Published online Apr 7, 2013. doi: 10.3748/wjg.v19.i13.2019
Revised: December 16, 2012
Accepted: December 22, 2012
Published online: April 7, 2013
Processing time: 178 Days and 19.1 Hours
Recent research has shown that microRNA (miRNA), which is involved in almost every step of gastric carcinogenesis, has broad prospective application in diagnosis and therapy of gastric carcinoma. Eastern Asia (South Korea, Japan and China) has the highest incidence of gastric cancer in the world. There were 988 000 new cases of gastric cancer worldwide and 736 000 deaths in 2008. Approximately 60% of the cases of gastric cancer are found in East Asia (mainly China). We herein provide a brief review of the clinical applications of miRNA, which include the following aspects: (1) miRNA may serve as a potential new generation of tumor markers; (2) a complete miRNA expression profile is highly specific, can reflect the evolutionary lineage and differentiation of tumors, and be used to carry out diversity analysis; (3) detecting specific miRNA expression in peripheral blood will become a new method for diagnosis of gastric cancer; (4) miRNA can predict prognosis of gastric cancer; (5) miRNA has predictive value in determining chemotherapy and radiotherapy resistance; and (6) miRNA could be a type of innovative drug. Finally, we focus on assessing the value of miRNA from laboratory to clinical application and the challenges it faces in East Asia.
- Citation: Gao M, Yin H, Fei ZW. Clinical application of microRNA in gastric cancer in Eastern Asian area. World J Gastroenterol 2013; 19(13): 2019-2027
- URL: https://www.wjgnet.com/1007-9327/full/v19/i13/2019.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i13.2019
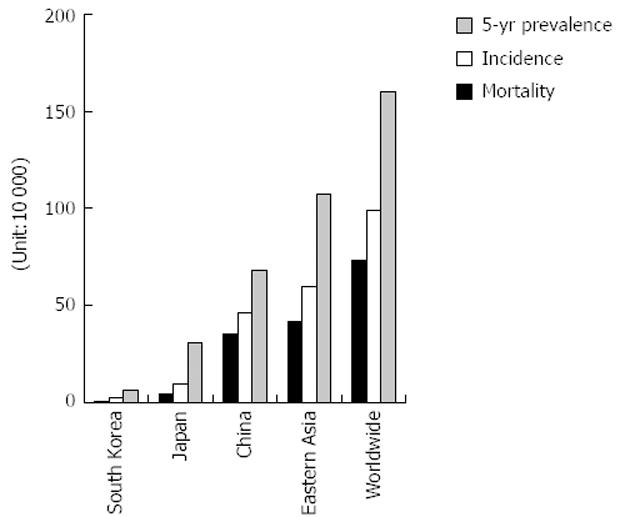
Gastric cancer is a leading disease in Eastern Asia (South Korea, Japan and China) (Figure 1). The incidence and mortality of gastric cancer in East Asian areas rank respectively the second and the third among the most common cancers worldwide[1,2]. According to World Health Organization[3] statistics, there were 988 000 new cases of gastric cancer worldwide and 736 000 deaths in 2008. Approximately 60% of the cases are found in East Asia (mainly China). In China, approximately two-thirds of patients develop advanced or metastatic disease, and more than half have recurrent disease after curative surgery. The median survival time for these patients is only 6-9 mo[4-6]. Several reasons restrict the diagnosis and treatment of gastric cancer: (1) limited diagnostic measures for early detection; (2) weak prognostic value of outcome; (3) poor effect of surgery or cytotoxic cell treatment for advanced disease; and (4) lack of biomarkers for targeted therapy. The discovery of microRNA (miRNA) may change the above-mentioned difficulties, and improve the level of diagnosis and treatment of gastric cancer.
miRNAs include 20-24 nucleotides and are a class of noncoding small molecular single chain RNAs, and have highly conservative, temporal and tissue-specific characteristics[7-9]. Through complete or incomplete base pairing with target gene mRNA, RNA-induced silencing complex degrades mRNA or blocks its translation, and regulates target gene expression at the post-transcriptional level[10]. They exist widely in eukaryotic organisms and regulate cell proliferation, differentiation and apoptosis. Although the tissues of the body appear malignant, specific miRNAs are overexpressed or underexpressed in different tumors and at different stages, which implies a correlation with occurrence and development of tumor and prognosis[11-13]. Further study of the relation of miRNA and gastric cancer could provide new applications in early tumor detection, monitoring, prognosis, gene therapy, and resolving chemotherapy resistance.
Specific tumor markers are often ideal screening tools. Existing clinical tumor markers [such as carcinoembryonic antigen (CEA), cancer antigen (CA)19-9, and CA72-4] for gastric cancer lack specificity and sensitivity[14]. miRNA may serve as a potential new generation of tumor markers for the following reasons[15-17]: (1) good tissue specificity - Rosenfeld et al[18] have detected unknown sources of miRNA in order to make clear its sources, and its specificity is 90%; (2) expression of miRNA in tumors differs significantly from that in normal tissues; (3) miRNA participates in tumor occurrence and development; (4) expression of miRNA has stage specificity - the same tumors at different stages have different expression profiles[19,20]; and (5) miRNA in fresh tissues, paraffin-embedded tissues, and cells and peripheral blood shows good stability[21].
Detection methods of miRNA and miRNA expression in gastric cancer: miRNA is a good tumor marker in clinical application[22,23]. At present, the detection method for miRNA has become mature. By deep sequencing, we can discover unknown miRNAs, and miRNA chips can be used for identification of the differences in miRNAs between the study and control groups. Finally, they can be verified by real-time quantitative polymerase chain reaction (qPCR)[24].
In diagnosis of gastric cancer, a single miRNA is often characterized by poor specificity or sensitivity, but a complete miRNA expression profile is highly specific, can reflect the evolutionary lineage and differentiation of tumors, and be used to carry out diversity analysis[25,26]. Through horizontal comparison of gastric carcinoma with adjacent normal tissues, we have found specific expression of miRNAs in cancer tissues. Further longitudinal comparison at different tumor stages has enabled us to identify the different miRNAs at each stage and to complete final tumor diagnosis and staging[27,28].
We acquired gastric cancer miRNA expression profiles from numerous Chinese and international study groups from 2008 to 2012[29-36] (Table 1). These results differed considerably and lacked stability and consistency for the following reasons: (1) differences in miRNA chips and software; (2) individual differences between races and patients; (3) differences in collected specimen standards; (4) differences in sample size; and (5) miRNA expression profile differences for different cancer types and stages. According to the above expression profiling, we confirmed several reliable miRNAs in the multiple experiments which had 1.5 fold differential expressions between gastric cancer and normal gastric tissues. We are looking forward to having a large sample multi-center study or even international cooperation to compare complete miRNA expression profiling based on different pathological types and stages of gastric cancer. In particular, countries like China, Japan and South Korea should cooperate using the same platform in complete standard miRNA expression profiling of gastric cancer in East Asian populations.
| Group | Method | Sample | Upregulated | Downregulated |
| Katada et al[29] | TaqManmiRNA assays + qRT-PCR | 42 undifferentiated gastric cancer and controls | miR-34b miR-34c miR-128a | miR-128b, miR-129, miR-148 |
| Guo et al[30] | Microarray | 3 gastric cancers and adjacent normal tissues | miR-20b, miR-20a, miR-17, miR-106a, miR-18a, miR-21, miR-106b, miR-18b, miR-421, miR-340, miR-19a, miR-658 | miR-768-3p, miR-378, miR-31, miR-139-5p, miR-195, miR-497, miR-133b, |
| Yao et al[31] | Microarray + qRT-PCR | 10 gastric cancers and adjacent normal tissue | miR-223, miR-106b, miR-147, miR-34a, miR-130b, miR-106a, miR-18a, miR-17, miR-98, miR-616,miR-181a-2, miR-185, miR-1259, miR-601, miR-196a, miR-221, miR-302f, miR-340, miR-337-3p, miR-520c-3p, miR-575 and miR-138 | miR-638, miR-378 |
| Luo et al[32] | Microarray + qRT-PCR | 24 gastric cancers | MiR-26b, miR-30a-5p, miR-212, miR-320, miR-379, miR-518b, miR-409-3b | MiR-9, miR-19b, miR-155, miR-188, miR-197, miR-338, miR-370, miR-383, miR-433, miR-490, miR-503, miR-545, miR-551a, miR-567, miR-575, miR-611, miR-630, miR-649, miR-652 |
| Ueda et al[33] | Microarray + qRT-PCR | 353 (184 gastric cancers 169 controls) | miR-181d, miR-181a-1, miR-181a-2, miR-181c, miR-181b-1, miR-181b-2, miR-21, miR-25, miR-92-1, miR-92-2, miR-93, miR-17-5p, miR-106a, miR-20b, miR-135a-1, miR-135a-2, miR-425-5p, miR-106b, miR-20a, miR-19b-1, miR-19b-2 | miR-148a, miR-148b, miR-375, miR-29b-1, miR-29b-2, miR-29c, miR-152, miR-218-2miR-451, miR-30d |
| Tsukamoto et al[34] | Microarray (470) + qRT-PCR | 22 gastric cancers | miR-18a, miR-106a, miR-17, miR-146a, miR-93, miR-19a, miR-20a, miR-20b, miR-25, miR-15b, miR-425, miR-92a, miR-194, miR-10a, miR-222, miR-7, miR-106b, miR-320a, miR-21, miR-34a, miR-19b, miR-103, miR-215, miR-192, miR-429, miR-27a, miR-223, miR-23a, miR-107, miR-200b, miR-24, miR-15a, miR-16 | miR-375, miR-29c, miR-148a, miR-30a-5p, miR-30e-5p, miR-638 |
| Li et al[35] | TaqManmiRNA assays + qRT-PCR | 30 gastric cancer and controls | miR-223, miR-21, miR-23b, miR-222, miR-25, miR-23a, miR-221, miR-107, miR-103, miR-99a, miR-100, miR-125b, miR-92, miR-146a, miR-214 and miR-191, | let-7a, miR-126, miR-210, miR-181b, miR-197, miR-30aa-5p |
| Carvalho et al[36] | Microarray + qRT-PCR | 76 gastric cancers | miR-582-5p, miR-151-5p, miR-296-5p, miR-30b, miR-513-5p, miR-335, miR-576-5p, miR-219-2-3p, miR-331-5p, miR-889, miR-152, miR-992, miR-93, miR-519c, miR-599, miR-520a-5p, miR-631, miR-550, miR-136, miR-22, miR-515-5p, miR-127-3p, miR-374a, miR-181a, miR-192, miR-532-3p, miR-30d, miR-640, miR-425, miR-92b, miR-501-5p, miR-514, miR-576-3p, miR-519e, miR-149, miR-219-1-3p, miR-424, miR-220, miR-96, miR-218-2, miR-649, miR-215, miR-182, miR-122, miR-524-3p, miR-187, miR-526b, miR-770-5p, miR-545, miR-200b, miR-9, miR-141, miR-579, miR-493, miR-137, miR-216a, miR-503, miR-126, miR-23b, miR-99b, miR-101, miR-323-3p, miR-25, miR-92a-1, miR-429 | miR-451, miR-502-3p, miR-101 miR-33a, miR-516a-3p/miR-516b |
Change in miRNA expression is an early event during the development of gastric tumor[37,38]. Tracking the changes in miRNA expression profiling in the relevant gastric tissues might enable early tumor diagnosis. Traditional methods for the detection of gastric cancer are endoscopy and biopsy. A minimally invasive examination method would be helpful for screening and early detection of cancer in high-risk populations[39]. Detection of tumor in the peripheral blood of patients with specific miRNA expression level has been a research hotspot in recent years, which will perhaps become a new method for diagnosis of gastric cancer[40-42].
Researchers have shown that 90% of plasma miRNA is based on protein-miRNA complex formation. As tumor markers in peripheral blood, miRNAs have the following advantages: (1) miRNAs exist in great volume in peripheral blood[43,44]; (2) miRNAs can resist enzymatic digestion[45-47]; (3) miRNAs have strong resistance to the external environment; and (4) miRNAs show abnormal expression in tumor patients’ serum[48].
In the past two years, Japanese and Chinese researchers have investigated miRNA in the peripheral blood of patients with gastric cancer[49-55] (Table 2) and have obtained some positive results. For example, by comparing serum of 61 gastric cancer patients with that of 61 healthy persons, Liu et al[49] found that the expression of miR-378 in the gastric cancer group was significantly higher than that in the healthy group. The area under the receiver-operating characteristic curve was 0.861 (95%CI: 0.766-0.928), and sensitivity/specificity was 87.5%/70.7%, respectively. Similarly, after investigating the peripheral serum in 69 gastric cancer patients and 30 healthy volunteers by qRT-PCR, Tsujiura et al[55] found that the plasma concentrations of miRNAs (miR-106a and miR-106b) were significantly higher in the patients than in the controls, whereas let-7a concentration was lower in the patients, in which the area under the curve (AUC) for miR-106a and let-7a was 0.879, and sensitivity/specificity was 85.5%/80.0%, respectively. These miRNAs could become ideal tumor markers for gastric cancer. In addition, Liu et al[49] observed that the plasma miRNAs (miR-1, miR-20a, miR-27a, miR-34, and miR-423-5P) in the gastric cancer patients had significantly higher expression than in the control group (164 gastric cancer patients vs 127 healthy individuals). The AUC was 0.879 (95%CI: 0.822-0.936). It is interesting that, in the same sample, they also compared the AUC values of CEA and CA19-9 which were only 0.503 and 0.600, respectively. The results show that miRNA has some advantages as a tumor marker. We have found that miRNAs have good sensitivity and specificity for gastric cancer and are promising tumor markers. However, at present, some factors still limit their clinical diagnostic applications[56]: (1) relative difficulty of detection (in quality and quantity); (2) lack of a unified testing platform and standardization; (3) plasma miRNA source and release mechanism are not clear; and (4) differences in expression of tissue and peripheral blood miRNA still exist[57]. Thus, searching and identifying specific miRNAs for the diagnosis of gastric cancer is the first task that must be undertaken. Establishment of a suitable standard testing system for clinical application, including quality control, and diagnostic threshold determination are still issues that require some work. There is a high incidence of gastric cancer in East Asian countries. High-risk populations could be screened by miRNAs, which should be able to increase the detection rate of early gastric cancer and improve the effects of treatment.
| Group | Sample | MicroRNA | AUC | Method | Sensitivity/specificity (%) |
| Liu et al[49] | 61 GC/61 C | miR-378↑ | 0.861 | qRT-PCR | 87.5/70.7 |
| Liu et al[50] | 164 GC/127 C | (miR-1, miR-20a, miR-27a, miR-34, miR-423-5P)↑ | 0.879 | Microarray + qRT-PCR | 79.3/86.5 |
| 0.831 | |||||
| Konish et al[51] | 56 GC/30 C | miR-451↑ | 0.96 | Microarray + qRT-PCR | 96.0/100 |
| miR-486↑ | 0.92 | 86.0/97.0 | |||
| Song et al[52] | 82 GC/82 C | miR-221, miR-744 and miR-376c↑ | NA | qRT-PCR | 82.4/58.8 |
| Zhou et al[53] | 90 GC/27 C | miR-106a↑ | 0.684 | Microarray + qRT-PCR | 48.2/90.2 |
| miR-17↑ | 0.743 | 51.9/92.7 | |||
| Wang et al[54] | 174 GC/39 C | miR-21↑ | 0.81 | Microarray + qRT-PCR | 56.7/94.9 |
| Tsujiura et al[55] | 69 GC/30 C | miR-106b↑ | 0.72 | Microarray + qRT-PCR | NA |
| miR-106a↑, let-7a↓ | 0.879 | 85.5/80.0 |
Predicting patient survival time, disease progression, prognostic outcome or response to treatment is challenging. Because of the stability and specificity of expression in tissues and circulation, miRNA may be regarded as a forecasting tool for disease outcomes. A lot of the literature suggests that miRNAs have a close relationship with survival time of gastric cancer patients, disease stage, tumor recurrence, and lymph node metastasis. Li et al[58] have shown that a seven-miRNA signature (miR-10b, miR-21, miR-223, miR-338, let-7a, miR-30a-5p, and miR-126) is an independent predictor of overall survival [hazard ratio (HR) = 3.046; P = 0.015] and relapse-free survival (HR = 3.337; P = 0.012). It can predict the prognosis of the patient in relation to tumor stage, cytological subtypes, and Lauren classification[59,60]. In gastric cancer, many Chinese and other research teams have discovered a number of miRNAs that play a role as a predictor. For example, high expression of miRNA-20b, miRNA-150, miRNA-142-5p[61], miRNA-375 and miRNA-214[62] and low expression of miRNA-451, let7g, miRNA-433 and miRNA-125-5p[63] are associated with short survival time. High levels of miRNA-27a and miRNA-650[64] and low levels of miRNA-126[65], miRNA-146a[66], miRNA-148[67], miRNA-218[68], miRNA-335[69] and miRNA-429[70] indicate lymph node metastasis. Patients with overexpression of miRNA-375, miRNA-451, miRNA-199-3p and miRNA-195[71] and decreased expression of miRNA-142-5p are more likely to relapse. High levels of miRNA-221[72] and decreased levels of miRNA-126, miRNA-148a and miRNA-218 indicate advanced gastric cancer. High expression of miRNA-223, miRNA-148a[73] and miRNA-107[74] and reduced expression of miRNA-610[75], miRNA-200b[76] and miRNA-7[77] were associated with invasion and metastasis (Table 3). Therefore, many potential predictors have proved useful for judging the prognosis of gastric cancer patients and are the basis for targeted therapy. However, researching miRNAs as prognostic factors involves small sample sets, high volume of work in validation, and research of independent cohorts, all of which are required before assays for miRNAs can be used clinically.
| Potential predictive role of miRNA | ||
| MiRNAs of high expression | MiRNAs of low expression | |
| Short survival time | miRNA-20b, miRNA-150[29], miRNA-142-5p[61], miRNA-375, miRNA-214[62] | miRNA-451, let7g1, miRNA-4331[33], miRNA-125-5p3[63] |
| Lymph node metastasis | miRNA-27a[29], miRNA-650[64] | miRNA-126[65], miRNA-146a[66], miRNA-148[67], miRNA-218[68], miRNA-335[69], miRNA-429[70] |
| Relapse | miRNA-375[61], miRNA-4512, miRNA-199-3p2, miRNA-1952[71] | miRNA-142-5p[61] |
| Advanced gastric cancer | miRNA-2214[72] | miRNA-126[65], miRNA-148a[67], miRNA-218[68] |
| Invasion, metastasis | miRNA-223[35], miRNA-148a[73], miRNA-107[74] | miRNA-610[75], miRNA-200b5[76], miRNA-7[77] |
Resistance to chemotherapy and radiotherapy is a major obstacle to improving the survival of the patients with gastric cancer[78-80]. We can predict the occurrence of resistance to chemotherapy and radiotherapy[81-83] (Table 4) through detecting the miRNA expression profile of the patients. Through investigating drug resistance to cisplatin and 5-fluorouracil in 90 patients with gastric cancer and comparing patients’ miRNA expression before and after chemotherapy, Kim et al[81] found that high expression of let-7g, miR-342, miR-16, miR-181, miR-1 and miR-34 indicated sensitivity to chemotherapy, and high expression of miR-518f, miR-520a, miR-520d, miR-519e, miR-363 and miR-517 indicated resistance to chemotherapy. By predicting miRNAs, we used a new method for choosing chemotherapy regimen and monitoring its effects, and even reversing the chemotherapy resistance through transfecting specific pre-miRNA. miRNA-15b and miRNA-16 are downregulated severely in the multi-drug resistant gastric carcinoma cell line SGC7901/VCR. By improving miRNA-15b and miRNA-16 expression levels, sensitivity to vincristine was enhanced. Chen et al[84] transfected miRNA-200c into SGC7901/DDP gastric cancer cells, which increased sensitivity to DDP, 5-fluorouracil, paclitaxel and doxorubicin. The same situation occurred in radiotherapy on gastric cancer by transfection into AS-miRNA-221/222, which down-regulated the miRNA-221/222 expression in gastric cancer cell line SGC7901. Zhang et al found that the survival rate of cancer cell was significantly lower than that in the control group. Radiosensitivity was promoted through 0-6 Gy irradiation. In addition, Bandres et al[85] transfected cancer cells with pre-miRNA-451, which improved expression of miRNA-451 in AGS gastric cancer cells. Under 0-4 Gy irradiation, the effect of treatment was significantly better than that in the control group.
| Upregulated | Downregulated | |
| Chemosensitivity | (let-7g, miR-342, miR-16, miR-181, miR-1, miR-34)1[81] | |
| Chemoresistance | (miR-518f, miR-520a, miR-520d, miR-519e, miR-363, miR-517)1[81] | (miR-196a, miR-200family, miR-338, miR-126, miR-31, miR-98, let-7g, miR-7)2[82] miR-15b, miR-16[83] |
| Radiosensitivity | miR-451[85] | |
| Radioresistance | miR-221/222[97] |
miRNAs are regulatory factors for gene expression and act as a control center in the process of tumor development[86,87]. miRNAs can modulate protein expression and affect multiple information pathways[88]. miRNAs will be more effective than coding genes as a biological treatment of tumor target molecules. The basic strategy of current treatment based on miRNAs is to adopt gene knockout to inhibit or downregulate the expression level of oncogene miRNAs. On the contrary, for anti-oncogenes, we used the method of gene knock-in to introduce foreign miRNAs, increase the expression level, and achieve the purpose of tumor treatment. The following strategies were used: administration of small molecule drugs for inhibiting miRNA, e.g., anti-miRNA oligonucleotides (AMOs) following base pairing rules, competitively blocked the miRNA with target gene interaction[89], such as locked nucleic acid[90-91]; miRNA sponges, e.g., the adsorption with miRNA could not combine with the natural target[92]; and miR-Mask[93], miRNA inhibitors and so on. miRNA expression is often increased by using viruses as a carrier to introduce a specific miRNA or miRNA mimics and finally upregulate miRNA and inhibit the tumor[94].
A number of Eastern Asian researchers followed the above principle of miRNA-mediated treatment and achieved good results in vitro and in animal experiments. For example, miRNA-221/222 is upregulated in gastric cancer cell line SGC7901. By transfecting AS-miR-221/222 2000 into cancer cells with liposomes, miRNA-221/222 is knocked out. This inhibits gastric cancer cell growth and invasion. Their target molecule is PTEN[95]. MiR-516-3p has been transfected into the gastric scirrhous carcinoma cell line 44AS3 with liposomes, was significantly overexpressed, and finally inhibited cancer cell growth, invasion and metastasis[96]. Similar results were obtained in a nude mouse transplantation model of human gastric cancer. Zhang et al[97] through AMOs, knocked down the originally high expression of miRNA-21 and caused the proliferation of gastric cancer cells to slow and apoptosis to increase visibly. In addition, Ji et al[98] have added a miRNA-34 analog to p53 mutated gastric cancer cell lines to restore its function and upregulate its expression, which inhibited cell growth and maintained them at phase G1.
Many studies based on treatment of gastric cancer by miRNA have shown good results. In particular, Chinese, Japanese and South Korean researchers have attempted this. However, we still have several major obstacles to overcome. First, the multi-targeting nature of miRNAs brings the risk of unconscious off-target effects. Second, the expression of target genes may often be regulated by multiple miRNAs, which could greatly reduce the effect of treatment based on a specific miRNA. Finally, we still lack good specificity and an efficient miRNA delivery system for treatment[99,100].
miRNAs are involved in almost all stages of gastric carcinogenesis, and may have broad applications in early diagnosis of gastric carcinoma, prognosis, detection of radiotherapy and chemotherapy efficacy, and be a new target for treatment. However, studies based on the clinical application of miRNAs for gastric cancer still lack reliable and exact data from large multi-center studies. In recent years, miRNAs have been a focus of biomedical research. New miRNAs have been discovered and research techniques constantly updated. It will be a great challenge to integrate new data and establish standard procedures. For diagnosis, we need unified standards and testing platforms. For treatment, we need better-designed small-molecule drugs based on a well detailed and more accurate medication carrier without toxic side effects. We look forward to further studies of miRNAs improving their clinical applications for the diagnosis and treatment of gastric cancer. East Asia, as an area with a high incidence of gastric cancer, should undertake more studies for the application of miRNAs in gastric cancer.
P- Reviewers Ziogas D, Sakurazawa N S- Editor Song XX L- Editor A E- Editor Zhang DN
| 1. | Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893-2917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11128] [Cited by in RCA: 11836] [Article Influence: 845.4] [Reference Citation Analysis (4)] |
| 2. | Yang L. Incidence and mortality of gastric cancer in China. World J Gastroenterol. 2006;12:17-20. [PubMed] |
| 3. | Available from: http: //globocan.iarc.fr. |
| 4. | Verdecchia A, Corazziari I, Gatta G, Lisi D, Faivre J, Forman D. Explaining gastric cancer survival differences among European countries. Int J Cancer. 2004;109:737-741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 68] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 5. | Bonenkamp JJ, Hermans J, Sasako M, van de Velde CJ, Welvaart K, Songun I, Meyer S, Plukker JT, Van Elk P, Obertop H. Extended lymph-node dissection for gastric cancer. N Engl J Med. 1999;340:908-914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1158] [Cited by in RCA: 1069] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 6. | Kaneko S, Yoshimura T. Time trend analysis of gastric cancer incidence in Japan by histological types, 1975-1989. Br J Cancer. 2001;84:400-405. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 106] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 7. | Finnegan EF, Pasquinelli AE. MicroRNA biogenesis: regulating the regulators. Crit Rev Biochem Mol Biol. 2013;48:51-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 226] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 8. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25833] [Cited by in RCA: 27842] [Article Influence: 1325.8] [Reference Citation Analysis (0)] |
| 9. | Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8672] [Cited by in RCA: 8875] [Article Influence: 277.3] [Reference Citation Analysis (0)] |
| 10. | Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776-780. [PubMed] |
| 11. | Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857-866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5705] [Cited by in RCA: 6030] [Article Influence: 317.4] [Reference Citation Analysis (0)] |
| 12. | Chen B, Li H, Zeng X, Yang P, Liu X, Zhao X, Liang S. Roles of microRNA on cancer cell metabolism. J Transl Med. 2012;10:228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2908] [Cited by in RCA: 3110] [Article Influence: 148.1] [Reference Citation Analysis (0)] |
| 13. | Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999-3004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2908] [Cited by in RCA: 3110] [Article Influence: 148.1] [Reference Citation Analysis (0)] |
| 14. | Liu X, Cai H, Wang Y. Prognostic significance of tumour markers in Chinese patients with gastric cancer. ANZ J Surg. 2012;Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 15. | Pignot G, Cizeron-Clairac G, Vacher S, Susini A, Tozlu S, Vieillefond A, Zerbib M, Lidereau R, Debre B, Amsellem-Ouazana D. microRNA expression profile in a large series of bladder tumors: Identification of a 3-miRNA signature associated with aggressiveness of muscle-invasive bladder cancer. Int J Cancer. 2012;Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 145] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 16. | Dettmer M, Vogetseder A, Durso MB, Moch H, Komminoth P, Perren A, Nikiforov YE, Nikiforova MN. MicroRNA expression array identifies novel diagnostic markers for conventional and oncocytic follicular thyroid carcinomas. J Clin Endocrinol Metab. 2013;98:E1-E7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 85] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 17. | Heneghan HM, Miller N, Kerin MJ. MiRNAs as biomarkers and therapeutic targets in cancer. Curr Opin Pharmacol. 2010;10:543-550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 174] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 18. | Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, Benjamin H, Shabes N, Tabak S, Levy A. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462-469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 728] [Cited by in RCA: 720] [Article Influence: 42.4] [Reference Citation Analysis (0)] |
| 19. | Malumbres R, Sarosiek KA, Cubedo E, Ruiz JW, Jiang X, Gascoyne RD, Tibshirani R, Lossos IS. Differentiation stage-specific expression of microRNAs in B lymphocytes and diffuse large B-cell lymphomas. Blood. 2009;113:3754-3764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 198] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 20. | Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348-2360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 825] [Cited by in RCA: 900] [Article Influence: 52.9] [Reference Citation Analysis (0)] |
| 21. | Liu BR, Xie L. MiRNA: A new cancer biomarker. Linchuang Zhongliuxue Zazhi. 2010;15:1-5. |
| 22. | Nuovo GJ, Elton TS, Nana-Sinkam P, Volinia S, Croce CM, Schmittgen TD. A methodology for the combined in situ analyses of the precursor and mature forms of microRNAs and correlation with their putative targets. Nat Protoc. 2009;4:107-115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 107] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 23. | Schotte D, Akbari Moqadam F, Lange-Turenhout EA, Chen C, van Ijcken WF, Pieters R, den Boer ML. Discovery of new microRNAs by small RNAome deep sequencing in childhood acute lymphoblastic leukemia. Leukemia. 2011;25:1389-1399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 73] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 24. | Mei Q, Li X, Meng Y, Wu Z, Guo M, Zhao Y, Fu X, Han W. A facile and specific assay for quantifying microRNA by an optimized RT-qPCR approach. PLoS One. 2012;7:e46890. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 55] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 25. | Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7723] [Cited by in RCA: 7370] [Article Influence: 368.5] [Reference Citation Analysis (0)] |
| 26. | Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257-2261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4471] [Cited by in RCA: 4521] [Article Influence: 237.9] [Reference Citation Analysis (0)] |
| 27. | Qi P, Gao CF. MiRNA and research progress in hepatocellularcarcinoma. Zhongguo Shengwu Gongcheng Zazhi. 2008;28:94-101. |
| 28. | Jia H, Xu Y. MicroRNA and gastric cancer. Yixue Zongshu. 2009;5:670-672. |
| 29. | Katada T, Ishiguro H, Kuwabara Y, Kimura M, Mitui A, Mori Y, Ogawa R, Harata K, Fujii Y. microRNA expression profile in undifferentiated gastric cancer. Int J Oncol. 2009;34:537-542. [PubMed] |
| 30. | Guo J, Miao Y, Xiao B, Huan R, Jiang Z, Meng D, Wang Y. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol. 2009;24:652-657. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 343] [Cited by in RCA: 370] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 31. | Yao Y, Suo AL, Li ZF, Liu LY, Tian T, Ni L, Zhang WG, Nan KJ, Song TS, Huang C. MicroRNA profiling of human gastric cancer. Mol Med Rep. 2009;2:963-970. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 147] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 32. | Luo H, Zhang H, Zhang Z, Zhang X, Ning B, Guo J, Nie N, Liu B, Wu X. Down-regulated miR-9 and miR-433 in human gastric carcinoma. J Exp Clin Cancer Res. 2009;28:82. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 148] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 33. | Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 644] [Cited by in RCA: 676] [Article Influence: 45.1] [Reference Citation Analysis (0)] |
| 34. | Tsukamoto Y, Nakada C, Noguchi T, Tanigawa M, Nguyen LT, Uchida T, Hijiya N, Matsuura K, Fujioka T, Seto M. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res. 2010;70:2339-2349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 320] [Cited by in RCA: 346] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 35. | Li X, Zhang Y, Zhang H, Liu X, Gong T, Li M, Sun L, Ji G, Shi Y, Han Z. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res. 2011;9:824-833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 293] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 36. | Carvalho J, van Grieken NC, Pereira PM, Sousa S, Tijssen M, Buffart TE, Diosdado B, Grabsch H, Santos MA, Meijer G. Lack of microRNA-101 causes E-cadherin functional deregulation through EZH2 up-regulation in intestinal gastric cancer. J Pathol. 2012;228:31-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 86] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 37. | Jiao LR, Frampton AE, Jacob J, Pellegrino L, Krell J, Giamas G, Tsim N, Vlavianos P, Cohen P, Ahmad R. MicroRNAs targeting oncogenes are down-regulated in pancreatic malignant transformation from benign tumors. PLoS One. 2012;7:e32068. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 113] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 38. | Park SM, Shell S, Radjabi AR, Schickel R, Feig C, Boyerinas B, Dinulescu DM, Lengyel E, Peter ME. Let-7 prevents early cancer progression by suppressing expression of the embryonic gene HMGA2. Cell Cycle. 2007;6:2585-2590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 178] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 39. | Keller A, Leidinger P, Gislefoss R, Haugen A, Langseth H, Staehler P, Lenhof HP, Meese E. Stable serum miRNA profiles as potential tool for non-invasive lung cancer diagnosis. RNA Biol. 2011;8:506-516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 113] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 40. | Selth LA, Tilley WD, Butler LM. Circulating microRNAs: macro-utility as markers of prostate cancer? Endocr Relat Cancer. 2012;19:R99-R113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 41. | Sun Y, Zhang K, Fan G, Li J. Identification of circulating microRNAs as biomarkers in cancers: what have we got? Clin Chem Lab Med. 2012;50:2121-2126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 42. | Bianchi F, Nicassio F, Veronesi G, di Fiore PP. Circulating microRNAs: next-generation biomarkers for early lung cancer detection. Ecancermedicalscience. 2012;6:246. [PubMed] |
| 43. | Tsang JC, Lo YM. Circulating nucleic acids in plasma/serum. Pathology. 2007;39:197-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 122] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 44. | Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997-1006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3218] [Cited by in RCA: 3554] [Article Influence: 209.1] [Reference Citation Analysis (0)] |
| 45. | Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513-10518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5636] [Cited by in RCA: 6313] [Article Influence: 371.4] [Reference Citation Analysis (0)] |
| 46. | Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17:879-887. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1041] [Cited by in RCA: 1034] [Article Influence: 51.7] [Reference Citation Analysis (0)] |
| 47. | Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8246] [Cited by in RCA: 9797] [Article Influence: 544.3] [Reference Citation Analysis (0)] |
| 48. | Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672-675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1258] [Cited by in RCA: 1333] [Article Influence: 78.4] [Reference Citation Analysis (0)] |
| 49. | Liu H, Zhu L, Liu B, Yang L, Meng X, Zhang W, Ma Y, Xiao H. Genome-wide microRNA profiles identify miR-378 as a serum biomarker for early detection of gastric cancer. Cancer Lett. 2012;316:196-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 208] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 50. | Liu R, Zhang C, Hu Z, Li G, Wang C, Yang C, Huang D, Chen X, Zhang H, Zhuang R. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer. 2011;47:784-791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 308] [Cited by in RCA: 328] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 51. | Zhou H, Guo JM, Lou YR, Zhang XJ, Zhong FD, Jiang Z, Cheng J, Xiao BX. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using microRNA as a marker. J Mol Med (Berl). 2010;88:709-717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 115] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 52. | Song MY, Pan KF, Su HJ, Zhang L, Ma JL, Li JY, Yuasa Y, Kang D, Kim YS, You WC. Identification of serum microRNAs as novel non-invasive biomarkers for early detection of gastric cancer. PLoS One. 2012;7:e33608. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 159] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 53. | Konishi H, Ichikawa D, Komatsu S, Shiozaki A, Tsujiura M, Takeshita H, Morimura R, Nagata H, Arita T, Kawaguchi T. Detection of gastric cancer-associated microRNAs on microRNA microarray comparing pre- and post-operative plasma. Br J Cancer. 2012;106:740-747. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 160] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 54. | Wang B, Zhang Q. The expression and clinical significance of circulating microRNA-21 in serum of five solid tumors. J Cancer Res Clin Oncol. 2012;138:1659-1666. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 173] [Cited by in RCA: 179] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 55. | Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, Kosuga T, Konishi H, Morimura R, Deguchi K, Fujiwara H. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer. 2010;102:1174-1179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 449] [Cited by in RCA: 509] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 56. | Song JH, Meltzer SJ. MicroRNAs in pathogenesis, diagnosis, and treatment of gastroesophageal cancers. Gastroenterology. 2012;143:35-47.e2. [PubMed] |
| 57. | Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, Mayr A, Weger S, Oberhollenzer F, Bonora E. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res. 2010;107:810-817. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1196] [Cited by in RCA: 1135] [Article Influence: 75.7] [Reference Citation Analysis (0)] |
| 58. | Li X, Zhang Y, Zhang Y, Ding J, Wu K, Fan D. Survival prediction of gastric cancer by a seven-microRNA signature. Gut. 2010;59:579-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 268] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 59. | Fu X, Han Y, Wu Y, Zhu X, Lu X, Mao F, Wang X, He X, Zhao Y, Zhao Y. Prognostic role of microRNA-21 in various carcinomas: a systematic review and meta-analysis. Eur J Clin Invest. 2011;41:1245-1253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 122] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 60. | Huang L, Lin JX, Yu YH, Zhang MY, Wang HY, Zheng M. Downregulation of six microRNAs is associated with advanced stage, lymph node metastasis and poor prognosis in small cell carcinoma of the cervix. PLoS One. 2012;7:e33762. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 100] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 61. | Zhang X, Yan Z, Zhang J, Gong L, Li W, Cui J, Liu Y, Gao Z, Li J, Shen L. Combination of hsa-miR-375 and hsa-miR-142-5p as a predictor for recurrence risk in gastric cancer patients following surgical resection. Ann Oncol. 2011;22:2257-2266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 134] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 62. | Xiong X, Ren HZ, Li MH, Mei JH, Wen JF, Zheng CL. Down-regulated miRNA-214 induces a cell cycle G1 arrest in gastric cancer cells by up-regulating the PTEN protein. Pathol Oncol Res. 2011;17:931-937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 47] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 63. | Nishida N, Mimori K, Fabbri M, Yokobori T, Sudo T, Tanaka F, Shibata K, Ishii H, Doki Y, Mori M. MicroRNA-125a-5p is an independent prognostic factor in gastric cancer and inhibits the proliferation of human gastric cancer cells in combination with trastuzumab. Clin Cancer Res. 2011;17:2725-2733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 206] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 64. | Zhang X, Zhu W, Zhang J, Huo S, Zhou L, Gu Z, Zhang M. MicroRNA-650 targets ING4 to promote gastric cancer tumorigenicity. Biochem Biophys Res Commun. 2010;395:275-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 85] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 65. | Feng R, Chen X, Yu Y, Su L, Yu B, Li J, Cai Q, Yan M, Liu B, Zhu Z. miR-126 functions as a tumour suppressor in human gastric cancer. Cancer Lett. 2010;298:50-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 231] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 66. | Kogo R, Mimori K, Tanaka F, Komune S, Mori M. Clinical significance of miR-146a in gastric cancer cases. Clin Cancer Res. 2011;17:4277-4284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 173] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 67. | Zheng B, Liang L, Wang C, Huang S, Cao X, Zha R, Liu L, Jia D, Tian Q, Wu J. MicroRNA-148a suppresses tumor cell invasion and metastasis by downregulating ROCK1 in gastric cancer. Clin Cancer Res. 2011;17:7574-7583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 232] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 68. | Tie J, Pan Y, Zhao L, Wu K, Liu J, Sun S, Guo X, Wang B, Gang Y, Zhang Y. MiR-218 inhibits invasion and metastasis of gastric cancer by targeting the Robo1 receptor. PLoS Genet. 2010;6:e1000879. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 350] [Cited by in RCA: 370] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 69. | Xu Y, Zhao F, Wang Z, Song Y, Luo Y, Zhang X, Jiang L, Sun Z, Miao Z, Xu H. MicroRNA-335 acts as a metastasis suppressor in gastric cancer by targeting Bcl-w and specificity protein 1. Oncogene. 2012;31:1398-1407. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 144] [Cited by in RCA: 161] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 70. | Sun T, Wang C, Xing J, Wu D. miR-429 modulates the expression of c-myc in human gastric carcinoma cells. Eur J Cancer. 2011;47:2552-2559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 74] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 71. | Brenner B, Hoshen MB, Purim O, David MB, Ashkenazi K, Marshak G, Kundel Y, Brenner R, Morgenstern S, Halpern M. MicroRNAs as a potential prognostic factor in gastric cancer. World J Gastroenterol. 2011;17:3976-3985. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 70] [Cited by in RCA: 80] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 72. | Liu K, Li G, Fan C, Diao Y, Wu B, Li J. Increased Expression of MicroRNA-221 in gastric cancer and its clinical significance. J Int Med Res. 2012;40:467-474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 59] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 73. | Tseng CW, Lin CC, Chen CN, Huang HC, Juan HF. Integrative network analysis reveals active microRNAs and their functions in gastric cancer. BMC Syst Biol. 2011;5:99. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 75] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 74. | Li X, Zhang Y, Shi Y, Dong G, Liang J, Han Y, Wang X, Zhao Q, Ding J, Wu K. MicroRNA-107, an oncogene microRNA that regulates tumour invasion and metastasis by targeting DICER1 in gastric cancer. J Cell Mol Med. 2011;15:1887-1895. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 94] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 75. | Wang J, Zhang J, Wu J, Luo D, Su K, Shi W, Liu J, Tian Y, Wei L. MicroRNA-610 inhibits the migration and invasion of gastric cancer cells by suppressing the expression of vasodilator-stimulated phosphoprotein. Eur J Cancer. 2012;48:1904-1913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 76. | Kurashige J, Kamohara H, Watanabe M, Hiyoshi Y, Iwatsuki M, Tanaka Y, Kinoshita K, Saito S, Baba Y, Baba H. MicroRNA-200b regulates cell proliferation, invasion, and migration by directly targeting ZEB2 in gastric carcinoma. Ann Surg Oncol. 2012;19 Suppl 3:S656-S664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 137] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 77. | Zhao X, Dou W, He L, Liang S, Tie J, Liu C, Li T, Lu Y, Mo P, Shi Y. MicroRNA-7 functions as an anti-metastatic microRNA in gastric cancer by targeting insulin-like growth factor-1 receptor. Oncogene. 2013;32:1363-1372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 185] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 78. | Linkous AG, Yazlovitskaya EM. Novel radiosensitizing anticancer therapeutics. Anticancer Res. 2012;32:2487-2499. [PubMed] |
| 79. | Gordon RR, Nelson PS. Cellular senescence and cancer chemotherapy resistance. Drug Resist Updat. 2012;15:123-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 118] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 80. | Perez-Plasencia C, Duenas-Gonzalez A. Can the state of cancer chemotherapy resistance be reverted by epigenetic therapy? Mol Cancer. 2006;5:27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 81. | Kim CH, Kim HK, Rettig RL, Kim J, Lee ET, Aprelikova O, Choi IJ, Munroe DJ, Green JE. miRNA signature associated with outcome of gastric cancer patients following chemotherapy. BMC Med Genomics. 2011;4:79. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 117] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 82. | Wu XM, Shao XQ, Meng XX, Zhang XN, Zhu L, Liu SX, Lin J, Xiao HS. Genome-wide analysis of microRNA and mRNA expression signatures in hydroxycamptothecin-resistant gastric cancer cells. Acta Pharmacol Sin. 2011;32:259-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 41] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 83. | Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, Hong L, Liu J, Fan D. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123:372-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 531] [Cited by in RCA: 561] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 84. | Chen Y, Zuo J, Liu Y, Gao H, Liu W. Inhibitory effects of miRNA-200c on chemotherapy-resistance and cell proliferation of gastric cancer SGC7901/DDP cells. Chin J Cancer. 2010;29:1006-1011. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 85. | Bandres E, Bitarte N, Arias F, Agorreta J, Fortes P, Agirre X, Zarate R, Diaz-Gonzalez JA, Ramirez N, Sola JJ. microRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clin Cancer Res. 2009;15:2281-2290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 284] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 86. | Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435-443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1009] [Cited by in RCA: 973] [Article Influence: 51.2] [Reference Citation Analysis (0)] |
| 87. | Lujambio A, Calin GA, Villanueva A, Ropero S, Sánchez-Céspedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci USA. 2008;105:13556-13561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 795] [Cited by in RCA: 822] [Article Influence: 48.4] [Reference Citation Analysis (0)] |
| 88. | Sotiropoulou G, Pampalakis G, Lianidou E, Mourelatos Z. Emerging roles of microRNAs as molecular switches in the integrated circuit of the cancer cell. RNA. 2009;15:1443-1461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 126] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 89. | Weiler J, Hunziker J, Hall J. Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene Ther. 2006;13:496-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 274] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 90. | Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, Berger U. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896-899. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1268] [Cited by in RCA: 1281] [Article Influence: 75.4] [Reference Citation Analysis (0)] |
| 91. | Tachibana A, Yamada Y, Ida H, Saito S, Tanabe T. LidNA, a novel miRNA inhibitor constructed with unmodified DNA. FEBS Lett. 2012;586:1529-1532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 92. | Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721-726. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1744] [Cited by in RCA: 1652] [Article Influence: 91.8] [Reference Citation Analysis (0)] |
| 93. | Wang Z. The principles of MiRNA-masking antisense oligonucleotides technology. Methods Mol Biol. 2011;676:43-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 76] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 94. | Cheng N, Wang KH, Sun SH. Application perspectives of microRNA-based human cancer therapy. J Mol Diagn Ther. 2010;2:6. |
| 95. | Chun-Zhi Z, Lei H, An-Ling Z, Yan-Chao F, Xiao Y, Guang-Xiu W, Zhi-Fan J, Pei-Yu P, Qing-Yu Z, Chun-Sheng K. MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer. 2010;10:367. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 275] [Cited by in RCA: 321] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 96. | Takei Y, Takigahira M, Mihara K, Tarumi Y, Yanagihara K. The metastasis-associated microRNA miR-516a-3p is a novel therapeutic target for inhibiting peritoneal dissemination of human scirrhous gastric cancer. Cancer Res. 2011;71:1442-1453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 79] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 97. | Zhang Z, Li Z, Gao C, Chen P, Chen J, Liu W, Xiao S, Lu H. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Invest. 2008;88:1358-1366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 339] [Cited by in RCA: 375] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 98. | Ji Q, Hao X, Meng Y, Zhang M, Desano J, Fan D, Xu L. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer. 2008;8:266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 298] [Cited by in RCA: 321] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 99. | Wu Y, Crawford M, Yu B, Mao Y, Nana-Sinkam SP, Lee LJ. MicroRNA delivery by cationic lipoplexes for lung cancer therapy. Mol Pharm. 2011;8:1381-1389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 143] [Cited by in RCA: 139] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 100. | Yang X, Haurigot V, Zhou S, Luo G, Couto LB. Inhibition of hepatitis C virus replication using adeno-associated virus vector delivery of an exogenous anti-hepatitis C virus microRNA cluster. Hepatology. 2010;52:1877-1887. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 2.5] [Reference Citation Analysis (0)] |