Published online Apr 28, 2013. doi: 10.3748/wjg.v19.i16.2449
Revised: March 6, 2013
Accepted: March 22, 2013
Published online: April 28, 2013
Processing time: 153 Days and 11.9 Hours
Interleukin-6 (IL-6) is a pleiotropic cytokine which is expressed in many inflammatory cells in response to different types of stimuli, regulating a number of biological processes. The IL-6 gene is polymorphic in both the 5’ and 3’ flanking regions and more than 150 single nucleotide polymorphisms have been identified so far. Genetic polymorphisms of IL-6 may affect the outcomes of several diseases, where the presence of high levels of circulating IL-6 have been correlated to the stage and/or the progression of the disease itself. The -174 G/C polymorphism is a frequent polymorphism, that is located in the upstream regulatory region of the IL-6 gene and affects IL-6 production. However, the data in the literature on the genetic association between the -174 G/C polymorphism and some specific liver diseases characterized by different etiologies are still controversial. In particular, most of the studies are quite unanimous in describing a correlation between the presence of the high-producer genotype and a worse evolution of the chronic liver disease. This is valid for patients with hepatitis C virus (HCV)-related chronic hepatitis and liver cirrhosis and hepatocellular carcinoma (HCC) whatever the etiology. Studies in hepatitis B virus-related chronic liver diseases are not conclusive, while specific populations like non alcoholic fatty liver disease/non-alcoholic steatohepatitis, autoimmune and human immunodeficiency virus/HCV co-infected patients show a higher prevalence of the low-producer genotype, probably due to the complexity of these clinical pictures. In this direction, a systematic revision of these data should shed more light on the role of this polymorphism in chronic liver diseases and HCC.
Core tip: Several studies suggested the possibility of an association between -174 interleukin-6 gene G/C polymorphism and some liver diseases however, the data in the literature are still controversial. This work aims to review the literature data on the role of this polymorphism and its possible biological function in chronic liver diseases and hepatocellular carcinoma.
- Citation: Giannitrapani L, Soresi M, Balasus D, Licata A, Montalto G. Genetic association of interleukin-6 polymorphism (-174 G/C) with chronic liver diseases and hepatocellular carcinoma. World J Gastroenterol 2013; 19(16): 2449-2455
- URL: https://www.wjgnet.com/1007-9327/full/v19/i16/2449.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i16.2449
In recent decades chronic liver diseases (chronic hepatitis, liver cirrhosis) and hepatocellular carcinoma have become more and more diffuse both in Western and in Eastern countries, representing an important problem for health systems worldwide[1-5]. Whatever the etiology, these diseases share a common pathogenetic mechanism which is linked to chronic inflammation[6]. Hepatotropic viruses, toxins and alcohol, metabolic liver disease or autoimmunity can be the triggers which, acting chronically in the liver, ultimately activate cellular pathways involving transcription factors of the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) family and signal transducer and activator of transcription 3 (STAT3), as well as cytokines such as interleukin-6 (IL-6) and IL-1α, etc.
In particular, IL-6 is a cytokine involved in the regulation of several cellular processes including proliferation and differentiation and plays a pivotal role in acute phase response and in the control of the balance between pro-inflammatory and anti-inflammatory pathways. The IL-6 gene is located on chromosome 7p21[7]. A number of studies indicated that the presence of a G/C single nucleotide polymorphism (SNP) at the promoter -174 of the IL-6 gene, one of the numerous known polymorphisms in the IL-6 gene, is related to the IL-6 gene transcription rate and, as a consequence, to the control of circulating IL-6 levels[8,9].
Subsequently, two phenotypes for this polymorphism were identified: the high-producer phenotype, including the -174 G/G and -174 G/C genotypes, characterized by higher circulating IL-6 levels; and the low-producer phenotype, including the -174 C/C genotype[8]. Genetic population studies have shown that there are ethnic differences in the frequency of the -174 G allele, with higher frequencies in non-Caucasian than in Caucasian populations[10,11].
High circulating levels of IL-6 have been documented in several clinical conditions (inflammatory, neoplastic diseases) and in particular in various liver diseases such as viral chronic hepatitis[12], alcoholic liver disease[13], liver cirrhosis and hepatocellular carcinoma (HCC)[14]. A small number of studies have investigated a possible correlation between the presence of the -174 G/C polymorphism, IL-6 circulating levels and the stage of disease[15]. However, the results of these studies are quite controversial. This work aims to review the literature data on the role of G/C base exchange at position -174 of the IL-6 gene and its possible biological function in chronic liver diseases and HCC.
Produced by a variety of cells such as macrophages, B and T cells and fibroblasts, IL-6 plays a central role in the inflammatory response associated with the course of chronic hepatitis due to hepatitis C virus (HCV)- and hepatitis B virus (HBV)-related infection[16,17].
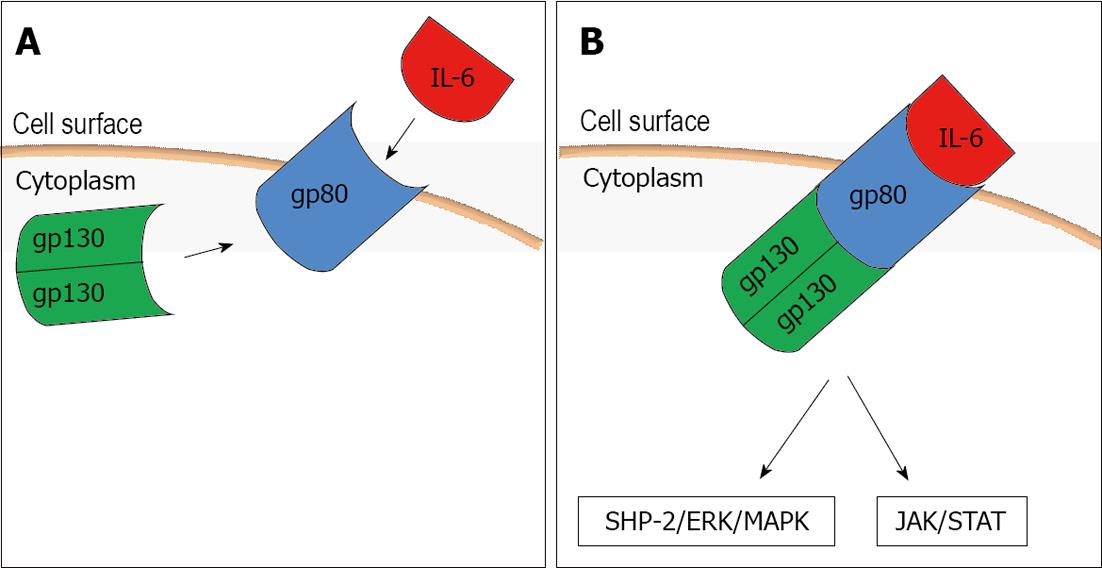
To mediate its biological effects it interacts with a receptor complex consisting of a specific ligand-binding protein (IL-6R, gp80) and a signal transduction protein (gp130) (Figure 1A). When IL-6 binds its cell surface receptor (IL-6R) on the hepatocyte a homodimer of the signal transduction receptor gp130 is recruited to the complex and it activates a janus kinase 1 which in turn triggers two main signaling pathways: the gp130 Tyr759-derived Src homology 2 domain-containing protein tyrosine phosphatase-2/extracellular-signal-regulated kinase/mitogen-activated protein kinase pathway and the gp130 YXXQ-mediated Janus associated kinase/signal transducer and activator of transcription pathway (Figure 1B). Interestingly, sIL-6R (soluble form of IL-6R) also binds with IL-6, and the IL-6-sIL-6R complex can then form a complex with gp130[18,19]. Through this receptor system IL-6 can influence various cell types and exert its multiple biological activities regulating immune response, acute phase response and inflammation.
During HCV infection, an altered production of cytokines seems to be related to viral persistence and to affect response to therapy. Barret et al[20] comparing various cytokine polymorphisms (including -174 G/C IL-6) in individuals with spontaneous viral clearance after HCV infection and in those with persistent viremia, reported that the CC genotype with low IL-6 production was associated with spontaneous viral clearance, while an association between the high IL-6 producer genotypes and persistent infection only became apparent when both genotypes (GG and GC) were combined. As regards the influence of the genetic background in individuals with HCV infection and response to the antiviral therapy, the most recent literature data have investigated the role of IL-28B polymorphisms as a predictor of the outcome of the commonly-used treatments[21]. However, because of the central importance of IL-6 as a mediator of the immune response to infectious agents, and considering that host genetic variation, and in particular haplotypes, may affect IL-6 expression, Yee et al[22] examined the contribution of haplotypes in the IL-6 gene to therapy for chronic HCV infection on sustained viral response (SVR). Among the SNPs genotyped and included in haplotype construction, the authors found some SNPs (including -174G/C, and in particular genotypes GG and GC) showing significant associations with a reduced likelihood of SVR.
These results are in contrast with previously reported ones published by Nattermanne et al[23] which, however, were obtained in another specific population, i.e., patients co-infected with both acute and chronic HCV and human immunodeficiency virus (HIV). The aim of this study was to evaluate whether IL-6 -174 G/C polymorphism could affect response to antiviral treatment in HCV-infected HIV-positive patients. The study group was compared to a group of HCV- and a group of HIV-monoinfected patients, as well as to a group of healthy individuals and no significant difference was found in the distribution of IL-6 genotypes between the study groups. However, the authors concluded that carriers of high-producer genotypes (genotypes IL-6 174 GG and 174 GC) had significantly higher SVR rates than patients with an IL-6 low-producer genotype (genotype 174 CC).
Another particular subgroup of HCV patients is the one with persistently normal or near normal alanine aminotransferases levels (PNALT), which for several years was supposed to have a milder course of disease, whereas it is now well known that in a few cases it can evolve to cirrhosis[24]. Among the studies evaluating genetic polymorphisms in chronic HCV carriers with PNALT, Falleti et al[25] evaluated the role of five IL-6 polymorphisms (among them -174 G/C) in modulating fibrosis progression in PNALT patients with chronic HCV infection. The principal point of interest in this study were the associations found between IL-6 polymorphisms and grading and staging increase during the follow-up of the patients with chronic viral hepatitis C and PNALT. In particular, grading increase appeared to be related to the presence of the G allele of the IL-6 -174G/C polymorphism, while the C allele seemed to be protective.
As cytokines also play a fundamental role in the immune response to HBV and HBV infection may have different forms of evolution (self-limited or persistent and progressive), IL-6 polymorphisms have also been studied to investigate a possible correlation between IL-6 promoter variants and chronic hepatitis B progression, infection evolution in adult patients and risk of HCC development. Unfortunately, the data reported by Park et al[26] are not conclusive because in their attempt to analyze additional polymorphisms in variants of genes implicated in chronic hepatitis B progression they found that Koreans and Caucasians had different genetic backgrounds in terms of the allele frequencies of the IL-6 promoter SNPs. In particular, in their study the allele frequencies reported in Caucasians (range: 0.40-0.45) were much higher than those found in Koreans (allele frequencies 0.002). The authors concluded that at least in their population, although IL-6 may have important functions in the progression of chronic HBV infection, its genetic variants probably do not influence the development of LC and HCC from chronic HBV infection, due to too low frequencies of IL-6 174 G/C.
Another attempt to correlate cytokine genetic polymorphism with hepatitis B infection evolution was made in a Brazilian population, but the study found no significant differences in the polymorphism of IL-6 -174 between the chronic HBV patient group and the self-limited infection group as regards alleles, genotypes or phenotypic expression[27].
Similarly, the study of a Japanese population by Migita et al[28] with the aim of characterizing cytokine gene polymorphisms in chronic HBV infection and their associations with HCC, was unable to show conclusive data about the role of IL-6 -174 because no polymorphisms were found at that position (Table 1).
| Ref. | Country | Ethnicity | Cases | Controls | Genotyping method | Association with chronic hepatitis/response to therapy |
| Barrett et al[20] | Ireland | Caucasian | 158 | - | PCR-SSP | Positive significant/- |
| Nattermann et al[23] | Germany | Caucasian | 210 | 100 | Cytokine genotyping tray | -/Uncertain |
| Falleti et al[25] | Italy | Caucasian | 121 | - | PCR-RFLP | Positive significant /- |
| Park et al[26] | South Korea | Asian | 1046 | - | PCR-SBE | NS/- |
| Ribeiro et al[27] | Brazil | American1 | 26 | 41 | PCR-SSP | NS/- |
Non-alcoholic fatty liver disease (NAFLD) includes a broad spectrum of clinic-pathological entities, including simple steatosis and non-alcoholic steatohepatitis (NASH), which can progress to advanced liver diseases[29]. Its pathogenesis is strictly linked to insulin resistance and to all the mechanisms described for the development of metabolic syndrome, and in this perspective an important role is played by genetic background[30,31]. It is well known that the balance between pro- and anti-inflammatory acting cytokines is fundamental in the control of hepatic and systemic insulin action, and as a consequence, in the development of NAFLD. In particular, serum levels of this cytokine correlate remarkably well with the presence of insulin resistance, and adipose tissue-derived IL-6 has been shown to regulate hepatic insulin resistance via up-regulation of suppressor of cytokine signaling 3[32]. However, the role of -174 G/C polymorphism in this population raises some questions. In fact, a study by Carulli et al[33] found that the IL-6 -174C variant, is significantly more prevalent in NAFLD than in healthy subjects, is associated with increased fasting insulin and homeostasis model assessment of insulin resistance, and is an independent predictor of NAFLD and NASH. This finding is in contrast with other studies which showed that the IL-6 -174G variant was associated with lipid abnormalities[34] and with diabetes in Caucasians as well as Pima Indians[35-38] and that the C allele at -174 position was unlikely to play a role in the development of type 2 diabetes mellitus in a Taiwanese population[39]. One possible explanation for these contradictory results can be found in the conclusion of a study on an experimental mouse model of ASH and NAFLD: IL-10-/- mice were prone to liver inflammatory response but resistant to steatosis and hepatocellular damage induced by ethanol or high-fat diet feeding, thanks to the elevation of inflammation-associated hepatic IL-6/STAT3 activation that subsequently down-regulated lipogenic genes, but up-regulated fatty acid oxidation-associated genes in the liver[40].
In an attempt to explain why only a minority of heavy drinkers develop alcoholic liver cirrhosis or alcohol use disorders, some genetic factors have been considered[41,42], such as polymorphisms of genes encoding cytokines. Several studies support the hypothesis of a pivotal role of ethanol-induced cytokine changes in contributing to alcohol pathogenesis in a number of tissues, including the liver[43-45]. Moreover, elevated serum concentrations of pro-inflammatory cytokines such as tumor necrosis factor-α, IL-1, IL-6 and IL-8 and decreased levels of anti-inflammatory cytokines like IL-10 have been shown in patients with this disease[43,46,47]. However, the only study on common polymorphisms in interleukin genes (including -174G/C IL-6) in a population of Spanish alcoholic patients did not find any statistically significant associations between any of the studied polymorphisms or the combinations of pro-inflammatory polymorphisms and the risk of alcoholic liver cirrhosis or alcohol abuse or dependence[48].
Autoimmune hepatitis (AIH), primary biliary cirrhosis (PBC), and primary sclerosing cholangitis represent the three main categories of autoimmune liver diseases. However, their etiology and possible environmental triggers still remain obscure even if it is well established that a complex genetic background contributes to disease susceptibility and severity. Several studies have established that genetic factors are involved in the pathogenesis of autoimmune liver diseases[49-52]. Among these studies, one in a Chinese population of patients with AIH and PBC found that frequency of IL-6 -174C was high and significantly increased in PBC patients compared with controls. This result supports the hypothesis that the IL-6 -174G/C polymorphism could contribute to the change in susceptibility to PBC in some subjects[53] (Table 2).
| Ref. | Country | Ethnicity | Cases | Controls | Genotyping method | Association |
| Carulli et al[33] | Italy | Caucasian | 79 | 114 | PCR-RFLP | Positive significant (NAFLD) |
| Fernández-Real et al[34] | Spain | Caucasian | 32 | - | PCR-RFLP | Positive significant (diabetes and lipid abnormalities) (G allele) |
| Marcos et al[48] | Spain | Caucasian | 258 | 101 | TaqMan genotyping | NS (alcoholic liver disease) |
| Fan et al[53] | China | Asian | 77 | - | PCR-RFLP | Positive significant (PBC) |
An interrelation between chronic inflammation and cancer has been suspected for a long time[54]. Many tumors occur in association with chronic infectious diseases and persistent inflammation increases the risk and accelerates the development of cancer[55-59]. HCC is one of the most clear examples of inflammation-related cancer[60,61]. It is a tumor that slowly progresses through a chronic inflammation state, triggered by exposure to various agents. The molecular links that connect inflammation and cancer are not completely known, although there is a consistent body of evidence pointing to the role of transcription factors such as NF-κB[62] and STAT3[63] and cytokines like IL-6[64] as well as other inflammatory mediators in HCC development. A first attempt to study the potential role of cytokine polymorphisms in determining the risk of HBV-related HCC was made in 2005 by Nieters et al[65], who examined the correlation between polymorphisms in Th1 and Th2 cytokine genes in a group of 250 patients with incident HCC and a group of 250 matched hospitalized controls in China: however, none of the study participants presented the C allele of the IL-6 -174 G/C polymorphism, therefore this polymorphism was not further investigated. Subsequently, a population-based case-control study of HCC, including 120 HCC patients and 230 matched control subjects, was conducted in non-Asian residents of Los Angeles County, California, into genetic polymorphisms in the cytokine genes and risk of HCC. The authors demonstrated that the GG IL-6 genotype showed the strongest influence on HCC risk among all the cytokine polymorphisms studied[66]. In a more recent study Falleti et al[25] investigated whether IL-6 polymorphisms could be associated with the occurrence of HCC in patients with liver cirrhosis, analyzing 219 consecutive patients who underwent liver transplantation for liver cirrhosis. They found a significant association between the presence of the low-producer genotype (-174 CC) and absence of HCC[67]. Finally, our group performed a study which aimed to evaluate the frequency of SNPs in the IL-6 promoter region at position -174 and IL-6 serum levels in a group of patients with HCC and underlying liver cirrhosis compared with a group of LC patients without HCC. We found that IL-6 serum levels were higher in G/G compared to C/C genotypes only in HCC; IL-6 serum levels in G carriers were higher in HCC versus LC patients while there were no differences for the C allele. IL-6 serum levels in HCC correlated with G carriers[15] (Table 3).
| Ref. | Country | Ethnicity | Cases | Controls | Genotyping method | Association with HCC |
| Nieters et al[65] | China | Asian | 250 | 250 | PCR-RFLP | NS |
| Ognjanovic et al[66] | United States | American1 | 120 | 230 | 5’nuclease Taqman allelic discrimination assay | Positive significant |
| Falleti et al[67] | Italy | Caucasian | 219 | - | PCR-RFLP | Positive significant |
| Giannitrapani et al[15] | Italy | Caucasian | 105 | - | PCR-RFLP | Positive significant |
The possibility of a genetic association between -174 G/C polymorphism and some specific liver diseases has been suggested by several studies which are quite unanimous in observing a correlation between the presence of the high-producer genotype (GG) and a worse evolution of the chronic disease. This has been observed in patients with HCV-related chronic hepatitis even with PNALT and in patients with liver cirrhosis and HCC whatever the etiology. Studies on HBV-related chronic hepatitis have not been conclusive because they were performed in populations (generally Asiatic) which have much lower frequencies of the -174 C allele than Caucasian populations. Finally, specific populations like NAFLD/NASH, autoimmune and HIV/HCV co-infected patients not achieving SVR showed a higher prevalence of the CC genotype, probably as a result of many other complex immunological, virological and host-related interrelations that cannot be explained by the presence of a unique SNP.
The authors are grateful to Carole Greenall (BA) for the English revision of this manuscript.
P- Reviewer Margarete O S- Editor Huang XZ L- Editor A E- Editor Xiong L
| 1. | Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558-567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1927] [Cited by in RCA: 1931] [Article Influence: 96.6] [Reference Citation Analysis (0)] |
| 2. | Averhoff FM, Glass N, Holtzman D. Global burden of hepatitis C: considerations for healthcare providers in the United States. Clin Infect Dis. 2012;55 Suppl 1:S10-S15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 215] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 3. | Spradling PR, Rupp L, Moorman AC, Lu M, Teshale EH, Gordon SC, Nakasato C, Boscarino JA, Henkle EM, Nerenz DR. Hepatitis B and C virus infection among 1.2 million persons with access to care: factors associated with testing and infection prevalence. Clin Infect Dis. 2012;55:1047-1055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 138] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 4. | Ott JJ, Stevens GA, Groeger J, Wiersma ST. Global epidemiology of hepatitis B virus infection: new estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine. 2012;30:2212-2219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1217] [Cited by in RCA: 1328] [Article Influence: 102.2] [Reference Citation Analysis (0)] |
| 5. | McGlynn KA, London WT. The global epidemiology of hepatocellular carcinoma: present and future. Clin Liver Dis. 2011;15:223-243, vii-x. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 360] [Cited by in RCA: 377] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 6. | Alison MR, Nicholson LJ, Lin WR. Chronic inflammation and hepatocellular carcinoma. Recent Results Cancer Res. 2011;185:135-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 53] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 7. | Bowcock AM, Kidd JR, Lathrop GM, Daneshvar L, May LT, Ray A, Sehgal PB, Kidd KK, Cavalli-Sforza LL. The human “interferon-beta 2/hepatocyte stimulating factor/interleukin-6” gene: DNA polymorphism studies and localization to chromosome 7p21. Genomics. 1988;3:8-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 108] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 8. | Fishman D, Faulds G, Jeffery R, Mohamed-Ali V, Yudkin JS, Humphries S, Woo P. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest. 1998;102:1369-1376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1591] [Cited by in RCA: 1663] [Article Influence: 61.6] [Reference Citation Analysis (0)] |
| 9. | Olomolaiye O, Wood NA, Bidwell JL. A novel polymorphism in the human IL-6 promoter. Eur J Immunogenet. 1998;25:267. [RCA] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 10. | Cox ED, Hoffmann SC, DiMercurio BS, Wesley RA, Harlan DM, Kirk AD, Blair PJ. Cytokine polymorphic analyses indicate ethnic differences in the allelic distribution of interleukin-2 and interleukin-6. Transplantation. 2001;72:720-726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 124] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 11. | Meenagh A, Williams F, Ross OA, Patterson C, Gorodezky C, Hammond M, Leheny WA, Middleton D. Frequency of cytokine polymorphisms in populations from western Europe, Africa, Asia, the Middle East and South America. Hum Immunol. 2002;63:1055-1061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 130] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 12. | Spanakis NE, Garinis GA, Alexopoulos EC, Patrinos GP, Menounos PG, Sklavounou A, Manolis EN, Gorgoulis VG, Valis D. Cytokine serum levels in patients with chronic HCV infection. J Clin Lab Anal. 2002;16:40-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 13. | Martinez F, Abril ER, Earnest DL, Watson RR. Ethanol and cytokine secretion. Alcohol. 1992;9:455-458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 52] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 14. | Soresi M, Giannitrapani L, D’Antona F, Florena AM, La Spada E, Terranova A, Cervello M, D’Alessandro N, Montalto G. Interleukin-6 and its soluble receptor in patients with liver cirrhosis and hepatocellular carcinoma. World J Gastroenterol. 2006;12:2563-2568. [PubMed] |
| 15. | Giannitrapani L, Soresi M, Giacalone A, Campagna ME, Marasà M, Cervello M, Marasà S, Montalto G. IL-6 -174G/C polymorphism and IL-6 serum levels in patients with liver cirrhosis and hepatocellular carcinoma. OMICS. 2011;15:183-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 16. | Tacke RS, Tosello-Trampont A, Nguyen V, Mullins DW, Hahn YS. Extracellular hepatitis C virus core protein activates STAT3 in human monocytes/macrophages/dendritic cells via an IL-6 autocrine pathway. J Biol Chem. 2011;286:10847-10855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 81] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 17. | Kao JT, Lai HC, Tsai SM, Lin PC, Chuang PH, Yu CJ, Cheng KS, Su WP, Hsu PN, Peng CY. Rather than interleukin-27, interleukin-6 expresses positive correlation with liver severity in naïve hepatitis B infection patients. Liver Int. 2012;32:928-936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 41] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 18. | Heinrich PC, Behrmann I, Haan S, Hermanns HM, Müller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. 2003;374:1-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2296] [Cited by in RCA: 2407] [Article Influence: 109.4] [Reference Citation Analysis (0)] |
| 19. | Mihara M, Hashizume M, Yoshida H, Suzuki M, Shiina M. IL-6/IL-6 receptor system and its role in physiological and pathological conditions. Clin Sci (Lond). 2012;122:143-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 461] [Cited by in RCA: 628] [Article Influence: 44.9] [Reference Citation Analysis (0)] |
| 20. | Barrett S, Collins M, Kenny C, Ryan E, Keane CO, Crowe J. Polymorphisms in tumour necrosis factor-alpha, transforming growth factor-beta, interleukin-10, interleukin-6, interferon-gamma, and outcome of hepatitis C virus infection. J Med Virol. 2003;71:212-218. [PubMed] |
| 21. | Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2776] [Cited by in RCA: 2723] [Article Influence: 170.2] [Reference Citation Analysis (0)] |
| 22. | Yee LJ, Im K, Borg B, Yang H, Liang TJ. Interleukin-6 haplotypes and the response to therapy of chronic hepatitis C virus infection. Genes Immun. 2009;10:365-372. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 23. | Nattermann J, Vogel M, Berg T, Danta M, Axel B, Mayr C, Bruno R, Tural C, Klausen G, Clotet B. Effect of the interleukin-6 C174G gene polymorphism on treatment of acute and chronic hepatitis C in human immunodeficiency virus coinfected patients. Hepatology. 2007;46:1016-1025. [PubMed] |
| 24. | Alberti A, Benvegnù L, Boccato S, Ferrari A, Sebastiani G. Natural history of initially mild chronic hepatitis C. Dig Liver Dis. 2004;36:646-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 35] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 25. | Falleti E, Fabris C, Vandelli C, Colletta C, Cussigh A, Smirne C, Fontanini E, Cmet S, Minisini R, Bitetto D. Genetic polymorphisms of interleukin-6 modulate fibrosis progression in mild chronic hepatitis C. Hum Immunol. 2010;71:999-1004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 26. | Park BL, Lee HS, Kim YJ, Kim JY, Jung JH, Kim LH, Shin HD. Association between interleukin 6 promoter variants and chronic hepatitis B progression. Exp Mol Med. 2003;35:76-82. [PubMed] |
| 27. | Ribeiro CS, Visentainer JE, Moliterno RA. Association of cytokine genetic polymorphism with hepatitis B infection evolution in adult patients. Mem Inst Oswaldo Cruz. 2007;102:435-440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 28. | Migita K, Miyazoe S, Maeda Y, Daikoku M, Abiru S, Ueki T, Yano K, Nagaoka S, Matsumoto T, Nakao K. Cytokine gene polymorphisms in Japanese patients with hepatitis B virus infection--association between TGF-beta1 polymorphisms and hepatocellular carcinoma. J Hepatol. 2005;42:505-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 120] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 29. | McCullough AJ. The clinical features, diagnosis and natural history of nonalcoholic fatty liver disease. Clin Liver Dis. 2004;8:521-533, viii. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 382] [Cited by in RCA: 397] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 30. | Wilfred de Alwis NM, Day CP. Genes and nonalcoholic fatty liver disease. Curr Diab Rep. 2008;8:156-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 53] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 31. | Younossi ZM, Baranova A, Ziegler K, Del Giacco L, Schlauch K, Born TL, Elariny H, Gorreta F, VanMeter A, Younoszai A. A genomic and proteomic study of the spectrum of nonalcoholic fatty liver disease. Hepatology. 2005;42:665-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 165] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 32. | Tilg H. The role of cytokines in non-alcoholic fatty liver disease. Dig Dis. 2010;28:179-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 180] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 33. | Carulli L, Canedi I, Rondinella S, Lombardini S, Ganazzi D, Fargion S, De Palma M, Lonardo A, Ricchi M, Bertolotti M. Genetic polymorphisms in non-alcoholic fatty liver disease: interleukin-6-174G/C polymorphism is associated with non-alcoholic steatohepatitis. Dig Liver Dis. 2009;41:823-828. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 64] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 34. | Fernández-Real JM, Broch M, Vendrell J, Richart C, Ricart W. Interleukin-6 gene polymorphism and lipid abnormalities in healthy subjects. J Clin Endocrinol Metab. 2000;85:1334-1339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 50] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 35. | Hamid YH, Rose CS, Urhammer SA, Glümer C, Nolsøe R, Kristiansen OP, Mandrup-Poulsen T, Borch-Johnsen K, Jorgensen T, Hansen T. Variations of the interleukin-6 promoter are associated with features of the metabolic syndrome in Caucasian Danes. Diabetologia. 2005;48:251-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 113] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 36. | Vozarova B, Fernández-Real JM, Knowler WC, Gallart L, Hanson RL, Gruber JD, Ricart W, Vendrell J, Richart C, Tataranni PA. The interleukin-6 (-174) G/C promoter polymorphism is associated with type-2 diabetes mellitus in Native Americans and Caucasians. Hum Genet. 2003;112:409-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 126] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 37. | Cardellini M, Perego L, D’Adamo M, Marini MA, Procopio C, Hribal ML, Andreozzi F, Frontoni S, Giacomelli M, Paganelli M. C-174G polymorphism in the promoter of the interleukin-6 gene is associated with insulin resistance. Diabetes Care. 2005;28:2007-2012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 57] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 38. | Illig T, Bongardt F, Schöpfer A, Müller-Scholze S, Rathmann W, Koenig W, Thorand B, Vollmert C, Holle R, Kolb H. Significant association of the interleukin-6 gene polymorphisms C-174G and A-598G with type 2 diabetes. J Clin Endocrinol Metab. 2004;89:5053-5058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 72] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 39. | Chang YH, Huang CN, Shiau MY. The C-174G promoter polymorphism of the interleukin-6 (IL-6) gene that affects insulin sensitivity in Caucasians is not involved in the pathogenesis of Taiwanese type 2 diabetes mellitus. Eur Cytokine Netw. 2004;15:117-119. [PubMed] |
| 40. | Miller AM, Wang H, Bertola A, Park O, Horiguchi N, Ki SH, Yin S, Lafdil F, Gao B. Inflammation-associated interleukin-6/signal transducer and activator of transcription 3 activation ameliorates alcoholic and nonalcoholic fatty liver diseases in interleukin-10-deficient mice. Hepatology. 2011;54:846-856. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 142] [Cited by in RCA: 144] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 41. | Reed T, Page WF, Viken RJ, Christian JC. Genetic predisposition to organ-specific endpoints of alcoholism. Alcohol Clin Exp Res. 1996;20:1528-1533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 115] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 42. | Stickel F, Osterreicher CH. The role of genetic polymorphisms in alcoholic liver disease. Alcohol Alcohol. 2006;41:209-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 74] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 43. | Crews FT, Bechara R, Brown LA, Guidot DM, Mandrekar P, Oak S, Qin L, Szabo G, Wheeler M, Zou J. Cytokines and alcohol. Alcohol Clin Exp Res. 2006;30:720-730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 297] [Cited by in RCA: 311] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 44. | Laso FJ, Vaquero JM, Almeida J, Marcos M, Orfao A. Chronic alcohol consumption is associated with changes in the distribution, immunophenotype, and the inflammatory cytokine secretion profile of circulating dendritic cells. Alcohol Clin Exp Res. 2007;31:846-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 26] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 45. | Laso FJ, Vaquero JM, Almeida J, Marcos M, Orfao A. Production of inflammatory cytokines by peripheral blood monocytes in chronic alcoholism: relationship with ethanol intake and liver disease. Cytometry B Clin Cytom. 2007;72:408-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 55] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 46. | McClain CJ, Song Z, Barve SS, Hill DB, Deaciuc I. Recent advances in alcoholic liver disease. IV. Dysregulated cytokine metabolism in alcoholic liver disease. Am J Physiol Gastrointest Liver Physiol. 2004;287:G497-G502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 185] [Article Influence: 8.8] [Reference Citation Analysis (1)] |
| 47. | Nanji AA, Jokelainen K, Rahemtulla A, Miao L, Fogt F, Matsumoto H, Tahan SR, Su GL. Activation of nuclear factor kappa B and cytokine imbalance in experimental alcoholic liver disease in the rat. Hepatology. 1999;30:934-943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 181] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 48. | Marcos M, Pastor I, González-Sarmiento R, Laso FJ. Common polymorphisms in interleukin genes (IL4, IL6, IL8 and IL12) are not associated with alcoholic liver disease or alcoholism in Spanish men. Cytokine. 2009;45:158-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 49. | Czaja AJ, Strettell MD, Thomson LJ, Santrach PJ, Moore SB, Donaldson PT, Williams R. Associations between alleles of the major histocompatibility complex and type 1 autoimmune hepatitis. Hepatology. 1997;25:317-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 188] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 50. | Pando M, Larriba J, Fernandez GC, Fainboim H, Ciocca M, Ramonet M, Badia I, Daruich J, Findor J, Tanno H. Pediatric and adult forms of type I autoimmune hepatitis in Argentina: evidence for differential genetic predisposition. Hepatology. 1999;30:1374-1380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 132] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 51. | Jones DE, Watt FE, Metcalf JV, Bassendine MF, James OF. Familial primary biliary cirrhosis reassessed: a geographically-based population study. J Hepatol. 1999;30:402-407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 144] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 52. | Mella JG, Roschmann E, Maier KP, Volk BA. Association of primary biliary cirrhosis with the allele HLA-DPB1*0301 in a German population. Hepatology. 1995;21:398-402. [PubMed] |
| 53. | Fan LY, Tu XQ, Zhu Y, Pfeiffer T, Feltens R, Stoecker W, Zhong RQ. Genetic association of cytokines polymorphisms with autoimmune hepatitis and primary biliary cirrhosis in the Chinese. World J Gastroenterol. 2005;11:2768-2772. [PubMed] |
| 54. | Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539-545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5245] [Cited by in RCA: 5765] [Article Influence: 240.2] [Reference Citation Analysis (0)] |
| 55. | Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860-867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10123] [Cited by in RCA: 11283] [Article Influence: 490.6] [Reference Citation Analysis (2)] |
| 56. | Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2609] [Cited by in RCA: 2790] [Article Influence: 146.8] [Reference Citation Analysis (0)] |
| 57. | Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8581] [Cited by in RCA: 8327] [Article Influence: 489.8] [Reference Citation Analysis (0)] |
| 58. | Kuper H, Adami HO, Trichopoulos D. Infections as a major preventable cause of human cancer. J Intern Med. 2000;248:171-183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 505] [Cited by in RCA: 510] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 59. | Farinati F, Cardin R, Cassaro M, Bortolami M, Nitti D, Tieppo C, Zaninotto G, Rugge M. Helicobacter pylori, inflammation, oxidative damage and gastric cancer: a morphological, biological and molecular pathway. Eur J Cancer Prev. 2008;17:195-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 62] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 60. | El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557-2576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3846] [Cited by in RCA: 4267] [Article Influence: 237.1] [Reference Citation Analysis (2)] |
| 61. | Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674-687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1461] [Cited by in RCA: 1568] [Article Influence: 82.5] [Reference Citation Analysis (0)] |
| 62. | Luedde T, Schwabe RF. NF-κB in the liver--linking injury, fibrosis and hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2011;8:108-118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1094] [Cited by in RCA: 1089] [Article Influence: 77.8] [Reference Citation Analysis (0)] |
| 63. | He G, Karin M. NF-κB and STAT3 - key players in liver inflammation and cancer. Cell Res. 2011;21:159-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 879] [Cited by in RCA: 951] [Article Influence: 67.9] [Reference Citation Analysis (0)] |
| 64. | Johnson C, Han Y, Hughart N, McCarra J, Alpini G, Meng F. Interleukin-6 and its receptor, key players in hepatobiliary inflammation and cancer. Transl Gastrointest Cancer. 2012;1:58-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 73] [Reference Citation Analysis (0)] |
| 65. | Nieters A, Yuan JM, Sun CL, Zhang ZQ, Stoehlmacher J, Govindarajan S, Yu MC. Effect of cytokine genotypes on the hepatitis B virus-hepatocellular carcinoma association. Cancer. 2005;103:740-748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 82] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 66. | Ognjanovic S, Yuan JM, Chaptman AK, Fan Y, Yu MC. Genetic polymorphisms in the cytokine genes and risk of hepatocellular carcinoma in low-risk non-Asians of USA. Carcinogenesis. 2009;30:758-762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 54] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 67. | Falleti E, Fabris C, Toniutto P, Fontanini E, Cussigh A, Bitetto D, Fumolo E, Fornasiere E, Bragagnini W, Pinato DJ. Interleukin-6 polymorphisms and gender: relationship with the occurrence of hepatocellular carcinoma in patients with end-stage liver disease. Oncology. 2009;77:304-313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 1.9] [Reference Citation Analysis (0)] |