Published online May 28, 2013. doi: 10.3748/wjg.v19.i20.3130
Revised: March 29, 2013
Accepted: April 3, 2013
Published online: May 28, 2013
Processing time: 82 Days and 0.4 Hours
AIM: To detect the expression of huCdc7 in colorectal cancer.
METHODS: The mRNA and protein expression of huCdc7 in 39 colorectal cancer tissue specimens and matched tumor-adjacent normal colorectal tissue specimens was detected by reverse transcription-polymerase chain reaction and immunohistochemistry, respectively.
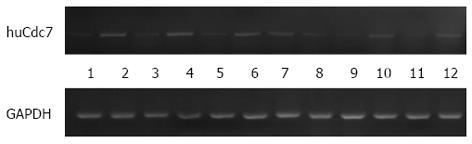
RESULTS: The relative expression level of huCdc7 mRNA in colorectal cancer was significantly higher than that in tumor-adjacent normal colorectal tissues (0.03675 ± 1.00 vs 0.01199 ± 0.44, P < 0.05). huCdc7-positive cells displayed brown granules in the nucleus. Tumor tissues contained many huCdc7-positive cells, whereas normal colorectal tissues contained very few positive cells.
CONCLUSION: huCdc7 may play an important role in the development and progression of colorectal cancer.
Core tip: huCdc7 is ubiquitously expressed in human tissues and can regulate DNA replication initiation. Abnormal expression or excessive activation of huCdc7 can promote excessive cell proliferation and cause tumorigenesis. Colorectal cancer is a common digestive tract tumor with a gradually increasing incidence. We found that huCdc7 may play an important role in the development and progression of colorectal cancer.
- Citation: Chen HJ, Zhu Z, Wang XL, Feng QL, Wu Q, Xu ZP, Wu J, Yu XF, Qian HL, Lu Q. Expression of huCdc7 in colorectal cancer. World J Gastroenterol 2013; 19(20): 3130-3133
- URL: https://www.wjgnet.com/1007-9327/full/v19/i20/3130.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i20.3130
huCdc7 is ubiquitously expressed in human tissues and can regulate DNA replication initiation. Abnormal expression or excessive activation of huCdc7 can promote excessive cell proliferation and cause tumorigenesis. Colorectal cancer is a common digestive tract tumor with a gradually increasing incidence. This study aimed to detect the expression of huCdc7 in colorectal cancer to provide a basis for the diagnosis and treatment of this malignancy.
Tissue specimens were collected from 39 patients with colorectal cancer who were surgically treated at our hospital. Colorectal cancer tissue specimens and matched tumor-adjacent normal colorectal tissue specimens (5 cm away from tumor tissue) were placed in liquid nitrogen immediately after surgical specimens were removed. The final diagnosis of colorectal cancer and tumor-adjacent normal colorectal tissues was made pathologically.
M-MLV reverse transcriptase (Promega, United States), Taq DNA polymerase (Takara, Japan), and dNTPs (Takara, Japan) were obtained commercially. The primers for the amplification of a 525-bp CDC7 gene fragment were: forward, 5’-GCT CAG CAG GAA AGG TGT TC-3’ and reverse, 5’-AGT TTG ATT GGG GCA CTT TG-3’. The primers for amplification of a 420-bp glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene fragment were: forward, 5’-GTC AGT GGT GGA CCT GAC CT- 3’ and reverse, 5’-AGG GGT CTA CAT GGC AAC TG-3’.
Preparation of total RNA: Total RNA was extracted using TRIzol reagent (GIBCO/BRL, United States). All centrifuge tubes, plastic and glasswares and water used for RNA extraction were treated to create an RNAse-free environment. A homogenizer was baked at 200 °C for 4 h to remove RNAse and then cooled. Tissue samples were frozen in liquid nitrogen, pulverized into powder, and placed in the homogenizer containing TRIzol reagent. After homogenization for several minutes, the homogenized sample was transferred to an RNAse-free centrifuge tube. Following the addition of chloroform, the tube was centrifuged at 4 °C. The upper aqueous phase was transferred to an RNAse-free centrifuge tube, and isopropanol was added to precipitate the RNA. The tube was centrifuged at 4 °C, and the pellet was washed twice with 75% ethanol and dissolved in RNAse-free deionized water. The purity of extracted RNA was assessed by measuring the A260/A280 ratio (1.7-2.0) using an ultraviolet spectrophotometer. 3-morpholinopropanesulfonic acid-formaldehyde denaturing agarose gel electrophoresis was performed to check if the extracted RNA was degraded. To remove the contamination of genomic DNA, RNA sample was treated with RNAse-free DNA enzyme I (Ambion, United States).
cDNA synthesis: Two micrograms of total RNA were mixed with 1 μL of random primers (50 pmol/L, 1 μL) and 15 μL of diethylpyrocarbonate-treated water, placed at 70 °C for 3 min and then immediately placed on ice for 5 min. The following components were then added: 5 μL of 5 × buffer, 1 μL of 10 mmol/L dNTP, 1 μL of M-MLV reverse transcriptase (200 U/μL), and 0.75 μL of RNAse inhibitor (40 U/μL, TaKaRa). The mixture was incubated at 42 °C for 2 h.
Semiquantitative reverse transcription-polymerase chain reaction: The huCdc7 gene was amplified using cDNA as the template. GAPDH was used as a control. Polymerase chain reaction (PCR) conditions were as follows: denaturation at 94 °C for 3 min; 35 cycles (25 cycles for GAPDH) of denaturation at 94 °C for 30 s, annealing at 58.5 °C for 30 s, and extension at 72 °C for 40 s; and a final extension at 72 °C for 5 min. PCR products were resolved by 2% agarose gel electrophoresis. Band densities were analyzed using the FR-980 bio-electrophoresis image analysis system.
Real-time PCR: The CFX96 real-time PCR detection system (BIO-RAD, United States) was used to detect the expression of genes of interest in tumor tissues and matched tumor-adjacent normal tissues. PCR reaction was performed in a 20-μL system consisting of 10 μL of SYBR Premix EX Taq, 0.4 μL of each primer (10 μm), 2 μL of DNA, and 7.2 μL of ddH2O. GAPDH was used as an internal control. The experiment was repeated three times to ensure the reliability of the results. The expression level of the huCdc7 gene was calculated using the following formulas: Cdc7 ∆Ct = mean Cdc7Ct - mean GAPDHCt, and Cdc7 ∆∆Ct = Cdc7 ∆Ct cancer tissue - Cdc7 ∆Ct tumor-adjacent tissue. Relative expression level of the huCdc7 gene was calculated using the 2-Cdc7 ∆∆Ct method.
Streptavidin-peroxidase immunohistochemical staining was performed using a commercial kit according to the manufacturer’s instructions.
Statistical analysis were performed using SPSS 10.0 software. Means between two groups were compared using the t test. P values < 0.05 were considered statistically significant.
The relative expression levels of huCdc7 mRNA in colorectal cancer and tumor-adjacent normal colorectal tissues were 0.03675 ± 1.00 and 0.01199 ± 0.44, respectively. Statistical analysis indicated that the expression level of huCdc7 mRNA was significantly higher in colorectal cancer than in tumor-adjacent normal colorectal tissues (P < 0.05) (Figure 1).
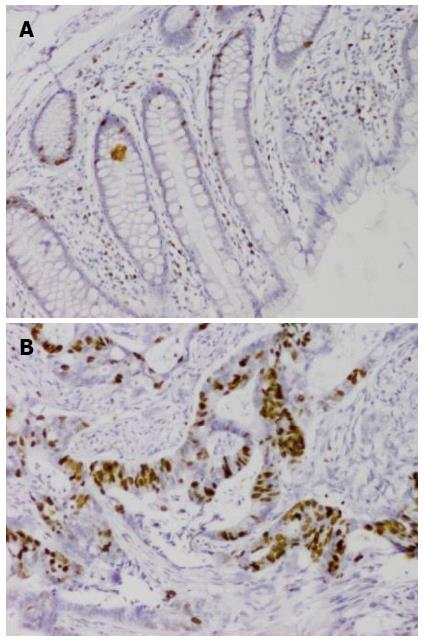
huCdc7-positive cells displayed brown granules in the nucleus. Tumor tissues contained many huCdc7-positive cells, whereas normal colorectal tissues contained very few positive cells (Figure 2).
Cdc7 is a serine/threonine kinase, and huCdc7 is expressed in all human tissues[1]. By forming complex with other molecules in the nucleus, huCdc7 can phosphorylate and activate chromosome-binding minichromosome maintenance complex (MCM) proteins. The MCM family has multiple members, including MCM2, MCM4 and MCM6. huCdc7 has the strongest ability to phosphorylate MCM2[2]. On one hand, MCMs function as helicase, a component of cell cycle initiation complex[3]. On the other hand, MCMs can act as important regulatory factors for S phase checkpoints to control cell cycle progression.
It is still unclear how huCdc7 mediates these processes. The expression of huCdc7 is tightly controlled by some factors and auxiliary proteins in normal cell cycle and maintained in a dynamic equilibrium state. In tumor cells, huCdc7 is abnormally expressed and excessively activated due to cell cycle disturbances. Hess et al[4] found that huCdc7 was overexpressed in tumor cells and excessive expression of huCdc7 promoted excessive MCM2 activation and abnormal proliferation of tumor cells. In addition, they found that huCdc7 was overexpressed in metastatic tumor cells, suggesting that tumor metastasis may be closely related to abnormal high huCdc7 expression. A previous study has revealed that CDC7 kinase is a predictor of survival and a novel therapeutic target in epithelial ovarian carcinoma[5]. Similar findings have also been reported in some studies on lymphoma[6].
In this study, we utilized semi-quantitative reverse transcription-PCR and immunohistochemistry to determine the expression of huCdc7 in colorectal cancer and tumor-adjacent normal colorectal tissues. We found that huCdc7 mRNA expression was significantly higher in colorectal cancer than in normal colorectal tissues. huCdc7 is a conservative serine/threonine kinase that is indispensable for DNA replication initiation. Abnormal high expression of huCdc7 will promote DNA replication, cause abnormal cell proliferation, and thereby lead to the occurrence of tumors.
To investigate the relationship between huCdc7 and tumor development and progression, Montagnoli et al[7] used the siRNA interference technology to suppress high huCdc7 expression in tumor cells. They found that the phosphorylation levels of MCM2 activation sites were greatly decreased, DNA replication initiation in tumor cells was restrained, and tumor cell growth slowed down. In addition, some researchers believe that high huCdc7 expression in tumor cells is related to p53 inactivation[8].
A study on ovarian cancer revealed that CDC7 kinase can predict survival and serve as a target for cancer treatment. Kim et al[9] found that siRNA-mediated inhibition of huCdc7 reduced the phosphorylation level of the N-terminus of MCM4. As a result, Cdc 45 could not be positioned to the chromosome and replication initiation complex could not form. This inhibited, to a certain extent, tumor cell proliferation. Thus, inhibition of huCdc7 activity can effectively inhibit tumor cell growth and promote tumor cell apoptosis, without damage to normal cells. A recent study has identified PHA-767491 as an inhibitor of huCdc7[10], which may be used to down-regulate abnormal huCdc7 expression to inhibit abnormal DNA replication and cell cycle progression in tumors.
Small molecule compounds that can interfere with the activity of CDC7 have been developed and proved to be effective in inhibiting tumor growth in animal models. Unprecedented attention has been paid to the inhibition of CDC7 kinase activity for suppressing DNA replication in tumor cells and inducing tumor cell apoptosis. Further elucidation of the role of huCdc7 in colorectal cancer may improve the treatment of this malignancy.
huCdc7 is ubiquitously expressed in human tissues and can regulate DNA replication initiation. Abnormal expression or excessive activation of huCdc7 can promote excessive cell proliferation and cause tumorigenesis. Colorectal cancer is a common digestive tract tumor and has a gradually increasing incidence.
In this study, tissue specimens were collected from 39 surgically treated patients with colorectal cancer. Colorectal cancer tissue specimens and matched tumor-adjacent normal colorectal tissue specimens (5 cm away from tumor tissue) were placed in liquid nitrogen immediately after surgical specimens were removed. The final diagnosis of colorectal cancer and tumor-adjacent normal colorectal tissues was made pathologically.
The relative expression level of huCdc7 mRNA in colorectal cancer was significantly higher than that in tumor-adjacent normal colorectal tissue. huCdc7-positive cells displayed brown granules in the nucleus. Tumor tissue contained many huCdc7-positive cells, whereas normal colorectal tissue contained very few positive cells.
The authors found that huCdc7 may play an important role in the development and progression of colorectal cancer.
In the manuscript, the authors detected the expression of huCdc7 in colorectal cancer. The data is interesting. The manuscript is short, but informative.
P- Reviewers Buyse M, Hoff PM, Mitchell P S- Editor Wang JL L- Editor Ma JY E- Editor Li JY
| 1. | Jiang W, Hunter T. Identification and characterization of a human protein kinase related to budding yeast Cdc7p. Proc Natl Acad Sci USA. 1997;94:14320-14325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 61] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 2. | Masai H, Matsui E, You Z, Ishimi Y, Tamai K, Arai K. Human Cdc7-related kinase complex. In vitro phosphorylation of MCM by concerted actions of Cdks and Cdc7 and that of a criticial threonine residue of Cdc7 bY Cdks. J Biol Chem. 2000;275:29042-29052. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 128] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 3. | Forsburg SL. Eukaryotic MCM proteins: beyond replication initiation. Microbiol Mol Biol Rev. 2004;68:109-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 410] [Cited by in RCA: 427] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 4. | Hess GF, Drong RF, Weiland KL, Slightom JL, Sclafani RA, Hollingsworth RE. A human homolog of the yeast CDC7 gene is overexpressed in some tumors and transformed cell lines. Gene. 1998;211:133-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 50] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 5. | Kulkarni AA, Kingsbury SR, Tudzarova S, Hong HK, Loddo M, Rashid M, Rodriguez-Acebes S, Prevost AT, Ledermann JA, Stoeber K. Cdc7 kinase is a predictor of survival and a novel therapeutic target in epithelial ovarian carcinoma. Clin Cancer Res. 2009;15:2417-2425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 61] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 6. | Hou Y, Wang HQ, Fu K, Zhang HL, Qian ZZ, Qiu LH, Li W, Zhou SY, Li LF, Hao XS. Expression of Cdc7 and mcm2 as a marker for proliferation and prognosis in diffuse large B cell lymphoma. Zhonghua Zhongliu Zazhi. 2011;33:911-915. [PubMed] |
| 7. | Montagnoli A, Tenca P, Sola F, Carpani D, Brotherton D, Albanese C, Santocanale C. Cdc7 inhibition reveals a p53-dependent replication checkpoint that is defective in cancer cells. Cancer Res. 2004;64:7110-7116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 114] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 8. | Bonte D, Lindvall C, Liu H, Dykema K, Furge K, Weinreich M. Cdc7-Dbf4 kinase overexpression in multiple cancers and tumor cell lines is correlated with p53 inactivation. Neoplasia. 2008;10:920-931. [PubMed] |
| 9. | Kim BJ, Lee H. Importin-beta mediates Cdc7 nuclear import by binding to the kinase insert II domain, which can be antagonized by importin-alpha. J Biol Chem. 2006;281:12041-12049. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 10. | Montagnoli A, Valsasina B, Croci V, Menichincheri M, Rainoldi S, Marchesi V, Tibolla M, Tenca P, Brotherton D, Albanese C. A Cdc7 kinase inhibitor restricts initiation of DNA replication and has antitumor activity. Nat Chem Biol. 2008;4:357-365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 170] [Article Influence: 10.0] [Reference Citation Analysis (0)] |