Published online May 28, 2013. doi: 10.3748/wjg.v19.i20.2985
Revised: March 29, 2013
Accepted: April 10, 2013
Published online: May 28, 2013
Processing time: 123 Days and 14.9 Hours
MicroRNAs, a key class of gene expression regulators, have emerged as crucial players in various biological processes such as cellular proliferation and differentiation, development and apoptosis. In addition, microRNAs are coming to light as crucial regulators of innate and adaptive immune responses, and their abnormal expression and/or function in the immune system have been linked to multiple human diseases including inflammatory disorders, such as inflammatory bowel disease, and cancers. In this review, we discuss our current understanding of microRNAs with a focus on their role and mode of action in regulating the immune system during inflammation and carcinogenesis.
Core tip: MicroRNAs (miRNAs), a key class of gene expression regulators, have emerged as crucial players in various biological processes such as cellular proliferation and differentiation, development and apoptosis. A better understanding of the function of miRNAs is providing new insights into the molecular basis of human pathologies, and new biomarkers for disease diagnosis and therapy.
- Citation: Raisch J, Darfeuille-Michaud A, Nguyen HTT. Role of microRNAs in the immune system, inflammation and cancer. World J Gastroenterol 2013; 19(20): 2985-2996
- URL: https://www.wjgnet.com/1007-9327/full/v19/i20/2985.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i20.2985
MicroRNAs (miRNAs, miR) are small (approximately 20-22 nucleotides), non-coding RNAs that post-transcriptionally regulate gene expression by binding to the 3′-untranslated region of target mRNAs, leading to mRNA degradation or translational inhibition[1].
Since the identification of the first miRNA, lin-4, in Caenorabditis elegans in 1993[2,3], thousands of miRNA genes have been identified in animal and plant genomes[4]. As a class, miRNAs account for about 1%-2% of genes in worms, flies, and mammals[5]. Each miRNA can target hundreds of mRNAs within a given cell type, and a single mRNA is often the target of multiple miRNAs, and thus over half of the human transcriptome is predicted to be under miRNA regulation, embedding this post-transcriptional control pathway within nearly every biological process[5].
Given its fundamental biological roles, it is not surprising that miRNA expression is tightly controlled and that its deregulation can lead to various diseases. In this review, we summarize our current knowledge about the physiological role of miRNAs in mammalian biology and the manner in which miRNA activities contribute to diseases including inflammatory disorders and cancer.
Our knowledge of miRNA biogenesis and regulation has been greatly expanded in recent years[1]. The canonical miRNA biogenesis takes place in a multi-step process and involves two RNAse III endonucleases, Dicer and Drosha. MiRNAs are encoded by genomic DNA and are most commonly transcribed by RNA polymerase II, which generates a primary miRNA (pri-miRNA) transcript. Within the primary transcripts, miRNAs form stem-loop structures, which contain the mature miRNA as part of an imperfectly paired double-stranded stem connected by a short terminal loop. Pri-miRNAs are then processed by a microprocessor complex, a multiprotein complex with the two core components, Drosha and Di George Syndrome critical region 8 (DGCR8)[6-8]. This results in the formation of a hairpin-shaped RNA molecule of 70-100 bp called miRNA precursor or pre-miRNA, which is then exported into the cytoplasm in a process involving the nucleocytoplasmic shuttle Exportin-5 and in a Ran-GTP-dependent manner[9-11]. In cytoplasm, the pre-miRNA hairpin is cleaved by the endonuclease DICER into an imperfect miRNA:miRNA* duplex of 21-23 nucleotides in length[12]. After separation of the two strands of the duplex, one of the strands (the mature miRNA) is transferred into an Argonote (Ago) protein located in the RNA-induced silencing complex (RISC or miRISC), which is involved in the repression of gene expression by leading miRNAs to specific target mRNAs, whereas the other strand (the star-strand) is degraded. It has been shown that strand selection and RISC assembly in mammals are accomplished by a complex that contains Dicer, Ago and the double-stranded RNA binding protein TRBP[13-15]. MiRNAs target mRNAs by interacting with sites of imperfect complementarity. Short “seed” sequences at the 5′-ends of miRNAs (nucleotides 2-8) are critical, and in some cases fully sufficient, for target selection[16,17].
Although there have been recent advances in our knowledge of the biogenesis of the miRNA pathway, relatively little is known about the mechanisms regulating the activity of the pathway’s components. Several recent studies indicate that the regulation of miRNA expression and function occurs at three levels: transcription, processing and subcellular localization[17,18].
The first, and one of the most important, mechanisms controlling miRNA abundance is the regulation of pri-miRNA transcription, which could be positively or negatively regulated by different factors such as transcription factors, enhancers, silencers and epigenetic modification in miRNA promoters[16]. For example, the oncogene c-myc can bind to the promoter of the miR-17-5p cluster, thereby up-regulating expression of the miRNAs encoded by the cluster[19,20]. Similarly, the tumour suppressor p53 has been shown to upregulate the transcription of miR-34 family members, inhibiting important factors of cell proliferation and survival, such as Bcl2 and Cdk4 and 6[21-24]. A region of miRNA genes is located within CpG islands involving the epigenetic control of miRNA transcription. It is estimated from recent works that 5%-10% of mammalian miRNAs are epigenetically regulated[19,25-27].
Several post-transcriptional regulatory mechanisms that affect miRNA processing at different stages, from the pri-miRNA transcripts to the delivery of mature miRNAs to their target mRNAs, have recently been investigated[18]. For example, p53 can form a complex with Drosha, which increases the processing of pri-miRNAs to pre-miRNAs[28]. Histone deacetylase I can enhance pri-miRNA processing by deacetylating the protein DGCR8 of the microprocessor complex[29]. Cytokines such as interferons have been shown to inhibit Dicer expression, decreasing the processing of pre-miRNAs[30].
The immune system has evolved to maintain self-tolerance and to recognize efficiently specific pathogens. The innate immune system acts as a first protector providing an immediate response to pathogens, and propagation of the innate response activates the adaptive immune system. Both innate and adaptive immune responses are highly regulated, and recent studies have shed light on the role of miRNAs in this intricate system[31,32]. The role of miRNAs in immune responses will be discussed in this section.
The innate immune system is activated via recognition of pathogen-associated molecular patterns by toll-like receptors (TLRs)[33], which will recruit adaptor proteins to the receptor, followed by activation of downstream signalling pathways such as the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) pathway[34]. This signal transduction ultimately leads to induction of immune gene expression.
The first study examining the effect of lipopolysaccharide (LPS)-mediated activation of TLR signalling on miRNA production identified miR-155, miR-146a and miR-132, which are induced in human macrophages by LPS[35]. Further analysis showed that miR-155 is induced by LPS, cytokine IFN-β and various TLR ligands in murine macrophages[36,37]. MiR-155, once induced, is involved in the activation of tumor necrosis factor-α (TNF-α) and interleukin-6 (IL-6), enzyme linked immunosorbent assay pathway via targeting the Fas-associated death domain protein, I B kinase epsilon, and receptor (TNF receptor superfamily)-interacting serine-threonine kinase 1[37]. MiR-155 plays a role in the innate immune response by regulating suppressor of cytokine signalling (SOCS)-1, a negative regulator of dendritic cell antigen-presenting capacity[37-39]. Likewise, miR-155-deficient dendritic cells exhibit impaired antigen presentation and therefore are unable to activate T cells to promote inflammation[39]. One study demonstrated that in human myeloid-derived DCs, knockdown of miR-155 expression significantly increased protein expression of the pro-inflammatory cytokine IL-1[40]. The same study also showed that miR-155 directly inhibited expression of the pro-inflammatory signalling protein TAK1-binding protein 2 (TAB2, also known as MAP3K7IP2), which could be a mechanism underlying its anti-inflammatory property[40]. In contrast, other studies have shown that miR-155 can enhance inflammatory responses. Overexpression of miR-155 in mouse bone marrow leads to a myeloproliferative phenotype that is similar to that observed transiently after LPS stimulation[41]. MiR-155 can negatively regulate SHIP1, an important negative regulator of phosphoinositide 3-kinase (PI3K) and the downstream AKT pathway[42,43]. SHIP1, which is similar to SOCS1, is a negative regulator of TLR4 signaling[44], and hence repression of SHIP1 by miR-155 may counter this negative regulation and increase downstream AKT signalling.
Like miR-155, miR-146a is induced by LPS, TNF-α and IL-1β in a NF-κB-dependent manner. MiR-146a in turn inhibits expression of two components of the TLR4 signaling pathway, IL-1 receptor associated kinase and TNF receptor-associated factor-6[35]. Thus, miR-146a functions as a negative feedback regulator of the TLR/NF-κB pathway. MiR-155 and miR-146 expression is increased in macrophages in response to LPS stimulation, while miR-125b expression is decreased. MiR-125b can target TNF-α mRNA, and a decrease in its expression leads to elevated TNF-α production and consequently increased inflammatory response[37].
Macrophage inflammatory response to infection involves the upregulation of several miRNAs, such as miR-21, miR-9 and miR-147[45-47]. These miRNAs can also be induced by TLR signaling, and can negatively regulate activation of inflammatory pathways in myeloid cells. MiR-9 represses NF-κB subunit 1 (NFKB1/p50 unit) and helps to maintain a constant level of NF-κB1 protein expression during TLR4-mediated activation of monocytes and neutrophils[46]. MiR-147 has been shown to attenuate TLR2, TLR3 and TLR4-mediated production of inflammatory proteins such as TNF-α and IL-6[47]. Induction of miR-21 inhibits PDCD4, an IL-10 inhibitor, thereby derepressing IL-10. IL-10 in turn inhibits miR-155, allowing SHIP1 to be derepressed and inhibit TLR signaling[45,48]. Hence, immune responses are highly regulated by TLRs-mediated upregulation of different miRNAs.
In addition to miRNA induction by TLR signaling, recent studies have also reported inflammatory repression, such as miR-155 repression, in response to anti-inflammatory cytokine IL-10[49].
In addition to their role in regulating the innate immune system, miRNAs have been implicated in adaptive immunity by controlling the development and activation of T and B cells.
Specific miRNA expression profiles have been reported in different T cell subsets and stages of development[50-52], suggesting that miRNA-mediated regulation of signaling networks in T cells, and probably other immune cells, is dynamic and highly regulated. Interestingly, miRNA profiling in naive, effector and memory CD8+ T cells has revealed that a few highly expressed miRNAs are dynamically regulated during antigen-specific T-cell differentiation[52]. Mice exhibiting T-cell specific deletion of Dicer had lower numbers of mature T cells with abnormally developed T-cell subsets than wild-type mice, indicating that miRNAs are required for T cell development[53,54]. Two specific miRNAs have been implicated in T cell development, and probably account for some of the phenotype of Dicer-deficient T cells. The miR-17-92 cluster suppresses expression of pro-apoptotic proteins, including BCL-2-interacting mediators of cell death (BIM or BCL2L11) and phosphatase and tensin homologue. This miRNA cluster is thought to increase T cell survival during development and is expressed during the double negative 2 stage of thymopoiesis[55].
The role of miRNAs in the differentiation of T cells into distinct effector T helper cell subsets has been recently reported. It was demonstrated that miR-326 regulates differentiation of TH17 cells both in vitro and in vivo[56]. MiR-155 is implicated in regulatory T (Treg) cell formation and function, since forkhead box P3 (FOXP3), a transcription factor that is required for the development and function of Treg cells, may directly regulate the expression of this miRNA[57]. Furthermore, miRNA-155-deficient mice are immunodeficient, indicating the implication of miR-155 in homeostasis and the immune system[39]. Similarly, using genetic deletion and transgenic approaches, Thai and colleagues showed the important role of miR-155 in the mammalian immune system, specifically in regulating T helper cell differentiation and the germinal center reaction to produce an optimal T cell-dependent antibody response[58]. Certain miRNAs, such as the miR-17-92 cluster, might be involved in the development and function of T follicular helper cells (specialized T cells that provide selective signals to supporting geminal center B cells), which are essential for long-lived antibody responses[59,60]. In addition, miR-181a, which is increased during early T cell development and down-regulated in mature CD4 T cells such as Th1 and Th2 effector cells, can enhance TCR signaling strength by inhibiting multiple phosphatases that negatively regulate the TCR signaling cascade[61]. Finally, conditional deletion of Dicer or Drosha in Treg cells led to lethal autoimmune inflammatory disease, accompanied by impaired development or function of Treg cells, indicating the role of miRNAs in Treg cell biology[62-64].
Distinct miRNA profiles in naive, germinal central and post-germinal central B cells have been reported[65-67], suggesting the implication of miRNAs in B cell development and maturation. A pioneer study showed that miR-181 is highly expressed in B cells of mouse bone marrow, and its ectopic expression in hematopoietic stem and progenitor cells resulted in an increase in the percentage of B-lineage cells but not in T cells or myeloid cells[68], indicating the role of lineage-specific miRNAs in regulating lymphocyte development. Conditional deletion of Dicer in B cells completely arrested B cell development in mice, which is thought to be due to dysregulated expression of the pro-apoptotic protein BIM, probably during the selection of effective antigen receptors[69]. Notably, B cells lacking miR-17-92 family and Dicer-deficient B cells exhibited similar gene expression profiles[70], suggesting that this miRNA cluster could play a determining role in the regulation of B cell development.
Recent studies have explored the role of miR-150, a miRNA specifically expressed by mature lymphocytes, in B cell differentiation[51,71,72]. MiR-150 expression increases during B-cell maturation in bone marrow, and its constitutive expression blocked B cell development at the transition from the pro-B-cell to pre-B-cell developmental stage, leading to severe defects in the production of mature B cells[71]. MiR-150-deficient mice exhibited a 2-fold increase in splenic B-1 cell numbers, with a relative decrease in those of B-2 cells, but had no apparent defect in the development of other lymphoid-derived T- and B-cell types[72]. Mice expressing a miR-150 transgene early in life also had dramatically impaired B cell development with normal T cell levels. These defects in miR-150 gain- and loss-of-function were further shown to be due to dysregulation of c-Myb, a target of miR-150 and a transcription factor that controls multiple steps of lymphocyte development[72]. MiR-155-deficient B cells showed defects in antibody class switching and differentiation into plasma cells, resulting in an impaired humoral response to T cell-dependent antigenic stimulation[39,58,73]. The constitutive expression of miR-34a blocked B cell development at the pro-B to pre-B cell transition, leading to a reduction in mature B cells[74]. This block appeared to be mediated by miR-34a-inhibited expression of the transcription factor Foxp1[74], which is an essential regulator of B cell development[75]. Together, these studies show the important role of miRNAs in normal B cell development.
As miRNAs play a critical role in the regulation of the immune system, failure of miRNA regulation is associated with several human disorders such as inflammatory bowel disease (IBD) (Table 1), which is a chronic inflammatory gastrointestinal disorder. Although the etiology of IBD remains largely unknown, extensive studies in the last decades have suggested that it involves environmental and genetic factors that lead to dysfunction of the epithelial barrier with consequent deregulation of the mucosal immune system and responses to gut microbiota[76].
| Up-regulated | Down-regulated | Source, reference | |
| UC vs healthy | miR-16, miR-21, miR-23a, miR-24, miR-29a, miR-126, miR-195, let-7f, miR-21, miR-155 | miR-192, miR-375, miR-422b | Sigmoid colon[77] |
| miR-21, miR-155 | Colon[78] | ||
| miR-7, miR-31, miR-135b, miR-223, miR-29a, miR-29b, miR-126*, miR-127-3p, miR324-3p | miR-188-5p, miR-25, miR-320a, miR-346 | Colonic mucosa[79] | |
| (miR-196a, miR-29a, miR-29b, miR-126*, miR-127-3p, miR324-3p)1 | (miR-188-5p, miR-25, miR-320a, miR-346)2 | ||
| miR-28-5p, miR-151-5p, miR-199a-5p, miR-340*, miRplus-E1271, miR-103-2*, miR-362-3p, miR-532-3p | miR-505* | Peripheral blood[80] | |
| CD vs healthy | miR-9, miR-126, miR-130a, miR-181c, miR-375, miR-26a, miR-29b, miR-30b, miR-34c-5p, miR-126*, miR127-3p, miR-133b, miR-155, miR-196a, miR324-3p, miR-21, miR-22, miR-29c, miR-31, miR-106a, miR-146a, miR146b-3p, miR-150 | Colonic mucosa[79] | |
| (miR-9*, miR-30a*, miR-30c, miR-223 miR-25a, miR-29b, miR-30b, miR-34c-5p, miR-126*, miR127-3p, miR-133b, miR-155, miR-196a, miR324-3p, miR-21, miR-22, miR-29c, miR-31, miR-106a, miR-146a, miR146b-3p, miR-150)1 | |||
| UC vs CD | miR-199p-5a, miR-362-3p, miR-340*, miR-532-3p, miRplus-E1271 | miR-149*, miRplus-F1065 | Peripheral blood[80] |
| (miR-150, miR-196b, miR-199a-3p, miR-199b-5p, miR-223, miR-320a)2 | Colonic mucosa[79] | ||
| miR-28-5p, miR-103-2*, miR-149*, miR-151-5p, miR-340*, miR-532-3p, miRplus-E1153 | miR-505* | Peripheral blood[80] |
Distinguished miRNA expression profiles have been recently described in tissues of patients with active and inactive UC, CD, irritable bowel syndrome (IBS), infectious colitis (IC), and microscopic colitis (MC)[77]. Wu and colleagues demonstrated that active UC was associated with the differential expression of 11 miRNAs (3 significantly decreased and 8 significantly increased in UC tissues). MiR-192, the expression of which is decreased in active UC, was predominantly localized to colonic epithelial cells, and targeted macrophage inflammatory peptide (MIP)-2α, a chemokine expressed by epithelial cells[77]. In colonic epithelial cells, TNF-α-induced MIP-2α expression was inhibited by a miR-192 mimic. In contrast, miR-21 is significantly increased in patients with active UC compared to healthy subjects. In inactive UC patients, miR-375 and miR-422 expression was increased, while that of miR-192 was unaltered compared to healthy subjects[77]. Inactive UC showed similar expression levels of miR-375, miR-422b, and miR-23a to IBS and IC tissues. The miRNAs differently expressed in active UC were not dysregulated in MC and CD. This study highlights the specific miRNA expression patterns in active and inactive IBD tissues, and suggests that miRNAs could regulate expression of proteins implicated in the pathogenesis.
Another study showed the upregulated expression of several miRNAs in active UC compared to healthy colonic biopsies, suggesting that upregulation of miRNAs may be responsible for the development of intestinal inflammation in UC[78]. MiR-21 was found among the upregulated miRNAs, which is consistent with the findings of Takagi et al[78].
Of interest, Fasseu and colleagues identified restricted subsets of miRNAs abnormally expressed in inactive colonic mucosa of IBD patients[79]. This elegant study identified 14 (in UC) and 23 (in CD) miRNAs with significantly altered expression (> 5-fold increase or < 0.05-fold decrease) in quiescent colonic mucosa compared to healthy control tissues. Eight of the miRNAs (miR-26a, -29a, -29b, -30c, -126*, -127-3p, -196a, -324-3p) were commonly dysregulated in non-inflamed UC and CD. Six miRNAs (miR-196b, -199a-3p, -199b-5p, -320a, -150, -223) displayed significantly distinct dysregulation of expression between non-inflamed UC and CD colonic biopsies. Interestingly, several miRNA genes with dysregulated expression mapped within acknowledged IBD-susceptibility loci. In addition, significant dysregulated expression of four and five miRNAs specific to inflamed UC or CD tissues, respectively, compared to healthy controls was observed[79]. This study sheds light on the role of miRNAs as contributors to IBD susceptibility, in particular their implication in the onset and/or relapse of inflammation from quiescent mucosa of IBD patients.
There have been recent reports of differential miRNA expression profiles in the peripheral blood of IBD patients[80]. Four miRNAs (miR-199a5p, -362-3p, -532-3p and miRplus-E1271) were upregulated and one miRNA (miRplus-F1065) was downregulated in the peripheral blood of patients with active CD, but not inactive CD, compared to healthy controls[80]. Both active and inactive CD patients had increased expression of miR-340 and decreased expression of miR-149 in the blood. Expression of three miRNAs (miR-103-2, 262-3p, 532-3p) was increased in the blood of both active and inactive UC patients. In addition, a subset of 11 miRNAs can distinguish active CD from active UC[80]. This study importantly supports the evidence that distinct peripheral blood miRNA profiles in different circulating immune cell types are associated with IBD.
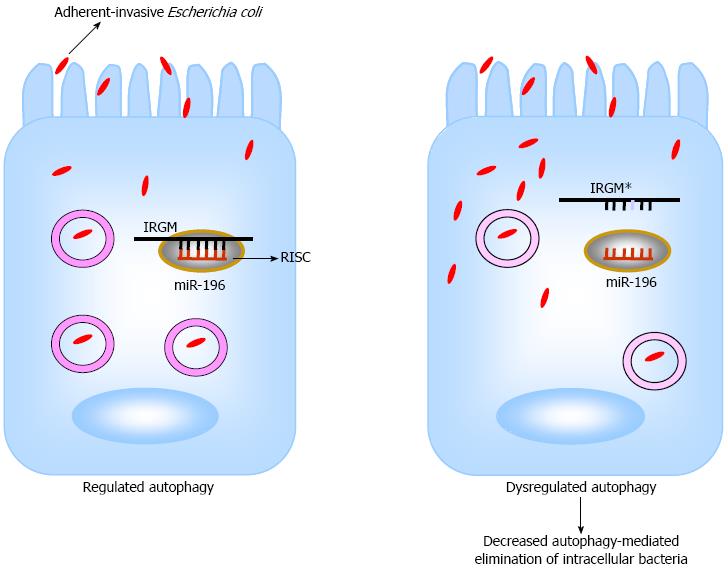
Efforts have been made to understand the mechanisms underlying the implication of miRNAs in the pathogenesis of IBD. The potential association between single nucleotide polymorphisms (SNPs) in pre-miRNA coding regions and IBD susceptibility has been analyzed. A study in a Japanese cohort of 170 UC patients and 403 healthy controls revealed the association of three SNPs (rs11614913, rs2910164, and rs3746444) in coding regions of pre-miR-196a2, pre-miR-146a and pre-miR-499[81]. Of particular interest, the CD-associated SNP C313T in immunity-related GTPase family, M (IRGM) gene caused a loss in binding of miR-196[82] (Figure 1). IRGM plays an important role in the immune system by its involvement in the autophagy process. In addition, miR-196 is overexpressed in the inflamed epithelium of CD patients and downregulates the IRGM protective variant (c.313C) but not the risk-associated allele (c.313T)[82]. Loss of tight regulation of IRGM expression by miR-196 resulted in defects in autophagy-mediated control of intracellular replication of adherent-invasive Escherichia coli (AIEC), leading to abnormal persistence of AIEC in host cells (Figure 1). This suggests that the association of IRGM with CD could arise from abnormal miRNA-mediated IRGM regulation, which affects the efficacy of autophagy, thereby contributing a synonymous polymorphism as a likely causal variant.
Intestinal microbiota is increasingly recognized as a risk for, and a causal factor of, IBD. Our recent studies showed that miRNAs are involved in the regulation of host gene expression by gut microbiota[83]. In another study, we showed that miRNAs play a role in determining the unique physiological characteristics of intestinal epithelial cells, such as their differentiation during migration along the crypt/villus axis[84]. In particular, expression of CD98, a transmembrane glycoprotein that regulates integrin signalling, cellular homeostasis and innate immune response in the gut[85], and its function are directly under the control of miRNAs during the differentiation of intestinal epithelial cells[86]. MiRNAs could also be involved in the upregulation of CD98 during intestinal inflammation and IBD[86]. The biological importance of miRNAs in the pathogenesis of IBD is becoming clearer, and targeting miRNAs in the gastrointestinal tract may be a promising approach for future therapeutic opportunities.
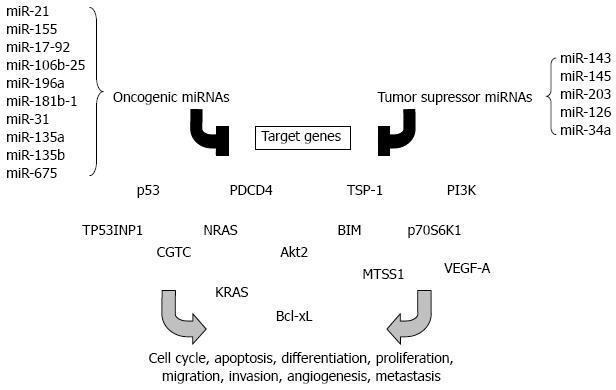
The transformation of a normal epithelium into a cancerous state involves modifications in several genes that are involved in different stages of carcinogenesis such as apoptosis, proliferation, limitless replicative potential of tumor cells, angiogenesis, migration and invasion[87]. Colorectal cancer (CRC) is one of the most common cancers worldwide. Its incidence is greater in industrial countries than in developing countries[88]. MiRNAs have been shown to play an important role in oncogenesis by regulating the expression of genes involved in cancer initiation, promotion and development[89]. Hundreds of miRNAs mapped to the human genome regions that are known to be altered in cancer, and a similar number of miRNAs are aberrantly expressed in cancerous tissues[90,91]. By analyzing miRNA expression profile (miRNome) of prostate, stomach, pancreas, lung, breast and colon tumors, Volinia and colleagues identified a solid cancer miRNA signature including those with well-characterized cancer association, such as miR-17-5p, miR-20a, miR-21, miR-92, miR-106a, and miR-155[92]. In particular, 21 miRNAs are up-regulated and 1 is down-regulated in colon tumors compared to normal tissue[92]. MiRNA profiles can identify different tissue and tumor types better than mRNA expression patterns, making them attractive targets for development as cancer biomarkers[93]. Distinguished miRNA profiles can even be found in the serum of patients with cancers. The functions of such circulating miRNAs have not been identified, but profiling of serum miRNAs might be a powerful approach for early cancer diagnosis. The cancer-associated miRNAs may function as oncogenes or tumor suppressors depending on their role in carcinogenesis. Some of the best examples of such miRNAs will be discussed in this section (Figure 2).
MiR-21 is one of the most up-regulated miRNAs in various cancers, including CRC[92,94], and was identified as an independent predictor of overall survival in the validation set containing tumor samples from 113 patients with CRC[95]. It has been shown that miR-21 is involved in invasion, intravasation and metastasis processes by targeting the tumor suppressor PDCD4[96], and in CRC tissues expression of miR-21 is inversely correlated with that of PDCD4 compared to normal tissue[97]. Shibuya and colleagues suggested that miR-21 expression may predict poor prognosis in CRC[98]. Likewise, these authors also examined the prognostic value of miR-155 in CRC since its expression is up-regulated in tumor tissues compared to normal adjacent tissues from CRC patients[98]. MiR-155 was previously shown to target the tumor protein 53-induced nuclear protein 1 (TP53INP1), a pro-apoptotic stress-induced p53 target, and significant reduction or loss of TP53INP1 expression was detected during adenocarcinoma progression[99].
MiR-17-92 and miR-106b-25 clusters are known to be up-regulated in CRC stromal tissues compared with normal stroma[100]. They include, respectively, multiple mature miRNAs, miR-17, miR-18a, miR-19a, miR-20a, miR-19b1, miR-92-1[101], and miR-106b, miR-93 and miR-25[102]. These miRNA clusters play an important role during carcinogenesis[92,100,103,104]. An anti-apoptotic effect of miR-17-92 appears to be one of the mechanisms underlying its procarcinogenic role in CRC development and progression[105]. Abrogation of miR-92a leads to cell apoptosis, and there is a correlation between the miR-17-92 overexpression in tumors of CRC patients and the downregulated expression of BIM, a member of the Bcl-2 family that promotes apoptosis[105]. Some works have reported that there is an interconnection between the expression of miR-17-92 cluster and angiogenesis, which occurs later in tumor development and is one of the most important stages in carcinogenesis. Dews and colleagues demonstrated that the anti-angiogenic factors thrombospondin-1 (tsp-1) and connective tissue growth factor (CTGC) are down-regulated by this cluster in intestinal epithelial cells expressing constitutively the oncogene c-myc[106], which was shown to be involved in regulation of miR-17-92 expression[20]. MiR-18 targets tsp-1 and miR-19 modulates the expression of CTGF[107].
Other miRNAs have also been identified as causal factors in colon carcinogenesis. For example, miR-196a had higher expression level in CRC tissues than in normal epithelial tissues[108]. MiR-196a exerts a pro-oncogenic influence in CRC as a high level of its expression promotes the oncogenic phenotype of colorectal cancer cells such as increased cell detachment, migration and invasion[109]. MiR-31 is often up-regulated in CRC and its high expression associated with advanced tumor stage but the clinical significance is unclear[110]. MiR-181b-1, miR-135a, miR-135b, miR-675 are also known to be up-regulated in CRC tumors[111]. MiR-135a is able to promote the growth and invasion of CRC cells by targeting the metastasis suppressor 1[112].
Mir-143 and miR-145 are among the best examples of tumor suppressor miRNAs. The expression of these miRNAs is down-regulated in CRC tumors, and in other cancers such as breast, prostate, cervical and lymphoid cancer[113-115]. Many studies have reported that down-regulation of miR-143 and miR-145 correlates with poor prognosis[110,115,116]. The expression and post-transcriptional maturation of these miRNAs were recently shown to be enhanced by the tumor suppressor p53 in response to DNA damages in CRC cell lines[28,117]. In particular, miR-143 is involved in inhibition of oncogene KRAS expression[118]. MiR-145 is reported to inhibit tumor growth and angiogenesis by directly targeting p70S6K1[117], which is activated by mTOR, and its overexpression in cancer cells induces tumor angiogenesis[119-121]. Another study reported that this miRNA is able to inhibit tumor growth and angiogenesis in breast cancer by targeting N-RAS and VEGF-A, which are key players in carcinogenesis[122].
It was recently demonstrated that miR-34a is down-regulated in colon tumors and also in circulating blood[123]. Furthermore, ectopic expression of miR-34a in CRC cell line reduces cell proliferation, demonstrating that this miRNA has a tumor suppressive function in colon carcinogenesis[124]. Several studies conducted in 2007 revealed that miR-34a can target p53, leading to apoptosis and cell cycle arrest[21-24,125]. MiR-203 is identified as another tumor suppressor miRNA. Its low expression was found in vitro in CRC cell lines and was correlated with tumor size in CRC. MiR-203 can inhibit proliferation of cancer cell lines[126]. Li et al[127] showed that miR-203 overexpression significantly decreased cell proliferation and survival and induced cell apoptosis in the p53-mutated CRC cells. The tumor suppressive role of miR-203 was mediated by negatively regulating Akt2 expression via mRNA degradation. In addition, overexpression of miR-203 decreased expression of the anti-apoptotic gene Bcl-xL, leading to a resistance to apoptosis[127]. MiR-126 is specifically expressed in endothelial cells and is known to be down-regulated in CRC compared to normal tissue via an unknown mechanism[128]. In vitro studies suggested that a loss in negative regulation of p85 subunit of PI3K by miR-126 could lead to a selective growth advantage during colon carcinogenesis[129].
MiRNAs are a class of gene regulators that have recently emerged as key players in the innate and adaptive immune system. Changes in miRNA expression are observed in many human diseases such as inflammatory bowel disease and cancers. Dysregulated miRNA expression profiles in IBD have been reported and could be used as diagnostic biomarkers but further studies are needed to examine the mechanism of their action in the etiopathogenesis of this disease and their clinical utility. Emerging evidence suggests that miRNAs play important roles in the pathogenesis of a limited range of human cancers. Some miRNAs may be directly involved in cancer development by controlling cell differentiation and apoptosis, while others may be involved in cancers by targeting cancer oncogenes and/or tumor suppressors. Given the critical role of miRNAs, current studies are focusing on their association with CRC incidence and prognosis and on the possibility of using circulating miRNAs or fecal miRNA expression as noninvasive early detection biomarkers. These data suggest that miRNAs may be potential molecular classifiers, early detection biomarkers, and therapeutic targets for CRC. Finally, miRNA-based cancer therapy has been limited to targeting a single miRNA[130,131]. However, it has been recently shown that the small molecule enoxacin, a fluoroquinolone used as an antibacterial compound, enhances the miRNA-processing machinery by binding to TRBP[132]. Thus, if most cancers are characterized by a dysregulation of global mature miRNA expression, restoration of the global miRNome may be an attractive approach in cancer therapy. In conclusion, a better understanding of the function of miRNAs is providing new insights into the molecular basis of human pathologies, and new biomarkers for disease diagnosis and therapy.
P- Reviewers Chi SG, Monticelli S S- Editor Gou SX L- Editor A E- Editor Zhang DN
| 1. | Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1888] [Cited by in RCA: 2001] [Article Influence: 125.1] [Reference Citation Analysis (0)] |
| 2. | Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855-862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2890] [Cited by in RCA: 2951] [Article Influence: 92.2] [Reference Citation Analysis (0)] |
| 3. | Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8672] [Cited by in RCA: 8875] [Article Influence: 277.3] [Reference Citation Analysis (0)] |
| 4. | Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152-D157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2680] [Cited by in RCA: 2786] [Article Influence: 185.7] [Reference Citation Analysis (0)] |
| 5. | Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14460] [Cited by in RCA: 16082] [Article Influence: 1005.1] [Reference Citation Analysis (2)] |
| 6. | Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3513] [Cited by in RCA: 3631] [Article Influence: 165.0] [Reference Citation Analysis (0)] |
| 7. | Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1964] [Cited by in RCA: 1987] [Article Influence: 94.6] [Reference Citation Analysis (0)] |
| 8. | Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1843] [Cited by in RCA: 1968] [Article Influence: 93.7] [Reference Citation Analysis (0)] |
| 9. | Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1918] [Cited by in RCA: 1888] [Article Influence: 89.9] [Reference Citation Analysis (0)] |
| 10. | Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011-3016. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1970] [Cited by in RCA: 2068] [Article Influence: 94.0] [Reference Citation Analysis (0)] |
| 11. | Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185-191. [PubMed] |
| 12. | Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1381] [Cited by in RCA: 1383] [Article Influence: 57.6] [Reference Citation Analysis (0)] |
| 13. | Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740-744. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1470] [Cited by in RCA: 1523] [Article Influence: 76.2] [Reference Citation Analysis (0)] |
| 14. | Haase AD, Jaskiewicz L, Zhang H, Lainé S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961-967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 487] [Cited by in RCA: 499] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 15. | Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631-640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1109] [Cited by in RCA: 1108] [Article Influence: 55.4] [Reference Citation Analysis (0)] |
| 16. | Rüegger S, Großhans H. MicroRNA turnover: when, how, and why. Trends Biochem Sci. 2012;37:436-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 211] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 17. | Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642-655. [PubMed] |
| 18. | Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. 2010;38:323-332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 442] [Cited by in RCA: 450] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 19. | Breving K, Esquela-Kerscher A. The complexities of microRNA regulation: miRandering around the rules. Int J Biochem Cell Biol. 2010;42:1316-1329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 183] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 20. | O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839-843. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2140] [Cited by in RCA: 2165] [Article Influence: 108.3] [Reference Citation Analysis (0)] |
| 21. | Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731-743. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1014] [Cited by in RCA: 1035] [Article Influence: 57.5] [Reference Citation Analysis (0)] |
| 22. | He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130-1134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2135] [Cited by in RCA: 2106] [Article Influence: 117.0] [Reference Citation Analysis (0)] |
| 23. | Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586-1593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 694] [Cited by in RCA: 738] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 24. | Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745-752. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1580] [Cited by in RCA: 1576] [Article Influence: 87.6] [Reference Citation Analysis (0)] |
| 25. | Brueckner B, Stresemann C, Kuner R, Mund C, Musch T, Meister M, Sültmann H, Lyko F. The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Cancer Res. 2007;67:1419-1423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 353] [Cited by in RCA: 342] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 26. | Han L, Witmer PD, Casey E, Valle D, Sukumar S. DNA methylation regulates MicroRNA expression. Cancer Biol Ther. 2007;6:1284-1288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 226] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 27. | Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, Tokino T. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123-4132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 496] [Cited by in RCA: 497] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 28. | Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529-533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 881] [Cited by in RCA: 898] [Article Influence: 56.1] [Reference Citation Analysis (0)] |
| 29. | Wada T, Kikuchi J, Furukawa Y. Histone deacetylase 1 enhances microRNA processing via deacetylation of DGCR8. EMBO Rep. 2012;13:142-149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 65] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 30. | Wiesen JL, Tomasi TB. Dicer is regulated by cellular stresses and interferons. Mol Immunol. 2009;46:1222-1228. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 166] [Cited by in RCA: 160] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 31. | Dalal SR, Kwon JH. The Role of MicroRNA in Inflammatory Bowel Disease. Gastroenterol Hepatol (N Y). 2010;6:714-722. [PubMed] |
| 32. | Lu LF, Liston A. MicroRNA in the immune system, microRNA as an immune system. Immunology. 2009;127:291-298. [PubMed] |
| 33. | Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4232] [Cited by in RCA: 4144] [Article Influence: 188.4] [Reference Citation Analysis (0)] |
| 34. | Dunne A, O’Neill LA. Adaptor usage and Toll-like receptor signaling specificity. FEBS Lett. 2005;579:3330-3335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 114] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 35. | Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481-12486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3121] [Cited by in RCA: 3548] [Article Influence: 186.7] [Reference Citation Analysis (0)] |
| 36. | O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104:1604-1609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1365] [Cited by in RCA: 1484] [Article Influence: 82.4] [Reference Citation Analysis (0)] |
| 37. | Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082-5089. [PubMed] |
| 38. | Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K, Loeb GB, Lee H, Yoshimura A, Rajewsky K. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80-91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 671] [Cited by in RCA: 677] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 39. | Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608-611. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1554] [Cited by in RCA: 1552] [Article Influence: 86.2] [Reference Citation Analysis (0)] |
| 40. | Ceppi M, Pereira PM, Dunand-Sauthier I, Barras E, Reith W, Santos MA, Pierre P. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc Natl Acad Sci USA. 2009;106:2735-2740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 549] [Cited by in RCA: 588] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 41. | O’Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585-594. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 537] [Cited by in RCA: 560] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 42. | Costinean S, Sandhu SK, Pedersen IM, Tili E, Trotta R, Perrotti D, Ciarlariello D, Neviani P, Harb J, Kauffman LR. Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein beta are targeted by miR-155 in B cells of Emicro-MiR-155 transgenic mice. Blood. 2009;114:1374-1382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 232] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 43. | O’Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci USA. 2009;106:7113-7118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 607] [Cited by in RCA: 676] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 44. | Sly LM, Rauh MJ, Kalesnikoff J, Song CH, Krystal G. LPS-induced upregulation of SHIP is essential for endotoxin tolerance. Immunity. 2004;21:227-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 234] [Cited by in RCA: 247] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 45. | Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O’Leary JJ, Ruan Q, Johnson DS, Chen Y, O’Neill LA. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2010;11:141-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 431] [Cited by in RCA: 459] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 46. | Bazzoni F, Rossato M, Fabbri M, Gaudiosi D, Mirolo M, Mori L, Tamassia N, Mantovani A, Cassatella MA, Locati M. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci USA. 2009;106:5282-5287. [PubMed] |
| 47. | Liu G, Friggeri A, Yang Y, Park YJ, Tsuruta Y, Abraham E. miR-147, a microRNA that is induced upon Toll-like receptor stimulation, regulates murine macrophage inflammatory responses. Proc Natl Acad Sci USA. 2009;106:15819-15824. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 321] [Cited by in RCA: 370] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 48. | O’Neill LA, Sheedy FJ, McCoy CE. MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat Rev Immunol. 2011;11:163-175. [PubMed] |
| 49. | McCoy CE, Sheedy FJ, Qualls JE, Doyle SL, Quinn SR, Murray PJ, O’Neill LA. IL-10 inhibits miR-155 induction by toll-like receptors. J Biol Chem. 2010;285:20492-20498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 207] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 50. | Merkerova M, Belickova M, Bruchova H. Differential expression of microRNAs in hematopoietic cell lineages. Eur J Haematol. 2008;81:304-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 198] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 51. | Monticelli S, Ansel KM, Xiao C, Socci ND, Krichevsky AM, Thai TH, Rajewsky N, Marks DS, Sander C, Rajewsky K. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 2005;6:R71. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 345] [Cited by in RCA: 358] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 52. | Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, Manjunath N. miRNA profiling of naive, effector and memory CD8 T cells. PLoS One. 2007;2:e1020. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 371] [Cited by in RCA: 404] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 53. | Cobb BS, Nesterova TB, Thompson E, Hertweck A, O’Connor E, Godwin J, Wilson CB, Brockdorff N, Fisher AG, Smale ST. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201:1367-1373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 396] [Cited by in RCA: 410] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 54. | Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261-269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 494] [Cited by in RCA: 508] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 55. | Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405-414. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1020] [Cited by in RCA: 993] [Article Influence: 58.4] [Reference Citation Analysis (0)] |
| 56. | Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, Li Z, Wu Z, Pei G. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10:1252-1259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 568] [Cited by in RCA: 618] [Article Influence: 38.6] [Reference Citation Analysis (0)] |
| 57. | Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007;445:936-940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 624] [Cited by in RCA: 673] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 58. | Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1145] [Cited by in RCA: 1199] [Article Influence: 66.6] [Reference Citation Analysis (0)] |
| 59. | Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006-1010. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1361] [Cited by in RCA: 1294] [Article Influence: 80.9] [Reference Citation Analysis (0)] |
| 60. | Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457-468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 873] [Cited by in RCA: 976] [Article Influence: 61.0] [Reference Citation Analysis (0)] |
| 61. | Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 967] [Cited by in RCA: 938] [Article Influence: 52.1] [Reference Citation Analysis (0)] |
| 62. | Chong MM, Rasmussen JP, Rudensky AY, Littman DR. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J Exp Med. 2008;205:2005-2017. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 308] [Cited by in RCA: 314] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 63. | Liston A, Lu LF, O’Carroll D, Tarakhovsky A, Rudensky AY. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J Exp Med. 2008;205:1993-2004. [PubMed] |
| 64. | Zhou X, Jeker LT, Fife BT, Zhu S, Anderson MS, McManus MT, Bluestone JA. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med. 2008;205:1983-1991. [PubMed] |
| 65. | Tan LP, Wang M, Robertus JL, Schakel RN, Gibcus JH, Diepstra A, Harms G, Peh SC, Reijmers RM, Pals ST. miRNA profiling of B-cell subsets: specific miRNA profile for germinal center B cells with variation between centroblasts and centrocytes. Lab Invest. 2009;89:708-716. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 88] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 66. | Basso K, Sumazin P, Morozov P, Schneider C, Maute RL, Kitagawa Y, Mandelbaum J, Haddad J, Chen CZ, Califano A. Identification of the human mature B cell miRNome. Immunity. 2009;30:744-752. [PubMed] |
| 67. | Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26-36. [PubMed] |
| 68. | Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2463] [Cited by in RCA: 2548] [Article Influence: 115.8] [Reference Citation Analysis (0)] |
| 69. | Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, Cobb BS, Merkenschlager M, Rajewsky N. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860-874. [PubMed] |
| 70. | Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875-886. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1370] [Cited by in RCA: 1311] [Article Influence: 77.1] [Reference Citation Analysis (1)] |
| 71. | Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci USA. 2007;104:7080-7085. [PubMed] |
| 72. | Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146-159. [PubMed] |
| 73. | Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847-859. [PubMed] |
| 74. | Rao DS, O’Connell RM, Chaudhuri AA, Garcia-Flores Y, Geiger TL, Baltimore D. MicroRNA-34a perturbs B lymphocyte development by repressing the forkhead box transcription factor Foxp1. Immunity. 2010;33:48-59. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 198] [Cited by in RCA: 197] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 75. | Hu H, Wang B, Borde M, Nardone J, Maika S, Allred L, Tucker PW, Rao A. Foxp1 is an essential transcriptional regulator of B cell development. Nat Immunol. 2006;7:819-826. [PubMed] |
| 76. | Blumberg R, Cho J, Lewis J, Wu G. Inflammatory bowel disease: an update on the fundamental biology and clinical management. Gastroenterology. 2011;140:1701-1703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 77. | Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, Brant SR, Chakravarti S, Kwon JH. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008;135:1624-1635.e24. [PubMed] |
| 78. | Takagi T, Naito Y, Mizushima K, Hirata I, Yagi N, Tomatsuri N, Ando T, Oyamada Y, Isozaki Y, Hongo H. Increased expression of microRNA in the inflamed colonic mucosa of patients with active ulcerative colitis. J Gastroenterol Hepatol. 2010;25 Suppl 1:S129-S133. [PubMed] |
| 79. | Fasseu M, Tréton X, Guichard C, Pedruzzi E, Cazals-Hatem D, Richard C, Aparicio T, Daniel F, Soulé JC, Moreau R. Identification of restricted subsets of mature microRNA abnormally expressed in inactive colonic mucosa of patients with inflammatory bowel disease. PLoS One. 2010;5:e13160. [PubMed] |
| 80. | Wu F, Guo NJ, Tian H, Marohn M, Gearhart S, Bayless TM, Brant SR, Kwon JH. Peripheral blood microRNAs distinguish active ulcerative colitis and Crohn’s disease. Inflamm Bowel Dis. 2011;17:241-250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 188] [Cited by in RCA: 191] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 81. | Okubo M, Tahara T, Shibata T, Yamashita H, Nakamura M, Yoshioka D, Yonemura J, Kamiya Y, Ishizuka T, Nakagawa Y. Association study of common genetic variants in pre-microRNAs in patients with ulcerative colitis. J Clin Immunol. 2011;31:69-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 67] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 82. | Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, Mari B, Barbry P, Mosnier JF, Hébuterne X. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nat Genet. 2011;43:242-245. [PubMed] |
| 83. | Dalmasso G, Nguyen HT, Yan Y, Laroui H, Charania MA, Ayyadurai S, Sitaraman SV, Merlin D. Microbiota modulate host gene expression via microRNAs. PLoS One. 2011;6:e19293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 143] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 84. | Dalmasso G, Nguyen HT, Yan Y, Laroui H, Srinivasan S, Sitaraman SV, Merlin D. MicroRNAs determine human intestinal epithelial cell fate. Differentiation. 2010;80:147-154. [PubMed] |
| 85. | Nguyen HT, Merlin D. Homeostatic and innate immune responses: role of the transmembrane glycoprotein CD98. Cell Mol Life Sci. 2012;69:3015-3026. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 86. | Nguyen HT, Dalmasso G, Yan Y, Laroui H, Dahan S, Mayer L, Sitaraman SV, Merlin D. MicroRNA-7 modulates CD98 expression during intestinal epithelial cell differentiation. J Biol Chem. 2010;285:1479-1489. [PubMed] |
| 87. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [PubMed] |
| 88. | Weitz J, Koch M, Debus J, Höhler T, Galle PR, Büchler MW. Colorectal cancer. Lancet. 2005;365:153-165. [PubMed] |
| 89. | Dalmay T, Edwards DR. MicroRNAs and the hallmarks of cancer. Oncogene. 2006;25:6170-6175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 275] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 90. | Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849-864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 730] [Cited by in RCA: 794] [Article Influence: 56.7] [Reference Citation Analysis (0)] |
| 91. | Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999-3004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2908] [Cited by in RCA: 3110] [Article Influence: 148.1] [Reference Citation Analysis (0)] |
| 92. | Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257-2261. [PubMed] |
| 93. | Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7723] [Cited by in RCA: 7370] [Article Influence: 368.5] [Reference Citation Analysis (0)] |
| 94. | Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647-658. [PubMed] |
| 95. | Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425-436. [PubMed] |
| 96. | Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128-2136. [PubMed] |
| 97. | Chang KH, Miller N, Kheirelseid EA, Ingoldsby H, Hennessy E, Curran CE, Curran S, Smith MJ, Regan M, McAnena OJ. MicroRNA-21 and PDCD4 expression in colorectal cancer. Eur J Surg Oncol. 2011;37:597-603. [PubMed] |
| 98. | Shibuya H, Iinuma H, Shimada R, Horiuchi A, Watanabe T. Clinicopathological and prognostic value of microRNA-21 and microRNA-155 in colorectal cancer. Oncology. 2010;79:313-320. [PubMed] |
| 99. | Gironella M, Seux M, Xie MJ, Cano C, Tomasini R, Gommeaux J, Garcia S, Nowak J, Yeung ML, Jeang KT. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci USA. 2007;104:16170-16175. [PubMed] |
| 100. | Nishida N, Nagahara M, Sato T, Mimori K, Sudo T, Tanaka F, Shibata K, Ishii H, Sugihara K, Doki Y. Microarray analysis of colorectal cancer stromal tissue reveals upregulation of two oncogenic miRNA clusters. Clin Cancer Res. 2012;18:3054-3070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 140] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 101. | Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219-230. [PubMed] |
| 102. | Hudson RS, Yi M, Esposito D, Glynn SA, Starks AM, Yang Y, Schetter AJ, Watkins SK, Hurwitz AA, Dorsey TH. MicroRNA-106b-25 cluster expression is associated with early disease recurrence and targets caspase-7 and focal adhesion in human prostate cancer. Oncogene. 2012;Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 101] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 103. | Petrocca F, Vecchione A, Croce CM. Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 2008;68:8191-8194. [PubMed] |
| 104. | Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217-222. [PubMed] |
| 105. | Tsuchida A, Ohno S, Wu W, Borjigin N, Fujita K, Aoki T, Ueda S, Takanashi M, Kuroda M. miR-92 is a key oncogenic component of the miR-17-92 cluster in colon cancer. Cancer Sci. 2011;102:2264-2271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 192] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 106. | Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060-1065. [PubMed] |
| 107. | Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008;79:581-588. [PubMed] |
| 108. | Motoyama K, Inoue H, Takatsuno Y, Tanaka F, Mimori K, Uetake H, Sugihara K, Mori M. Over- and under-expressed microRNAs in human colorectal cancer. Int J Oncol. 2009;34:1069-1075. [PubMed] |
| 109. | Schimanski CC, Frerichs K, Rahman F, Berger M, Lang H, Galle PR, Moehler M, Gockel I. High miR-196a levels promote the oncogenic phenotype of colorectal cancer cells. World J Gastroenterol. 2009;15:2089-2096. [PubMed] |
| 110. | Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, Nenutil R, Vyzula R. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397-402. [PubMed] |
| 111. | Schee K, Fodstad Ø, Flatmark K. MicroRNAs as biomarkers in colorectal cancer. Am J Pathol. 2010;177:1592-1599. [PubMed] |
| 112. | Zhou W, Li X, Liu F, Xiao Z, He M, Shen S, Liu S. MiR-135a promotes growth and invasion of colorectal cancer via metastasis suppressor 1 in vitro. Acta Biochim Biophys Sin (Shanghai). 2012;44:838-846. [PubMed] |
| 113. | Esquela-Kerscher A, Slack FJ. OncomiRs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259-269. [PubMed] |
| 114. | Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065-7070. [PubMed] |
| 115. | Michael MZ, O’ Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882-891. [PubMed] |
| 116. | Schepeler T, Reinert JT, Ostenfeld MS, Christensen LL, Silahtaroglu AN, Dyrskjøt L, Wiuf C, Sørensen FJ, Kruhøffer M, Laurberg S. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68:6416-6424. [PubMed] |
| 117. | Xu Q, Liu LZ, Qian X, Chen Q, Jiang Y, Li D, Lai L, Jiang BH. MiR-145 directly targets p70S6K1 in cancer cells to inhibit tumor growth and angiogenesis. Nucleic Acids Res. 2012;40:761-774. [PubMed] |
| 118. | Chen X, Guo X, Zhang H, Xiang Y, Chen J, Yin Y, Cai X, Wang K, Wang G, Ba Y. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28:1385-1392. [PubMed] |
| 119. | Liu LZ, Zheng JZ, Wang XR, Jiang BH. Endothelial p70 S6 kinase 1 in regulating tumor angiogenesis. Cancer Res. 2008;68:8183-8188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 42] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 120. | Skinner HD, Zheng JZ, Fang J, Agani F, Jiang BH. Vascular endothelial growth factor transcriptional activation is mediated by hypoxia-inducible factor 1alpha, HDM2, and p70S6K1 in response to phosphatidylinositol 3-kinase/AKT signaling. J Biol Chem. 2004;279:45643-45651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 164] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 121. | Li W, Tan D, Zhang Z, Liang JJ, Brown RE. Activation of Akt-mTOR-p70S6K pathway in angiogenesis in hepatocellular carcinoma. Oncol Rep. 2008;20:713-719. [PubMed] |
| 122. | Zou C, Xu Q, Mao F, Li D, Bian C, Liu LZ, Jiang Y, Chen X, Qi Y, Zhang X. MiR-145 inhibits tumor angiogenesis and growth by N-RAS and VEGF. Cell Cycle. 2012;11:2137-2145. [PubMed] |
| 123. | Nugent M, Miller N, Kerin MJ. Circulating miR-34a levels are reduced in colorectal cancer. J Surg Oncol. 2012;106:947-952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 64] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 124. | Tsuchiya N, Nakagama H. MicroRNA, SND1, and alterations in translational regulation in colon carcinogenesis. Mutat Res. 2010;693:94-100. [PubMed] |
| 125. | Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193-199. [PubMed] |
| 126. | Chiang Y, Song Y, Wang Z, Chen Y, Yue Z, Xu H, Xing C, Liu Z. Aberrant expression of miR-203 and its clinical significance in gastric and colorectal cancers. J Gastrointest Surg. 2011;15:63-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 78] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 127. | Li J, Chen Y, Zhao J, Kong F, Zhang Y. miR-203 reverses chemoresistance in p53-mutated colon cancer cells through downregulation of Akt2 expression. Cancer Lett. 2011;304:52-59. [PubMed] |
| 128. | Díaz R, Silva J, García JM, Lorenzo Y, García V, Peña C, Rodríguez R, Muñoz C, García F, Bonilla F. Deregulated expression of miR-106a predicts survival in human colon cancer patients. Genes Chromosomes Cancer. 2008;47:794-802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 183] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 129. | Guo C, Sah JF, Beard L, Willson JK, Markowitz SD, Guda K. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer. 2008;47:939-946. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 313] [Cited by in RCA: 329] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 130. | Bader AG, Brown D, Winkler M. The promise of microRNA replacement therapy. Cancer Res. 2010;70:7027-7030. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 489] [Cited by in RCA: 486] [Article Influence: 32.4] [Reference Citation Analysis (0)] |
| 131. | Duchaine TF, Slack FJ. RNA interference and micro RNA-oriented therapy in cancer: rationales, promises, and challenges. Curr Oncol. 2009;16:61-66. [PubMed] |
| 132. | Melo S, Villanueva A, Moutinho C, Davalos V, Spizzo R, Ivan C, Rossi S, Setien F, Casanovas O, Simo-Riudalbas L. Small molecule enoxacin is a cancer-specific growth inhibitor that acts by enhancing TAR RNA-binding protein 2-mediated microRNA processing. Proc Natl Acad Sci USA. 2011;108:4394-4399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 197] [Article Influence: 14.1] [Reference Citation Analysis (0)] |