minicircle vectors has been used to generate transgene-free iPSCs. Mouse embryonic fibroblasts were reprogrammed by repeated transfection with two plasmid constructs carrying the reprogramming factors; the first plasmid expressed c-Myc, while the second expressed the other three factors Oct4, Klf4 and Sox2[36]. Furthermore, experiments with non-integrating episomal vectors have also been successful in iPSC generation[16]. Similarly, minicircle vectors lack the bacterial origin of replication and antibiotic resistance gene and offer higher transfection efficiencies and more prolonged transgene expression as compared to regular plasmids[52]. Moreover, iPSCs have been established by the direct delivery of recombinant reprogramming proteins[38] and small molecules[39]. More recently, one research group has utilized synthetic mRNA molecules to reprogram human fibroblasts to pluripotency and stimulate them into myogenic cells[32]. However, reprogramming using modified RNAs is technically difficult, sensitive to reagents and requires labor-intensive procedures. The efficiency of iPSC induction using transgene-free methods is lower than that with retrovirus vectors, possibly due to low transduction efficiency and unstable expression. Therefore, it is essential to develop methods that require less time and have higher efficiency of reprogramming involving viral and transgene-free techniques to generate iPSCs.
Takahashi et al[7] used a combination of four nuclear reprogramming factors, such as Oct4, Sox2, c-Myc and Klf4, for generating iPSCs from mice and reported an efficiency of 0.02%. Simultaneously, the Thomson group used a slightly different combination of factors, namely Oct4, Sox2, Nanog and Lin28, to reprogram human somatic cells at a similar efficiency (0.02%)[9]. Subsequently, researchers have started to identify new reprogramming factors and usage of minimum factors for generating safe iPSCs. iPSCs have been established by 3 transcriptional factors without c-Myc (Oct3/4, Klf4, SOX2) at an efficiency of 0.002%[53,54]. It was also shown that using only Oct4 and Klf4 was enough to reprogram murine NSCs at an efficiency of 0.11%[55]. More recently, the forced expression of Oct4 alone was shown sufficient to reprogram murine NSCs, at a low efficiency of 0.014%[56]. However, the efficiency of iPSC generation has been significantly reduced with usage of minimum factors for generating safe iPSCs. The Oct4, Sox2, and Nanog genes code for transcription factors that activate the genes and signaling pathways responsible for the establishment and maintenance of the pluripotent state and repress the genes responsible for differentiation[57,58]. Others have reported that the expression of Oct4 and Sox2 genes is absolutely essential for iPSC generation. In addition, the products of the Nanog, c-Myc, Klf4 and Lin28 genes seem to act as catalysts which accelerate the reprogramming[59]. In Table 3, we have summarized the role of various reprogramming factors for iPSC generation[60-66].
| Reprogramming factors | Description | Function | Ref. |
| Oct4 | Octamer binding transcription factor 4 | This transcription factor plays a role in embryonic development, especially during early embryogenesis, and it is necessary for embryonic stem cell pluripotency | [7] |
| Sox2 | SRY box 2 | In embryonic stem cells, Sox2 and Oct3/4 often co-occupy target genes, including own promoters. These proteins cooperate regulatory feedback loops to maintain pluripotency | [60] |
| Klf4 | Kruppel-like factor 4 | This transcription factor plays a role in upregulation of pluripotency gene Nanog and the modification of chromatin structure to facilitate the binding of Oct3/4 and Sox2 to their sequences. Klf4 itself is an oncogenic factor. This gene is over expressed in a variety of tumor types associated with advanced cancer | [61-63] |
| c-Myc | Proto oncogene protein | An oncogene that induces global histone acetylation, allowing Oct3/4 and Sox2 to bind to their specific target loci | [60,63] |
| Nanog | Homeo box transcription factor | A transcription factor critically involved with self-renewal of undifferentiated embryonic stem cells | [64] |
| Lin28 | RNA binding protein Lin28 | The Lin28 gene codes for an RNA-binding protein that selectively blocks the processing of microRNAs of the let-7 family, and possibly certain other microRNAs in ESCs, to prevent their differentiation | [65,66] |
Recently, molecules have been used in combination with reprogramming factors to improve the efficiency of iPSC generation, including cotransduction of the catalytic subunit of human telomerase, human telomerase reverse transcriptase, along with SV40 large T antigen, or the repression of the Ink4a/Arf locus (encoding cell cycle-dependent kinase inhibitors), or repression of the p53/p21 pathway. These efforts have led to dramatic increases in the efficiency of reprogramming[10,67-69].
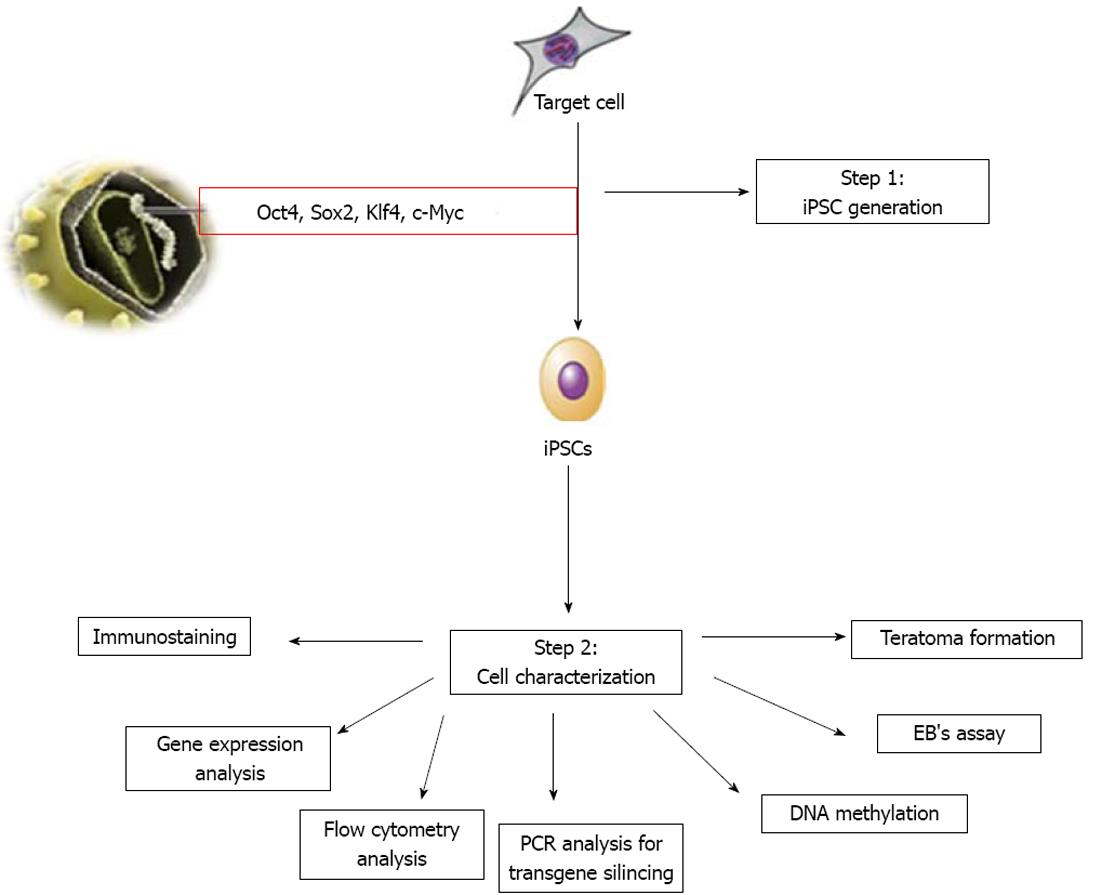
The hiPSCs generated can be characterized for their pluripotency, as shown in Figure 1. In addition, assessment of their epigenetic status, silencing of transgene expression and DNA fingerprinting need to be established for confirmation. Assessment of pluripotency of iPSCs can be performed by checking the expression of protein and genes of Oct4, Sox2, Nanog, as well as for SSEA-1 (mouse) or SSEA-3/-4 and TRA-1-60/-81 (human) using flow cytometry, immunocytochemistry and reverse transcription-polymerase chain reaction (PCR) methods[70]. The pluripotent nature of iPSCs is routinely tested by two methods. The first is to determine the in vitro differentiation ability of iPSCs, where iPSCs can be allowed to differentiate spontaneously in vitro to form embryoid bodies. These embryoid bodies can be assessed for three embryonic germ layers, i.e., mesoderm, endoderm and ectoderm. The second is to determine the in vivo differentiation ability of iPSCs[71], where iPSCs can be injected into adult immune-deficient mice (SCID mice). In the host animal, injected iPSCs can form tumors called teratomas. In addition to pluripotency assessment, it is important to confirm the silencing of exogenous transgene expression. PCR analysis can be used to demonstrate silencing of retro/lentiviral transgene expression using virus-specific primers[70]. Further, DNA fingerprinting can be performed to confirm iPSCs are genetically matched to their parental somatic cells. DNA methylation analysis of the Oct4, Sox2 and Nanog promoter regions using bisulfite sequencing can be used to reveal the different epigenetic states of the cells. Thus, the methylation status of promoter regions of pluripotency genes confirms successful reprogramming[70].
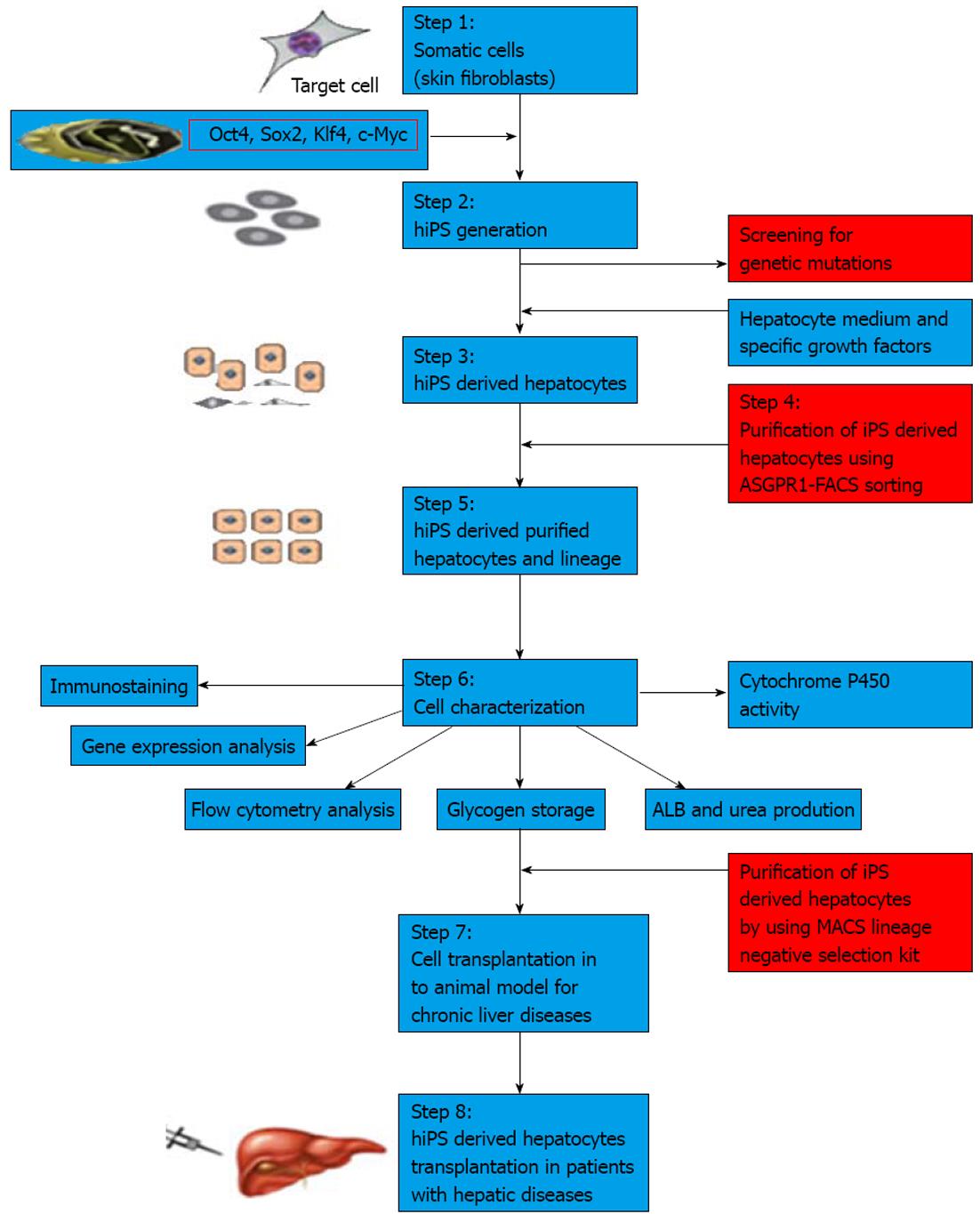
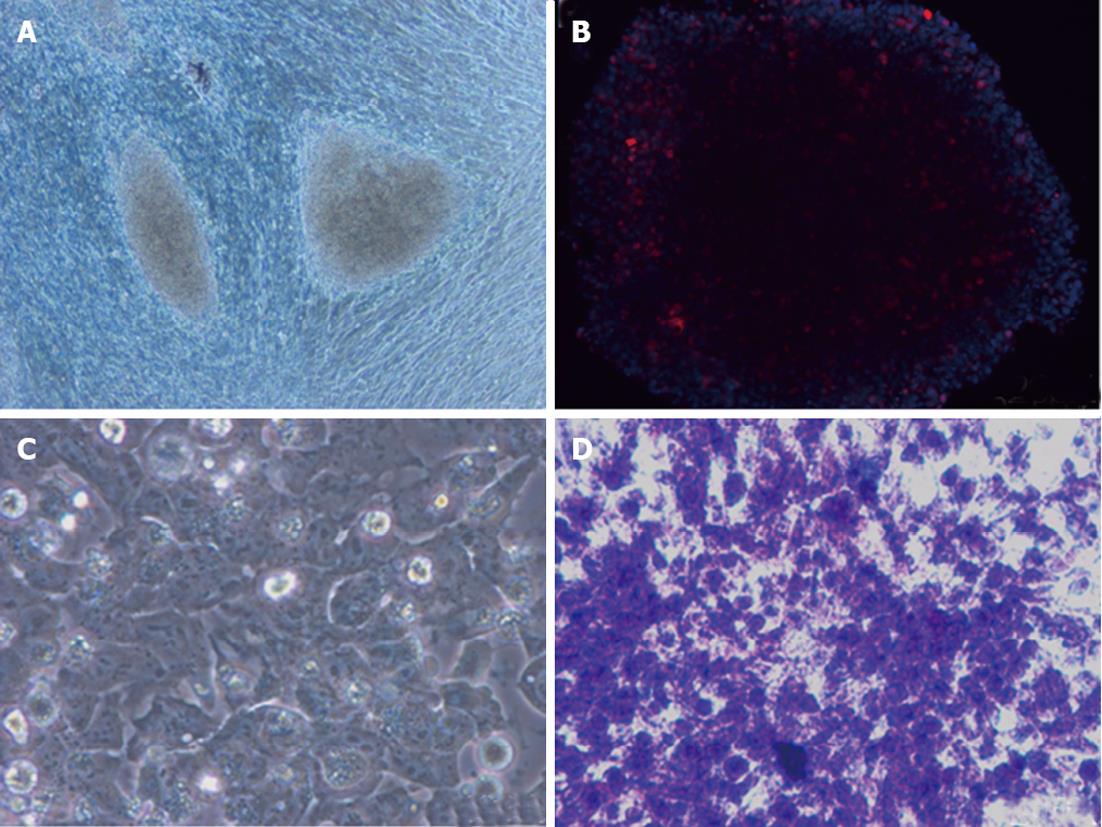
To date, many protocols have been used to differentiate iPSCs into desired cell types. However, different iPSC lines have different outcomes under identical culture conditions. iPSC lines have a propensity to produce certain lineages or cell types when allowed to differentiate spontaneously, indicating that choosing a proper clone is also essential in differentiating iPSCs into a specific lineage[72-74]. A major issue in differentiation is to obtain hepatocytes from pluripotent stem cells that have an adult phenotype, and which stably express liver-like functions and reflect those in vivo functions[75]. Recently, a number of protocols have been developed to derive hepatocytes from hiPSCs. These protocols for hepatocyte generation are hampered by inefficient differentiation and maturation that lead to low yield and heterogeneous cell populations in cultures[76]. Recently, a homogenous population of hepatocytes from pluripotent stem cells has been isolated by sorting for surface asialoglycoprotein receptor marker; however, these enriched cells are found to retain immature fetal liver characteristics[77]. In Table 4, we have summarized various protocols used to differentiate hepatocytes from iPSCs[78-84]. Even after enriching the hepatocytes from culture prior to transplantation, the risk of teratoma formation may arise due to the presence of a few undifferentiated iPSCs. Therefore, further enriching hepatocytes using negative selection against pluripotent cells could be useful to avoid teratoma formation. Figure 2 summarizes the strategy on differentiation of human iPSCs into hepatocytes. Figure 3 depicts the hepatocytes generated from hiPSCs in our laboratory.
| Ref. | Species | Differentiation protocol | Remarks |
| Sullivan et al[78] | Human | Activin A, Wnt3a (3 d), Activin A (2 d), DMSO (3 d), HGF, OSM (6 d) | Generated functional hepatocyte-like cells from human-iPSCs |
| Song et al[79] | Human | Activin A (3 d), FGF4, BMP-2 (4 d), HGF, KGF (6 d), OSM, Dex (5 d) then OSM, Dex, N2B27 (3 d) | iPSCs had fewer expressed liver-enriched genes compared with human hepatocytes |
| Si-Tayeb et al[80] | Human | Activin A (5 d), bFGF, BMP-4 (5 d), HGF (5 d), OSM (5 d) | Transplanted hepatocyte-like cells into the lobe of newborn mice and demonstrated homing of donor cells |
| Liu et al[81] | Human | Activin A (5 d), FGF4, HGF (5 d ), Single Quotes (lonza), FGF4, HGF, OSM, Dex (10 d) | Human hepatocyte-derived iPSCs are able to differentiate into functional hepatocytes |
| Takata et al[82] | Human | Activin A ( 3 d), HGF (5 d), OSM (5 d) | Generated hepatocyte-like cells from iPSCs using three growth factors in a short time |
| Gai et al[83] | Mouse | Activin A, Wnt3 (6 d), bFGF, DMSO (3 d), HGF, DMSO (9 d), HGF, OSM, DMSO (7 d) | Generated hepatocytes from iPSCs |
| Iwamuro et al[84] | Mouse | Activin A, bFGF (3 d), HGF (5 d) | Generated hepatocyte-like cells from iPSCs |
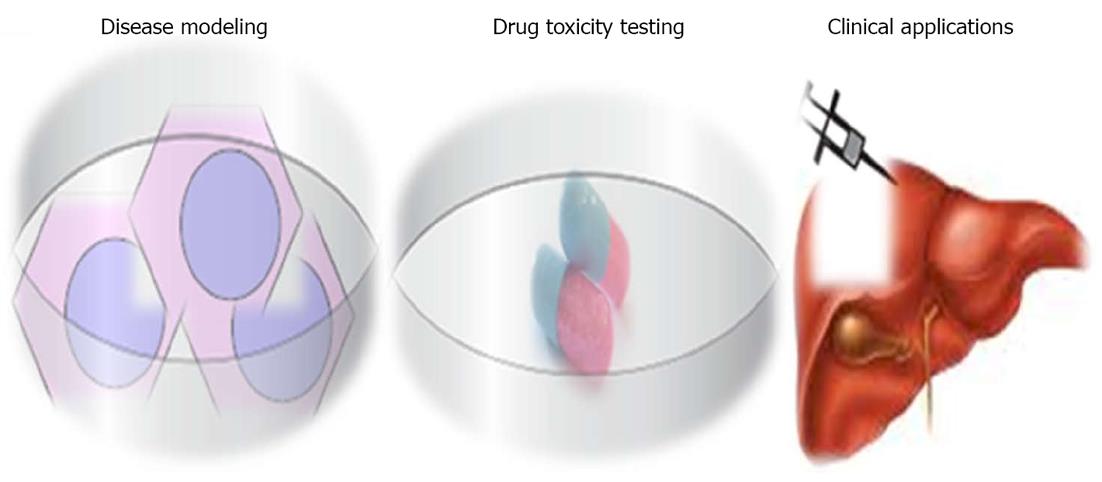
iPSCs represent a promising source of hepatocytes for a wide range of applications, including disease modeling, drug toxicity testing and cell transplantation (Figure 4).
iPSCs represent a novel tool for in vitro disease modeling. Traditionally, researchers rely on animal models, hepatic immortalized cell lines, or short-lived primary hepatocyte cultures to understand the mechanisms and pathogenesis of diseases and testing of drug candidates[85-87]. Each of these has limitations in functionality, reproducibility and availability. Disease-specific iPSCs derived from patients suffering from specific diseases may provide a more relevant model system because their properties closely resemble those found in the patient’s own system, without the need for genetic manipulation. Several groups have successfully derived a wide range of iPSCs from patients with diseases[88] and inherited liver diseases[21]. These cells can be used as models to study the pathogenesis, disease mechanism(s) and possible cure for liver disorders. Therefore, human iPSC-derived hepatocytes could generate more accurate predictions of human physiological responses than animal models. iPSC-derived hepatocytes will overcome these limitations and provide a reliable source of highly reproducible and readily available human hepatocytes for disease modeling in pre-clinical drug development.
Hepatotoxicity is the most common side effect of new candidate drugs under clinical trial, and is the leading cause of post approval drug recalls; for example, bromfenac and troglitazone[89]. The development of liver toxicity screening technologies utilizing iPSC-derived hepatocytes would allow investigation into the effects of single nucleotide polymorphisms on drug metabolism and toxicity[90]. An example of this is warfarin, a drug for which polymorphisms in cytochrome P-450 2C9 create problems with obtaining an appropriate pharmacotherapeutic range[91]. iPSC-derived hepatocytes could remain viable in culture for several months, enabling the assessment of acute and chronic toxicity of drugs due to their pluripotent ability. Drug toxicity assays will be performed in petri dishes which require small amounts of compound for a hepatic cytotoxicity profile. Terminally differentiated hepatocytes with cytochrome P-3A4 functional activity and scale-up of iPSC-derived hepatocytes will help in pharmaceutical industry drug toxicity applications.
Liver transplantation represents the only way to treat patients suffering from chronic liver failure, but this is associated with numerous problems, including shortage of donors, high cost, rejection and complications. Transplantation of hepatocytes derived from hiPSCs could represent an alternative cell source for liver failure and inborn liver diseases. The important issue is the generation of safe and functional cell types for therapy. Indeed, the cell sources of iPSCs influence the safety of the established iPSCs. It has been demonstrated that hiPSCs retain certain gene expressions of the parent cells, and this suggests that iPSCs of different origins may possess different capacities to differentiate. A complete study using various mouse iPSCs has demonstrated that the origin of the iPSCs has a profound influence on the tumor-forming propensities in a cell transplantation therapy model[92]. Mouse tail-tip fibroblast iPSCs (mesoderm origin) have shown the highest tumorigenic propensity, whereas gastric epithelial cells and hepatocyte iPSCs (both are endoderm origin) have shown lower propensities[93]. The recent evidence suggests that epigenetic memory of the somatic cell of origin is retained in the iPSCs, and that may influence their directed differentiation potential into blood cells[94,81] or hepatocytes[92]. In the mouse, iPSCs have been generated from derivatives of all three embryonic germ layers, including mesodermal fibroblasts, epithelial cells of endodermal origin and ectodermal keratinocytes, whereas human iPSCs have been produced from mesoderm (fibroblasts and blood cells) or ectoderm (keratinocytes and neural stem cells) and endoderm (hepatocytes)[81]. It is therefore extremely important to establish human iPSC lines of multiple origins and thoroughly examine the source impact on both the safety issues and their differentiation potentials.
Recently, it has been demonstrated that iPSC-derived hepatocytes can restore liver function in an animal model of liver failure[95]. These results indicate the utility of hiPSC-derived hepatocytes as an alternative treatment for patients with end-stage liver disease. Researchers investigated and analyzed the potential of hiPSC-derived hepatocytes to model inborn liver diseases such as α1-antitrypsin deficiency, familial hypercholesterolemia, glycogen storage disease type 1a, hereditary tyrosinemia, and Crigler-Najjar syndrome[6]. Genetic diseases of the liver modeled in hiPSC-derived human hepatocytes create new opportunities to develop autologous cell transplantation therapy to correct genetic defects in liver diseases.
The liver has a massive regenerative capacity. When liver regeneration is impaired, oval shaped cells emerge and are implicated in liver tissue repair[96]. These cells are derived from the canals of Hering, which are located in the periportal region of the liver and account for 0.3%-0.7% of the liver mass[97]. In rodents, these liver progenitor cells are called oval cells, while in humans they are known as hepatic progenitor cells[98]. These cells are phenotypically similar to fetal hepatoblasts and also have a bipotent differentiation potential. Oval cells or hepatic progenitors are difficult to isolate because of the lack of definitive markers. Various markers have been used to identify oval cells in adult liver, including liver stem cell and hematopoietic markers, such as OV6, Thy-1, CD34, c-kit, and Sca-1[99]. Hepatic progenitors have been isolated from fetal liver using the specific surface marker, epithelial cell adhesion molecule (EPCAM). These EPCAM+ cells showed positive for hepatic progenitor markers such as CD29, CD49f and CD90[86]. Clinical studies have identified and confirmed the efficacy of fetal liver hepatic progenitors in end-stage liver diseases[100]. However, the clinical application of this cell source is limited due to the difficulty in obtaining large numbers of fetal liver cells, as well as ethical and immune rejection issues. Another stem cell population found in the fetal liver is side population cells which represents another potential source of liver progenitor cells[101], but these cell numbers are very much fewer in fetal liver. There is increasing evidence in the literature suggesting that bone marrow is another source of hepatic progenitor cells[102,103]. Autologous bone marrow-derived stem cell transplantations have been performed in patients with liver diseases but it is difficult to assess overall clinical benefit from these therapies[104].
In support of a role in liver regeneration, oval cell activation has been detected in chronic liver injury caused by inflammation, chronic hepatic necrosis, chronic alcoholism induced cirrhosis and hepatitis models[105,106]. Although the full complements of signals required for oval cell activation are still unknown, both continuous metabolic stress and chemical hepatotoxic substances have been implicated as potential oval cell activators when hepatocyte proliferation is inhibited[105,106]. A recent study reported the production of a chemokine known as stromal derived factor-1α in the liver following tissue damage[107]. The role of liver stem cells in physiology, pathophysiology and therapy is not yet exactly known; therefore, it needs to be further investigated[108]. Although a number of successful techniques have been developed, stem cell-derived hepatocytes from adult, fetal and embryonic sources are found to retain immature fetal liver characteristics, which are not similar in primary hepatocyte functionality. Therefore, the elucidation of other key developmental factors and tissue culture environments, together with iPSC technology, are essential in order to obtain functional hepatocytes for hepatic applications.
Apart from the methods discussed above, overexpression of lineage-specific transcription factors in somatic cells is a new approach (direct conversion) to generate specific cell types including neurons, cardiomyocytes, blood progenitors and hepatocyte-like cells; as summarized in Table 5[109-112]. This method could be useful as an alternative approach for autologous cell-replacement therapies. Unlike iPSCs and ESCs, directly converted cells may not easily multiply in the lab since they do not have pluripotency properties. Therefore, this approach may have limitations. Choosing highly proliferative starting somatic cells is essential in the direct conversion approach.
| Ref. | Key factors | Direct converted cell type |
| Vierbuchen et al[109] | Brn2, Ascl1, and Myt1l | Transdifferentiated mouse fibroblasts into functional neuronal cells |
| Ieda et al[110] | Gata4, Mef2c, and Tbx5 | Transdifferentiated mouse dermal fibroblasts into cardiomyocyte-like cells |
| Szabo et al[111] | Oct4 | Transdifferentiated human fibroblast cells into hematopoietic progenitors |
| Huang et al[112] | Gata4, Hnf1α and Foxa3, and inactivation of p19Arf | Transdifferentiated mouse tail-tip fibroblasts into hepatocyte-like cell |
Thinking outside the liver explores the potential of iPSCs as an unlimited source for in vitro disease modeling and for drug toxicity studies and clinical applications. Patient-specific iPSCs or custom-made iPSCs may have future promising implications without immuno rejection. However, iPSC technology has several technical issues to be addressed such as generation of iPSCs without viral integration, elimination of tumor formation and genetic mutations that need to be eliminated before the cells are put to clinical applications. Despite limitations, iPSC-derived hepatocytes are a very promising population for cell therapies in hepatology.
P- Reviewers Gassler N, Tam PK S- Editor Cheng JX L- Editor Logan S E- Editor Xiong L
| 1. | Jozefczuk J, Prigione A, Chavez L, Adjaye J. Comparative analysis of human embryonic stem cell and induced pluripotent stem cell-derived hepatocyte-like cells reveals current drawbacks and possible strategies for improved differentiation. Stem Cells Dev. 2011;20:1259-1275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 58] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 2. | Herrera MB, Bruno S, Buttiglieri S, Tetta C, Gatti S, Deregibus MC, Bussolati B, Camussi G. Isolation and characterization of a stem cell population from adult human liver. Stem Cells. 2006;24:2840-2850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 309] [Cited by in RCA: 316] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 3. | Lázaro CA, Rhim JA, Yamada Y, Fausto N. Generation of hepatocytes from oval cell precursors in culture. Cancer Res. 1998;58:5514-5522. [PubMed] |
| 4. | Sahin MB, Schwartz RE, Buckley SM, Heremans Y, Chase L, Hu WS, Verfaillie CM. Isolation and characterization of a novel population of progenitor cells from unmanipulated rat liver. Liver Transpl. 2008;14:333-345. [PubMed] |
| 5. | Czyz J, Wiese C, Rolletschek A, Blyszczuk P, Cross M, Wobus AM. Potential of embryonic and adult stem cells in vitro. Biol Chem. 2003;384:1391-1409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 76] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 6. | Rashid ST, Corbineau S, Hannan N, Marciniak SJ, Miranda E, Alexander G, Huang-Doran I, Griffin J, Ahrlund-Richter L, Skepper J. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest. 2010;120:3127-3136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 498] [Cited by in RCA: 443] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 7. | Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17989] [Cited by in RCA: 18202] [Article Influence: 958.0] [Reference Citation Analysis (0)] |
| 8. | Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14327] [Cited by in RCA: 14322] [Article Influence: 842.5] [Reference Citation Analysis (0)] |
| 9. | Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917-1920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7589] [Cited by in RCA: 7249] [Article Influence: 402.7] [Reference Citation Analysis (0)] |
| 10. | Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2182] [Cited by in RCA: 2126] [Article Influence: 118.1] [Reference Citation Analysis (0)] |
| 11. | Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877-886. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1775] [Cited by in RCA: 1608] [Article Influence: 94.6] [Reference Citation Analysis (0)] |
| 12. | Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218-1221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1445] [Cited by in RCA: 1404] [Article Influence: 82.6] [Reference Citation Analysis (0)] |
| 13. | Ye Z, Zhan H, Mali P, Dowey S, Williams DM, Jang YY, Dang CV, Spivak JL, Moliterno AR, Cheng L. Human-induced pluripotent stem cells from blood cells of healthy donors and patients with acquired blood disorders. Blood. 2009;114:5473-5480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 285] [Cited by in RCA: 273] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 14. | Raya A, Rodríguez-Pizà I, Guenechea G, Vassena R, Navarro S, Barrero MJ, Consiglio A, Castellà M, Río P, Sleep E. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460:53-59. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 566] [Cited by in RCA: 512] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 15. | Ku S, Soragni E, Campau E, Thomas EA, Altun G, Laurent LC, Loring JF, Napierala M, Gottesfeld JM. Friedreich’s ataxia induced pluripotent stem cells model intergenerational GAA⋅TTC triplet repeat instability. Cell Stem Cell. 2010;7:631-637. [PubMed] |
| 16. | Lee G, Papapetrou EP, Kim H, Chambers SM, Tomishima MJ, Fasano CA, Ganat YM, Menon J, Shimizu F, Viale A. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402-406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 657] [Cited by in RCA: 651] [Article Influence: 40.7] [Reference Citation Analysis (0)] |
| 17. | Ebert AD, Yu J, Rose FF, Mattis VB, Lorson CL, Thomson JA, Svendsen CN. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277-280. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1169] [Cited by in RCA: 1039] [Article Influence: 64.9] [Reference Citation Analysis (0)] |
| 18. | Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964-977. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1202] [Cited by in RCA: 1071] [Article Influence: 66.9] [Reference Citation Analysis (0)] |
| 19. | Ye L, Chang JC, Lin C, Sun X, Yu J, Kan YW. Induced pluripotent stem cells offer new approach to therapy in thalassemia and sickle cell anemia and option in prenatal diagnosis in genetic diseases. Proc Natl Acad Sci USA. 2009;106:9826-9830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 149] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 20. | Carvajal-Vergara X, Sevilla A, D’Souza SL, Ang YS, Schaniel C, Lee DF, Yang L, Kaplan AD, Adler ED, Rozov R. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature. 2010;465:808-812. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 581] [Cited by in RCA: 514] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 21. | Ghodsizadeh A, Taei A, Totonchi M, Seifinejad A, Gourabi H, Pournasr B, Aghdami N, Malekzadeh R, Almadani N, Salekdeh GH. Generation of liver disease-specific induced pluripotent stem cells along with efficient differentiation to functional hepatocyte-like cells. Stem Cell Rev. 2010;6:622-632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 22. | Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH, Muotri AR. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527-539. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1082] [Cited by in RCA: 999] [Article Influence: 66.6] [Reference Citation Analysis (0)] |
| 23. | Zhang N, An MC, Montoro D, Ellerby LM. Characterization of Human Huntington’s Disease Cell Model from Induced Pluripotent Stem Cells. PLoS Curr. 2010;2:RRN1193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 193] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 24. | Stojkovic M, Lako M, Strachan T, Murdoch A. Derivation, growth and applications of human embryonic stem cells. Reproduction. 2004;128:259-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 85] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 25. | Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145-1147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11399] [Cited by in RCA: 10432] [Article Influence: 386.4] [Reference Citation Analysis (0)] |
| 26. | Drukker M, Benvenisty N. The immunogenicity of human embryonic stem-derived cells. Trends Biotechnol. 2004;22:136-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 145] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 27. | Rhind SM, Taylor JE, De Sousa PA, King TJ, McGarry M, Wilmut I. Human cloning: can it be made safe? Nat Rev Genet. 2003;4:855-864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 36] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 28. | Jaenisch R. Human cloning - the science and ethics of nuclear transplantation. N Engl J Med. 2004;351:2787-2791. [PubMed] |
| 29. | Cowan CA, Atienza J, Melton DA, Eggan K. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science. 2005;309:1369-1373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 689] [Cited by in RCA: 594] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 30. | Tada M, Takahama Y, Abe K, Nakatsuji N, Tada T. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr Biol. 2001;11:1553-1558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 662] [Cited by in RCA: 589] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 31. | Yoshida Y, Yamanaka S. iPS cells: a source of cardiac regeneration. J Mol Cell Cardiol. 2011;50:327-332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 113] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 32. | Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3332] [Cited by in RCA: 3082] [Article Influence: 171.2] [Reference Citation Analysis (0)] |
| 33. | Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415-419. [PubMed] |
| 34. | Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945-949. [PubMed] |
| 35. | Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:348-362. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1024] [Cited by in RCA: 966] [Article Influence: 60.4] [Reference Citation Analysis (0)] |
| 36. | Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949-953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1459] [Cited by in RCA: 1328] [Article Influence: 78.1] [Reference Citation Analysis (0)] |
| 37. | Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797-801. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1896] [Cited by in RCA: 1692] [Article Influence: 105.8] [Reference Citation Analysis (0)] |
| 38. | Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, Trauger S, Bien G, Yao S, Zhu Y. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1309] [Cited by in RCA: 1229] [Article Influence: 76.8] [Reference Citation Analysis (0)] |
| 39. | Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795-797. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1320] [Cited by in RCA: 1199] [Article Influence: 70.5] [Reference Citation Analysis (0)] |
| 40. | Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618-630. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2155] [Cited by in RCA: 1918] [Article Influence: 127.9] [Reference Citation Analysis (0)] |
| 41. | Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1254] [Cited by in RCA: 1239] [Article Influence: 72.9] [Reference Citation Analysis (0)] |
| 42. | Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2048] [Cited by in RCA: 1902] [Article Influence: 105.7] [Reference Citation Analysis (0)] |
| 43. | Sommer CA, Stadtfeld M, Murphy GJ, Hochedlinger K, Kotton DN, Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543-549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 500] [Cited by in RCA: 493] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 44. | Sommer CA, Sommer AG, Longmire TA, Christodoulou C, Thomas DD, Gostissa M, Alt FW, Murphy GJ, Kotton DN, Mostoslavsky G. Excision of reprogramming transgenes improves the differentiation potential of iPS cells generated with a single excisable vector. Stem Cells. 2010;28:64-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 187] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 45. | Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771-775. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 976] [Cited by in RCA: 855] [Article Influence: 53.4] [Reference Citation Analysis (0)] |
| 46. | Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hämäläinen R, Cowling R, Wang W, Liu P, Gertsenstein M. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766-770. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1423] [Cited by in RCA: 1263] [Article Influence: 78.9] [Reference Citation Analysis (0)] |
| 47. | Brambrink T, Foreman R, Welstead GG, Lengner CJ, Wernig M, Suh H, Jaenisch R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151-159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 672] [Cited by in RCA: 608] [Article Influence: 35.8] [Reference Citation Analysis (0)] |
| 48. | Maherali N, Ahfeldt T, Rigamonti A, Utikal J, Cowan C, Hochedlinger K. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008;3:340-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 417] [Cited by in RCA: 414] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 49. | Wernig M, Lengner CJ, Hanna J, Lodato MA, Steine E, Foreman R, Staerk J, Markoulaki S, Jaenisch R. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat Biotechnol. 2008;26:916-924. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 358] [Cited by in RCA: 327] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 50. | Okita K, Yamanaka S. Induction of pluripotency by defined factors. Exp Cell Res. 2010;316:2565-2570. [PubMed] |
| 51. | Hussein SM, Nagy K, Nagy A. Human induced pluripotent stem cells: the past, present, and future. Clin Pharmacol Ther. 2011;89:741-745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 52. | Jia F, Wilson KD, Sun N, Gupta DM, Huang M, Li Z, Panetta NJ, Chen ZY, Robbins RC, Kay MA. A nonviral minicircle vector for deriving human iPS cells. Nat Methods. 2010;7:197-199. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 575] [Cited by in RCA: 508] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 53. | Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2072] [Cited by in RCA: 1974] [Article Influence: 109.7] [Reference Citation Analysis (0)] |
| 54. | Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:10-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 481] [Cited by in RCA: 458] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 55. | Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, Araúzo-Bravo MJ, Ruau D, Han DW, Zenke M. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646-650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 723] [Cited by in RCA: 697] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 56. | Kim JB, Sebastiano V, Wu G, Araúzo-Bravo MJ, Sasse P, Gentile L, Ko K, Ruau D, Ehrich M, van den Boom D. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136:411-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 712] [Cited by in RCA: 690] [Article Influence: 43.1] [Reference Citation Analysis (0)] |
| 57. | Medvedev SP, Shevchenko AI, Mazurok NA, Zakiian SM. [OCT4 and NANOG are the key genes in the system of pluripotency maintenance in mammalian cells]. Genetika. 2008;44:1589-1608. [PubMed] [DOI] [Full Text] |
| 58. | Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625-635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 856] [Cited by in RCA: 868] [Article Influence: 48.2] [Reference Citation Analysis (0)] |
| 59. | Shevchenko AI, Medvedev SP, Mazurok NA, Zakiian SM. Induced pluripotent stem cells. Russ J Genet. 2009;45:139-146. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 60. | Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1695] [Cited by in RCA: 1696] [Article Influence: 77.1] [Reference Citation Analysis (0)] |
| 61. | Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol. 2005;7:1074-1082. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 408] [Cited by in RCA: 449] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 62. | Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E, Xu Y. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol. 2005;7:165-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 630] [Cited by in RCA: 684] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 63. | Yamanaka S. Strategies and new developments in the generation of patient-specific pluripotent stem cells. Cell Stem Cell. 2007;1:39-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 548] [Cited by in RCA: 523] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 64. | Field M, Alvarez A, Bushnev S, Sugaya K. Embryonic stem cell markers distinguishing cancer stem cells from normal human neuronal stem cell populations in malignant glioma patients. Clin Neurosurg. 2010;57:151-159. [PubMed] |
| 65. | Büssing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400-409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 454] [Cited by in RCA: 497] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 66. | Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97-100. [PubMed] |
| 67. | Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134-2139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 473] [Cited by in RCA: 493] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 68. | Li H, Collado M, Villasante A, Strati K, Ortega S, Cañamero M, Blasco MA, Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136-1139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 793] [Cited by in RCA: 767] [Article Influence: 47.9] [Reference Citation Analysis (0)] |
| 69. | Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145-1148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 654] [Cited by in RCA: 658] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 70. | Zaehres H, Kim JB, Schöler HR. Induced pluripotent stem cells. Methods Enzymol. 2010;476:309-325. [PubMed] |
| 71. | Ohnuki M, Takahashi K, Yamanaka S. Generation and characterization of human induced pluripotent stem cells. Curr Protoc Stem Cell Biol. 2009;Chapter 4:Unit 4A.2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 95] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 72. | Chang KH, Nelson AM, Fields PA, Hesson JL, Ulyanova T, Cao H, Nakamoto B, Ware CB, Papayannopoulou T. Diverse hematopoietic potentials of five human embryonic stem cell lines. Exp Cell Res. 2008;314:2930-2940. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 41] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 73. | Mikkola M, Olsson C, Palgi J, Ustinov J, Palomaki T, Horelli-Kuitunen N, Knuutila S, Lundin K, Otonkoski T, Tuuri T. Distinct differentiation characteristics of individual human embryonic stem cell lines. BMC Dev Biol. 2006;6:40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 58] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 74. | Buchholz DE, Hikita ST, Rowland TJ, Friedrich AM, Hinman CR, Johnson LV, Clegg DO. Derivation of functional retinal pigmented epithelium from induced pluripotent stem cells. Stem Cells. 2009;27:2427-2434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 305] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 75. | Andersson TB, Sundberg MI. Livers cells derived from human embryonic stem cells Progress and potential use in ADMET applications. Drug Discov Today: Technol. 2008;5:e143-e147. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 76. | Agarwal S, Holton KL, Lanza R. Efficient differentiation of functional hepatocytes from human embryonic stem cells. Stem Cells. 2008;26:1117-1127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 271] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 77. | Basma H, Soto-Gutiérrez A, Yannam GR, Liu L, Ito R, Yamamoto T, Ellis E, Carson SD, Sato S, Chen Y. Differentiation and transplantation of human embryonic stem cell-derived hepatocytes. Gastroenterology. 2009;136:990-999. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 416] [Cited by in RCA: 364] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 78. | Sullivan GJ, Hay DC, Park IH, Fletcher J, Hannoun Z, Payne CM, Dalgetty D, Black JR, Ross JA, Samuel K. Generation of functional human hepatic endoderm from human induced pluripotent stem cells. Hepatology. 2010;51:329-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 355] [Cited by in RCA: 313] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 79. | Song Z, Cai J, Liu Y, Zhao D, Yong J, Duo S, Song X, Guo Y, Zhao Y, Qin H. Efficient generation of hepatocyte-like cells from human induced pluripotent stem cells. Cell Res. 2009;19:1233-1242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 367] [Cited by in RCA: 366] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 80. | Si-Tayeb K, Noto FK, Nagaoka M, Li J, Battle MA, Duris C, North PE, Dalton S, Duncan SA. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 944] [Cited by in RCA: 953] [Article Influence: 63.5] [Reference Citation Analysis (0)] |
| 81. | Liu H, Ye Z, Kim Y, Sharkis S, Jang YY. Generation of endoderm-derived human induced pluripotent stem cells from primary hepatocytes. Hepatology. 2010;51:1810-1819. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 197] [Cited by in RCA: 174] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 82. | Takata A, Otsuka M, Kogiso T, Kojima K, Yoshikawa T, Tateishi R, Kato N, Shiina S, Yoshida H, Omata M. Direct differentiation of hepatic cells from human induced pluripotent stem cells using a limited number of cytokines. Hepatol Int. 2011;Feb 6 [Epub ahead of print]. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 83. | Gai H, Nguyen DM, Moon YJ, Aguila JR, Fink LM, Ward DC, Ma Y. Generation of murine hepatic lineage cells from induced pluripotent stem cells. Differentiation. 2010;79:171-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 34] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 84. | Iwamuro M, Komaki T, Kubota Y, Seita M, Kawamoto H, Yuasa T, Shahid JM, Hassan RA, Hassan WA, Nakaji S. Hepatic differentiation of mouse iPS cells in vitro. Cell Transplant. 2010;19:841-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 85. | Gómez-Lechón MJ, Donato MT, Castell JV, Jover R. Human hepatocytes as a tool for studying toxicity and drug metabolism. Curr Drug Metab. 2003;4:292-312. [PubMed] |
| 86. | Rao MS, Khan AA, Parveen N, Habeeb MA, Habibullah CM, Pande G. Characterization of hepatic progenitors from human fetal liver during second trimester. World J Gastroenterol. 2008;14:5730-5737. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 31] [Cited by in RCA: 32] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 87. | Wilkening S, Stahl F, Bader A. Comparison of primary human hepatocytes and hepatoma cell line Hepg2 with regard to their biotransformation properties. Drug Metab Dispos. 2003;31:1035-1042. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 613] [Cited by in RCA: 590] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 88. | Seifinejad A, Tabebordbar M, Baharvand H, Boyer LA, Salekdeh GH. Progress and promise towards safe induced pluripotent stem cells for therapy. Stem Cell Rev. 2010;6:297-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 49] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 89. | Andrade RJ, Robles M, Fernández-Castañer A, López-Ortega S, López-Vega MC, Lucena MI. Assessment of drug-induced hepatotoxicity in clinical practice: a challenge for gastroenterologists. World J Gastroenterol. 2007;13:329-340. [PubMed] |
| 90. | Dekant W. The role of biotransformation and bioactivation in toxicity. EXS. 2009;99:57-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 91. | Greenhough S, Medine CN, Hay DC. Pluripotent stem cell derived hepatocyte like cells and their potential in toxicity screening. Toxicology. 2010;278:250-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 47] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 92. | Miura K, Okada Y, Aoi T, Okada A, Takahashi K, Okita K, Nakagawa M, Koyanagi M, Tanabe K, Ohnuki M. Variation in the safety of induced pluripotent stem cell lines. Nat Biotechnol. 2009;27:743-745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 669] [Cited by in RCA: 640] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 93. | Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285-290. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1842] [Cited by in RCA: 1686] [Article Influence: 112.4] [Reference Citation Analysis (0)] |
| 94. | Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol. 2010;28:848-855. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 997] [Cited by in RCA: 913] [Article Influence: 60.9] [Reference Citation Analysis (0)] |
| 95. | Espejel S, Roll GR, McLaughlin KJ, Lee AY, Zhang JY, Laird DJ, Okita K, Yamanaka S, Willenbring H. Induced pluripotent stem cell-derived hepatocytes have the functional and proliferative capabilities needed for liver regeneration in mice. J Clin Invest. 2010;120:3120-3126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 134] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 96. | Fausto N. Liver regeneration and repair: hepatocytes, progenitor cells, and stem cells. Hepatology. 2004;39:1477-1487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 537] [Cited by in RCA: 517] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 97. | Schmelzer E, Wauthier E, Reid LM. The phenotypes of pluripotent human hepatic progenitors. Stem Cells. 2006;24:1852-1858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 265] [Cited by in RCA: 257] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 98. | Roskams TA, Theise ND, Balabaud C, Bhagat G, Bhathal PS, Bioulac-Sage P, Brunt EM, Crawford JM, Crosby HA, Desmet V. Nomenclature of the finer branches of the biliary tree: canals, ductules, and ductular reactions in human livers. Hepatology. 2004;39:1739-1745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 525] [Cited by in RCA: 524] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 99. | Terrace JD, Currie IS, Hay DC, Masson NM, Anderson RA, Forbes SJ, Parks RW, Ross JA. Progenitor cell characterization and location in the developing human liver. Stem Cells Dev. 2007;16:771-778. [PubMed] |
| 100. | Khan AA, Shaik MV, Parveen N, Rajendraprasad A, Aleem MA, Habeeb MA, Srinivas G, Raj TA, Tiwari SK, Kumaresan K. Human fetal liver-derived stem cell transplantation as supportive modality in the management of end-stage decompensated liver cirrhosis. Cell Transplant. 2010;19:409-418. [PubMed] |
| 101. | Challen GA, Little MH. A side order of stem cells: the SP phenotype. Stem Cells. 2006;24:3-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 374] [Cited by in RCA: 345] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 102. | Petersen BE, Bowen WC, Patrene KD, Mars WM, Sullivan AK, Murase N, Boggs SS, Greenberger JS, Goff JP. Bone marrow as a potential source of hepatic oval cells. Science. 1999;284:1168-1170. [PubMed] |
| 103. | Theise ND, Nimmakayalu M, Gardner R, Illei PB, Morgan G, Teperman L, Henegariu O, Krause DS. Liver from bone marrow in humans. Hepatology. 2000;32:11-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 900] [Cited by in RCA: 851] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 104. | Houlihan DD, Newsome PN. Critical review of clinical trials of bone marrow stem cells in liver disease. Gastroenterology. 2008;135:438-450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 131] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 105. | He ZP, Tan WQ, Tang YF, Zhang HJ, Feng MF. Activation, isolation, identification and in vitro proliferation of oval cells from adult rat livers. Cell Prolif. 2004;37:177-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 106. | Petersen BE, Goff JP, Greenberger JS, Michalopoulos GK. Hepatic oval cells express the hematopoietic stem cell marker Thy-1 in the rat. Hepatology. 1998;27:433-445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 271] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 107. | Hatch HM, Zheng D, Jorgensen ML, Petersen BE. SDF-1alpha/CXCR4: a mechanism for hepatic oval cell activation and bone marrow stem cell recruitment to the injured liver of rats. Cloning Stem Cells. 2002;4:339-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 175] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 108. | Sharma AD, Cantz T, Manns MP, Ott M. The role of stem cells in physiology, pathophysiology, and therapy of the liver. Stem Cell Rev. 2006;2:51-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 24] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 109. | Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035-1041. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2235] [Cited by in RCA: 2274] [Article Influence: 151.6] [Reference Citation Analysis (0)] |
| 110. | Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375-386. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2088] [Cited by in RCA: 1859] [Article Influence: 123.9] [Reference Citation Analysis (0)] |
| 111. | Szabo E, Rampalli S, Risueño RM, Schnerch A, Mitchell R, Fiebig-Comyn A, Levadoux-Martin M, Bhatia M. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468:521-526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 557] [Cited by in RCA: 530] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 112. | Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, Hu Y, Wang X, Hui L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386-389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 647] [Cited by in RCA: 661] [Article Influence: 47.2] [Reference Citation Analysis (0)] |