Published online Feb 14, 2012. doi: 10.3748/wjg.v18.i6.576
Revised: July 9, 2011
Accepted: November 9, 2011
Published online: February 14, 2012
AIM: To investigate the promoter region methylation status of retinoblastoma protein-interacting zinc finger gene 1 (RIZ1) in the human esophageal squamous cell carcinoma (ESCC) cell lines and tissues and verify the relationship between methylation of RIZ1 and oncogenesis, tumor progression and metastasis etc of ESCC.
METHODS: Methylation-specific polymerase chain reaction (MSP) was used to investigate the promoter region methylation status of RIZ1 in 6 ESCC cell lines. One cell line where RIZ1 promoter region methylation was detected was selected for the next study, where the cell line was treated with 5-aza-CdR. Real-time polymerase chain reaction was used to investigate its influence on the transcription of RIZ1. Experiments using frozen pathological specimens from 47 ESCC patients were performed using the same MSP methodology.
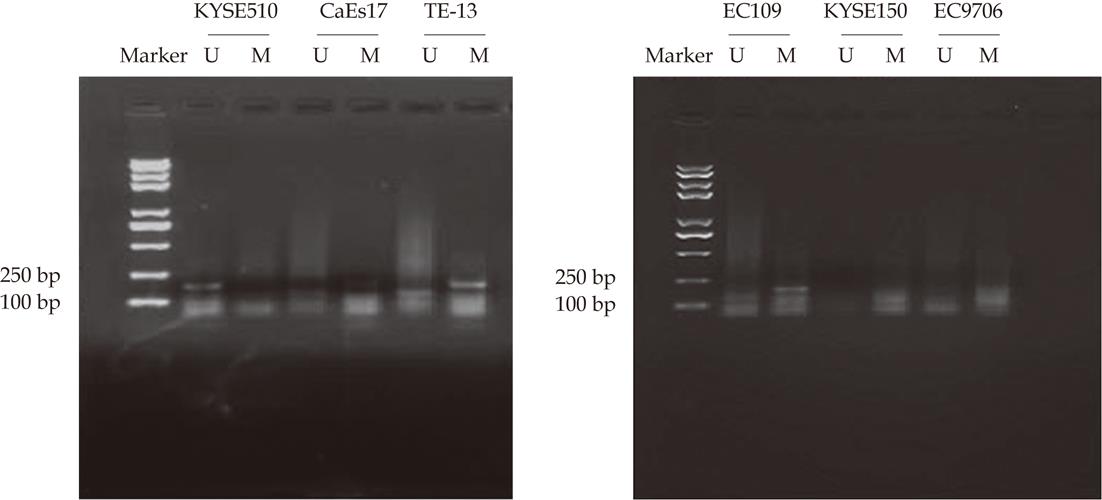
RESULTS: Promoter methylation of RIZ1 gene was detected in TE13, CaEs17 and EC109 cell lines and the cell line TE13 was chosen for further study. The expression of RIZ1 mRNA in TE-13 was up-regulated after treatment with 5-aza-CdR. The rate of methylation in carcinomas tissues was significantly higher than those in matched neighboring normal and distal ending normal tissue, and the deviation of data was statistically significant (χ2 = 24.136, P < 0.01). Analysis of the gender, age familial history, tumour deviation, tumour saturation, lymph gland displacement and clinical staging of 47 samples from ESCC patients showed that the fluctuation of data was not statistically significant.
CONCLUSION: Promoter methylation may play an important role in the epigenetic silencing of RIZ1 gene expression in human ESCC. RIZ1 is considered to be a potential tumor suppressor gene and may be a biological parameter for testing early stage human ESCC.
-
Citation: Dong SW, Zhang P, Liu YM, Cui YT, Wang S, Liang SJ, He Z, Sun P, Wang YG. Study on
RIZ1 gene promoter methylation status in human esophageal squamous cell carcinoma. World J Gastroenterol 2012; 18(6): 576-582 - URL: https://www.wjgnet.com/1007-9327/full/v18/i6/576.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i6.576
Esophageal cancer is one of the most aggressive malignancies with poor prognosis in the world. Esophageal squamous cell carcinoma (ESCC) is a major histological form of the disease, especially in the Northern part of China[1], which is different from Europe and America. Like other types of solid tumors, the development of ESCC is due to the accumulation of the abnormal expression of oncogenes and tumor suppressor genes (TSG). Several genetic alterations have been associated with the development of ESCC including p53 and p16 mutations, amplification of cyclin D, c-myc, and EGFR, and allelic loss on chromosomes[2-5]. In mammalian development, DNA methylation has an essential regulatory function which suppresses gene activity by changing chromatin structure[6,7]. It has become apparent that aberrant DNA methylation of promoter region CpG islands may serve as an alternate mechanism to genetic defects in the inactivation of TSG in human malignancies[8,9].
In recent years, researchers found that the retinoblastoma protein-interacting zinc finger gene (RIZ) maps to the distal short arm of human chromosome 1 (1p36), a region thought to harbor TSG for a variety of human cancers. The RIZ gene normally produces two protein products of different length, RIZ1 and RIZ2. RIZ1 contains the positive regulatory (positive regulatory domain I binding factor 1 and RIZ) domain, but RIZ2 lacks this domain[10]. In many human cancers, RIZ1 is considered a TSG because RIZ1 can induce G2-M arrest and apoptosis. Moreover, a knockout study showed that RIZ1 is a tumor susceptibility gene in mice[11]. The expression of RIZ1 is frequently silenced in many human malignant tumours, including carcinomas of the breast, prostate, and thyroid gland[12-14]. Recently, methylation of RIZ1 promoter CpG islands has been proposed as a common mechanism in inactivating RIZ1. Increasing clinical evidence reveals a positive correlation of reduced RIZ1 expression with increased risk for metastasis, indicating that RIZ1 may be a potential new TSG[12-14]. Although RIZ1 is a putative tumor suppressor in several cancer types, for instance breast cancer[15], gastric cancer[16], lung cancer[17] and so on, the role of RIZ1 in human ESCC has not been reported. In this study, we analyzed methylation status of the RIZ1 promoter and its relationship with RIZ1 mRNA expression in human ESCC cell lines. In addition, the study examined the relationship between methylation of the RIZ1 gene in the promoter region, oncogenesis, tumor progression, metastasis and hereditary factors etc of ESCC.
The human ESCC cell lines KYSE150, KYSE510, TE13, EC9706, CaEs17 and EC109 were provided by the Institute of Cellula Nervosa in Tianjin Huanhu Hospital and were cultured in recommended media RPMI1640 (GIBCO, HEPES 4.76 g/NaCO3 2.0 g/RPMI-1640 10.4 g/ddH2O 1000 mL) supplemented with 10% new-born bovine serum (GIBCO), 1 × L-glutamine and 1 × penicillin-streptomycin. Cells were maintained at 37 °C in a humidified environment with 5% CO2.
Carcinoma, matched adjacent normal (> 2 cm from the tumor) and distal ending normal (> 5 cm from the tumor) tissues were obtained in our department during surgical excision from 47 patients with ESCC. All specimens were placed in liquid nitrogen immediately after resection and stored at -80 °C until RNA or genomic DNA (gDNA) extraction. No patient had received chemotherapy or radiation therapy prior to surgery. All patients were confirmed to have ESCC by pathologic test.
gDNA from cell lines or ESCC frozen tissues was extracted by using a Dneasy kit (Biomiga). All extracted genomic DNA was treated with sodium bisulfite (Sigma) as reported previously. Briefly, 2 μg gDNA was denatured by 5.5 μL of 3 mol fresh NaOH (final concentration 0.3 mol/L) for 10 min at 37 °C. 30 μL of 10 mmol/L hydroquinone (Sigma) and 520 μL of 3 mol/L sodium bisulfite (pH 5.0) were added, away from light. The mixture was inverted, added to 200 μL liquid paraffin to prevent water evaporation and reagent oxidation, then incubated at 50 °C for 16 h. The modified DNA was purified using the Wizard DNA clean-up system (Promega). The purified DNA was treated again with NaOH and precipitated. DNA was resuspended in 20 μL of LoTE, 2 μL of which were subjected to polymerase chain reaction (PCR) amplification.
Methylation-specific primers were designed to cover 23 CpG dinucleotides numbered -124--103 (forward) and 32-52 (reverse). Similarly, unmethylation-specific primers were designed to cover 23 CpG dinucleotides numbered -123--103 (forward) and 32-52 (reverse). Primers specific for methylated DNA (forward 5’-GTGGTGGTTATTGGGCGACGGC-3’; reverse 5’-GCTATTTCGCCGACCCCGACG-3’) and unmethylated DNA (forward 5’-TGGTGGTTATTGGGTGATGGT-3’; reverse 5’-ACTATTTCACCAACCCCAAGA-3’) were added to the reaction and expected to generate 177-bp and 175-bp products, respectively. PCR conditions were 40 cycles of denaturation at 94 °C for 30 s, annealing at 68 °C for methylation-specific amplification or at 60 °C for unmethylation-specific amplification for 45 s and extension at 72 °C for 60 s, then sequencing of PCR products.
2 × 105 TE13 cancer cells were seeded into 6-well plates and treated with 10 μmol/L special DNA methyltransferase (DNMT) 5-aza-CdR (Sigma) for 3 d. The drug liquid was replaced every day, reagent was wiped out and incubation was continued routinely for 5 d. RNA was isolated, and real-time PCR was performed as described previously.
Total cellular and tissue RNA was isolated by Trizol (Invitrogen) reagent according to the manufacturer’s recommendations. Cellular RNA was isolated from 5 × 106 to 1 × 107 cells by 1 mL Trizol decomposition and tissues samples were ground into a fine powder using a mortar and pestle, and incubated in Trizol solution (100 g/L) for 15 min. Then 1/5 volume of chloroform was added. After vigorous agitation standing for 5 min, the inorganic phase was separated by centrifugation at 12000 g for 15 min at 4 °C; RNA was then precipitated in the presence of equivolume isopropanol and centrifuged at 12000 g for 10 min at 4 °C. RNA pellets were washed with 1 mL 75% ice-cold ethanol [diethypyrocarbonate (DEPC) treated] and centrifuged at 8000 g for 5 min at 4 °C then dissolved in DEPC-treated H2O. Total RNA was quantified and concentration determined using ultraviolet (UV) spectrophotometry (Beckman Coulter) by absorbency at 260/280 nm and 1.2% denaturing agarose gel. For real-time polymerase chain reaction (real-time quantitative PCR) analysis, 2 μg RNA was reverse transcribed using reverse transcriptase M-MLV (Takara), Ribonuclease inhibitor (Takara) and dNTP mixture (Takara), according to the manufacturer’s protocol; the cDNA templates were subjected to PCR amplification.
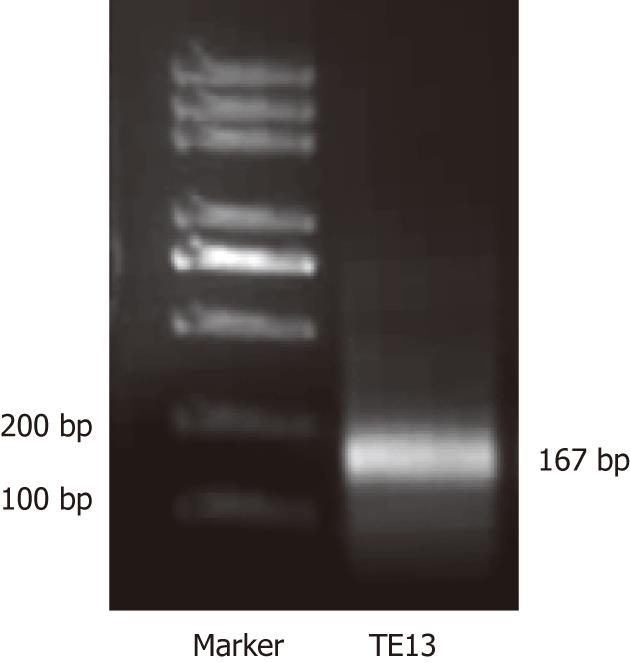
One μL cDNA from the TE13 cell line treated or not by 5-aza-CdR was used as the template to amplify specific fragments in 25 mL reaction mixture (10 × easy taq buffer 2.5 μL, 2 mol/L dNTP 2.5 μL, F primer 1 μL, R primer 1 μL, cDNA 1 μL, Easy taq 0.3 μL, ddH2O 16.7 μL) under the following conditions: denaturation at 94 °C for 3 min, 35 cycles at 94 °C for 30 s, at 56 °C for 30 s, at 72 °C for 20 s, then extensions at 72 °C for 10 min. The primer (10 μmol/L) sets were: RIZ1, forward 5’-TCTGCTGTTGACAAGACCC-3’, reverse 5’-GCATCAATGCACATCCATC-3’. The RIZ1 primer set yielded a band at 167 bp. 12 mL RT-PCR reaction product was analyzed by electrophoresis on a 12 g/L agarose gel. The electrophoresis images were scanned by UV spectrophotometer (Beckman Coulter). Sequencing of 0.75 μL of the resultant cDNA from TE13, which was mixed with 2 × SYBR Premix Ex Taq™ (Takara), was then performed. The primer (10 μmol/L) sets used were: RIZ1, forward 5’-TCTGCTGTTGACAAGACCC-3’, reverse 5’-GCATCAATGCACATCCATC -3’; GAPDH, forward 5’-GAAGGTGAAGGTCGGAGTC-3’, reverse 5’-GGGTGGAATCATATTGGAAC -3’. The amplifications were performed in LightCycler (Roche) real-time PCR system according to the manufacturer’s protocol. Each sample was run in triplicate for each gene. An initial denaturation step at 94 °C for 5 min was followed by 45 cycles of denaturation at 95 °C for 5 s, annealing at 59 °C for 20 s, extension at 72 °C for 10 s, then the solubility temperature curve assay was performed.
t-test was used to compare the measurement data, for instance the RIZ1 mRNA expression levels with primary ESCC and the adjacent and distal ending normal tissues by Real-time PCR. The relative quantitative results were analyzed by comparison of 2-averageΔΔCT× 100%. χ2 test was also used to estimate the enumeration data, for example the results. P values < 0.05 were considered statistically significant.
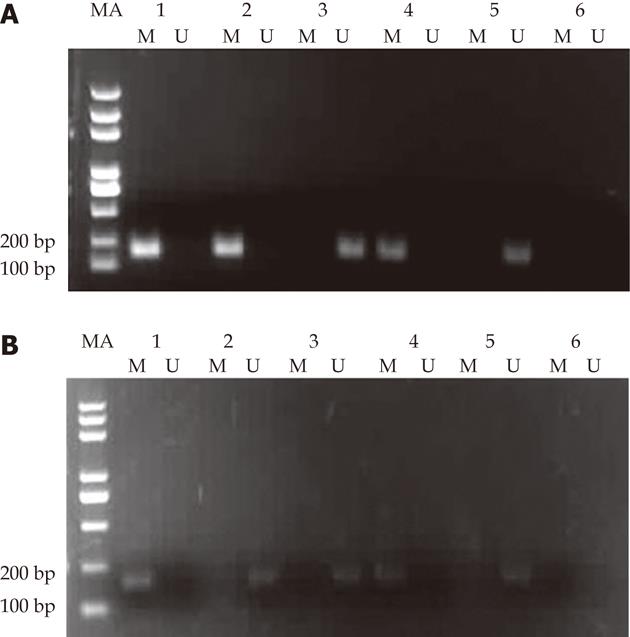
MSP analyses of RIZ1 promoter methylation status using genomic DNA extracted from 6 human ESCC cell lines, products of 177 bp and 175 bp were expected for methylated (M) and unmethylated (U) DNA. Promoter methylation of RIZ1 gene was detected in TE13, CaEs17, EC109 but not KYSE510, KYSE150 and EC9706 (Figure 1).
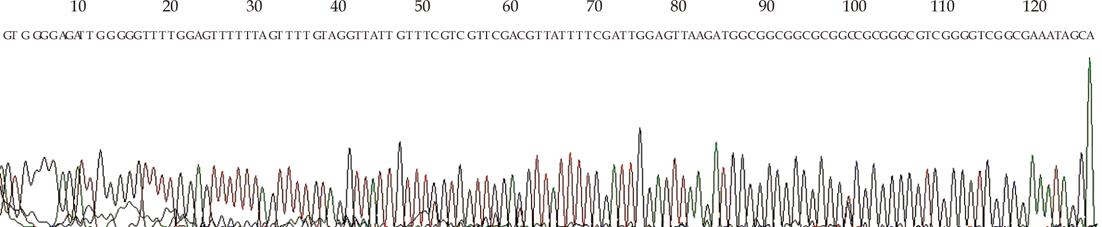
All cytosines were changed to thymines, except cytosines in CpG dinucleotide in M-sequences obtained from amplification using M-primer (Figure 2).
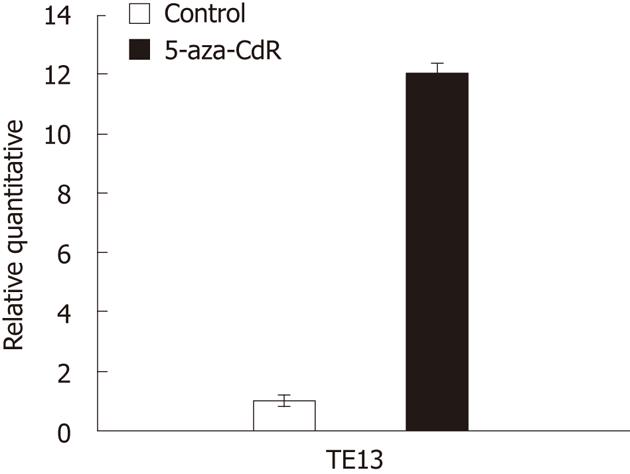
The expression of RIZ1 was higher after use of 5-aza-CdR than before its use (P < 0.01) (Figures 3-5).
Among 47 nonselective ESCC patients and matched adjacent normal and distal ending normal esophageal tissue, 26, 3 and 0 cases, respectively, exhibited methylation in the CpG island of the RIZ1 promoter. The corresponding methylation ratios were 55.3%, 6.4% and 0.0%. The rate of methylation in carcinomas tissues was significantly higher than that in matched adjacent normal and distal ending normal tissues, and the deviation of data was statistically significant (P < 0.01). The difference in methylation rates between matched adjacent normal and distal ending normal tissues possesses no statistical significance (P > 0.05). In the 3 samples where methylation was positive in matched neighbouring normal tissues, methylation also existed in the corresponding carcinoma tissues. MSP electrophoresis of ESCC patients with RIZ1 methylation positive amplification in both carcinomas and matched normal tissues is illustrated in Figure 6A, while that with RIZ1 methylation positive amplification only in carcinomas tissues but not in matched normal tissues is shown in Figure 6B.
The relation between analysis of the methylation state of the CpG island in the RIZ1 gene promoter by χ2 verification and the clinical information of the 47 sufferers of the ESCC is illustrated in Table 1. It can be seen that there was no correlation between the methylation state of the CpG island in the RIZ1 gene promoter of the 47 samples and the sufferers’ gender, age, familial history, tumour deviation, tumour saturation, lymph gland displacement and clinical stages, respectively. The fluctuation of data was not statistically significant (P > 0.05 in all groups) (Table 1 ).
| Factor | Methylation | Unmethylation | n | χ2 | P value |
| Gender | 0.340 | 0.560 | |||
| Male | 23 | 15 | 38 | ||
| Female | 7 | 2 | 9 | ||
| Age | 0.029 | 0.865 | |||
| > 50 | 14 | 20 | 34 | ||
| ≤ 50 | 5 | 8 | 13 | ||
| Family history | 0.151 | 0.698 | |||
| Present | 6 | 3 | 9 | ||
| Absent | 20 | 18 | 38 | ||
| Differentiation | 0.346 | 0.841 | |||
| Well | 6 | 9 | 15 | ||
| Moderately | 6 | 13 | 19 | ||
| Poorly | 4 | 9 | 13 | ||
| Depth of invasion | 1.420 | 0.492 | |||
| Mucosa and submuscosa | 4 | 5 | 9 | ||
| Muscle | 3 | 6 | 9 | ||
| Serosa | 16 | 13 | 29 | ||
| Lymph node metastasis | 3.489 | 0.062 | |||
| Present | 10 | 12 | 22 | ||
| Absent | 5 | 20 | 25 | ||
| Stage | 3.670 | 0.55 | |||
| I-II | 6 | 18 | 24 | ||
| III-IV | 12 | 11 | 23 |
Among the numerous genes that are known to be silenced in human cancers, for example p53, Syk, APC, BRCA1, etc.[18-20], RIZ1 is one of the few with a proven role in causing cancer as demonstrated. Whereas previous studies demonstrate reduced RIZ1 gene expression to be common in cancers, this study confirms that RIZ1 is commonly silenced by DNA methylation. The RIZ1 promoter has been demonstrated to have the characteristics of a CpG island, which suggests that RIZ1 is a target of inactivation by epigenetic mechanisms[21]. In prostate cancer, 42.6% of cancer cases were reported to have RIZ1 methylation, and was more frequent in patients with a high-grade malignancy[13]. In gastric adenocarcinoma, hypermethylation of RIZ1 was found in 69% of cancer tissues and in 21% of corresponding non-neoplastic mucosa[22]. In thyroid carcinoma Lal et al[14] reported that all of the 31 cancerous cases were methylated, and methylation was significantly frequent compared with normal thyroid tissues (33%). Du et al[21] reported that methylation of RIZ1 was detected in 44% (11/25) of breast cancer specimens and 62% (20/32) of liver cancer specimens. However, RIZ1 mutation has not been detected in these cancers[23]. Thus, DNA methylation may represent the preferred mechanism of RIZ1 inactivation in these cancers. Furthermore, because many types of human cancer cell lines exhibit reduced RIZ1 expression, we predict that RIZ1 gene methylation will be commonly found in many types of human cancer tissue. In a previous study, we found that, compared with normal tissues, the expression of RIZ1 mRNA was significantly lower in cancer tissues than in the adjacent non-cancerous tissues (P < 0.01). But in this study there was no significant difference between cancer tissues and the adjacent non-cancerous tissues (P = 0.067). No RIZ1 protein expression was observed in the cancer tissues, as it was 0% (0/12), while the expression level in the normal tissues was 66.67% (8/12). There was a statistically significant difference between RIZ1 mRNA and protein expression (P < 0.05) which indicates that RIZ1 expression may reduce the occurrence of ESCC. RIZ1 may be a candidate tumor suppressor in ESCC.
In this paper, we explored DNA methylation of RIZ1 in the promoter region among human ESCC cell lines, malignant human ESCC, its matched adjacent normal and distal ending normal tissues. MSP was used to detect the promoter region methylation status of RIZ1 gene in human ESCC cell lines including KYSE150, KYSE510, TE13, EC9706, CaEs17 and EC109. Promoter methylation of RIZ1 gene was detected in TE13, CaEs17 and EC109. TE13 was chosen for further research and treated with 5-aza-CdR. Real-time PCR shows us that the expression of RIZ1 mRNA in TE-13 was up-regulated after treatment by the drug. The results of study using ESCC pathological frozen specimens illustrate that the R1Z1 gene promoter of the ESCC possesses a 55.3% methylation positive ratio (26/47). RIZ1 methylation positive amplification was seen in 26 sets of carcinoma tissue from 47 sufferers and unmethylation positive amplification was seen in 21. In matched neighboring normal esophageal tissues, the RIZ1 methylation positive amplification appeared in 3 cases, while unmethylation positive amplification was found in 44 cases. Furthermore, in distal ending normal esophageal tissues, the methylation positive amplification did not exist, i.e., all 47 studied cases possessed unmethylation positive amplification. Among those 3 sufferers with RIZ1 methylation positive amplification in matched neighbouring normal esophageal tissues, the corresponding carcinoma tissue also contained RIZ1 methylation positive amplification. In the 47 sufferers of ESCC, 3 possessed methylation in carcinoma tissue and matched neighbouring normal tissue but possessed unmethylation in distal ending normal tissue. Twenty three sufferers showed methylation in carcinoma tissues, but unmethylation in matched neighbouring normal tissue and distal ending normal tissue. Twenty one sufferers possessed unmethylation in carcinoma, matched neighbouring normal and distal ending normal tissues. The rate of methylation of RIZ1 promoter in ESCC was higher than that of adjacent normal tissues (χ2 = 24.1, P < 0.01).
DNA methylation in the promoter region may play an important role in the epigenetic silencing of RIZ1 gene expression. However, the test to determine RIZ1 gene promoter methylation shows there is no methylation found in 2 other human ESCC cell lines. This means that dimunition of RIZ1 expression is not always triggered by promoter methylation. One such potential mechanism is that RIZ1 silencing could be caused by a defect in a certain transcription factor that normally activates the RIZ1 promoter. Another potential mechanism is mutation in the RIZ1 promoter. However, given the prevalence of DNA methylation, these other mechanisms are not likely to be commonly involved, and further research is required to find out the true mechanism. Furthermore, we did not observe any significant correlation between RIZ1 methylation and tumour grade. This may be attributable to a relatively small sample size and the complexity of the unselected patient population. Additional detailed studies using a patient cohort should be done needed to examine the value of RIZ1 methylation as a diagnostic or prognostic marker. Another explanation is that gene methylation exists in the early stage of ESCC, but not in the middle and late stages. Hence, there is a statistically insignificant discrepancy during the development of tumour and the displacement of lymph gland. We guess that the methylation of the CpG island in RIZ1 may be an important molecular mechanism during the appearance and development of ESCC in the early stage, and may become a biological parameter for testing early stage of ESCC.
The vital importance of the epigenetic changes on the generation and development of tumours has been thoroughly realized by human beings. Nowadays, there exists an expert database of DNA methylation for researchers (http://www.methdb.de). The DNA methylation abnormality of malignancy characterizes significantly according to: specifics of the tumour, specifics and reversibility of genes and tissues, etc. Methylation testing techniques possess high sensibility and specificity. Thereinto, the MSP technique can be applied for testing some small quantity of tissues sections, phlegm and urine, etc., as well as for clinical follow-up through quantitative analysis of relevant genes. Therefore, further research on the mechanisms of the tumour suppressor gene RIZ1 on esophageal cancer may show some new parameters of early diagnosis and prognosis evaluation for esophageal cancer. The DNMT, 5-aza-CdR, has been clinically applied for curing some solid tumors and some hematological diseases, such as myelodysplastic syndrome, acute myeloid leukemia, etc. A new therapy target for esophageal cancer may be found by strengthening the research on DNA methylation of the genes and the significance of application of 5-aza-CdR.
We thank Dr. Si-Cheng Zhao for verbal changes in the material.
Recently, methylation of the retinoblastoma protein-interacting zinc finger gene (RIZ1) promoter CpG islands has been proposed as a common mechanism in inactivating RIZ1. Increasing clinical evidence reveals a positive correlation of reduced RIZ1 expression with increased risk for metastasis, indicating that RIZ1 may be a potential new tumour suppressor gene (TSG). However, although RIZ1 is a putative tumour suppressor in several cancer types, for instance breast cancer, gastric cancer, lung cancer, the role of RIZ1 in human esophageal squamous cell carcinoma (ESCC) has not been reported.
In mammalian development, DNA methylation has an essential regulatory function of suppressing gene activity by changing chromatin structure. It has become apparent that aberrant DNA methylation of promoter region CpG islands may serve as an alternate mechanism to genetic defects in the inactivation of TSG in human malignancies.
China is a country with a high incidence of ESCC, and the pathological type is mainly squamous cell carcinoma, which is different from adenocarcinoma reported in other countries. The present study aimed to discover the effect and mechanism of the anti-cancer gene RIZ1 on ESCC. The results illustrate that low expression of RIZ1 in ESCC is relevant to promoter methylation. Methylation takes place in the early stage of carcinogenesis, and it may become a molecular biological parameter for early diagnosis.
The methylation of RIZ1 is the major reason for the low expression in ESCC. This could take place in early stages of ESCC, and is expected to be a molecular biological parameter for early diagnosis.
This paper reports RIZ1 promoter methylation in esophageal cancer in a usual way.
Peer reviewers: Roderick M Quiros, MD, FACS, Surgical Oncologist, Cancer Care Associates, 801 Ostrum Street, Bethlehem, PA 18015, United States; Haruhiko Sugimura, MD, PhD, Professor, Department of Pathology, Hamamatsu University School of Medicine, 1-20-1 Handayama, Higashi-ku, Hamamatsu, 431-3192, Japan
S- Editor Tian L L- Editor O’Neill M E- Editor Xiong L
| 1. | Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241-2252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2115] [Cited by in RCA: 2215] [Article Influence: 100.7] [Reference Citation Analysis (0)] |
| 2. | Chen BS, Wang MR, Xu X, Cai Y, Xu ZX, Han YL, Wu M. Transglutaminase-3, an esophageal cancer-related gene. Int J Cancer. 2000;88:862-865. [PubMed] |
| 3. | Metzger R, Schneider PM, Warnecke-Eberz U, Brabender J, Hölscher AH. Molecular biology of esophageal cancer. Onkologie. 2004;27:200-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 38] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 4. | Montesano R, Hollstein M, Hainaut P. Genetic alterations in esophageal cancer and their relevance to etiology and pathogenesis: a review. Int J Cancer. 1996;69:225-235. [PubMed] |
| 5. | Qin YR, Wang LD, Fan ZM, Kwong D, Guan XY. Comparative genomic hybridization analysis of genetic aberrations associated with development of esophageal squamous cell carcinoma in Henan, China. World J Gastroenterol. 2008;14:1828-1835. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 24] [Cited by in RCA: 24] [Article Influence: 1.4] [Reference Citation Analysis (9)] |
| 6. | Kass SU, Pruss D, Wolffe AP. How does DNA methylation repress transcription? Trends Genet. 1997;13:444-449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 370] [Cited by in RCA: 374] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 7. | Razin A, Cedar H. DNA methylation and gene expression. Microbiol Rev. 1991;55:451-458. [PubMed] |
| 8. | Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1657] [Cited by in RCA: 1606] [Article Influence: 61.8] [Reference Citation Analysis (0)] |
| 9. | Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225-3229. [PubMed] |
| 10. | Liu L, Shao G, Steele-Perkins G, Huang S. The retinoblastoma interacting zinc finger gene RIZ produces a PR domain-lacking product through an internal promoter. J Biol Chem. 1997;272:2984-2991. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 77] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 11. | Steele-Perkins G, Fang W, Yang XH, Van Gele M, Carling T, Gu J, Buyse IM, Fletcher JA, Liu J, Bronson R. Tumor formation and inactivation of RIZ1, an Rb-binding member of a nuclear protein-methyltransferase superfamily. Genes Dev. 2001;15:2250-2262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 149] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 12. | He L, Yu JX, Liu L, Buyse IM, Wang MS, Yang QC, Nakagawara A, Brodeur GM, Shi YE, Huang S. RIZ1, but not the alternative RIZ2 product of the same gene, is underexpressed in breast cancer, and forced RIZ1 expression causes G2-M cell cycle arrest and/or apoptosis. Cancer Res. 1998;58:4238-4244. [PubMed] |
| 13. | Hasegawa Y, Matsubara A, Teishima J, Seki M, Mita K, Usui T, Oue N, Yasui W. DNA methylation of the RIZ1 gene is associated with nuclear accumulation of p53 in prostate cancer. Cancer Sci. 2007;98:32-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 14. | Lal G, Padmanabha L, Smith BJ, Nicholson RM, Howe JR, O'Dorisio MS, Domann FE. RIZ1 is epigenetically inactivated by promoter hypermethylation in thyroid carcinoma. Cancer. 2006;107:2752-2759. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 34] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 15. | Gazzerro P, Abbondanza C, D'Arcangelo A, Rossi M, Medici N, Moncharmont B, Puca GA. Modulation of RIZ gene expression is associated to estradiol control of MCF-7 breast cancer cell proliferation. Exp Cell Res. 2006;312:340-349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 16. | Tokumaru Y, Nomoto S, Jerónimo C, Henrique R, Harden S, Trink B, Sidransky D. Biallelic inactivation of the RIZ1 gene in human gastric cancer. Oncogene. 2003;22:6954-6958. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 36] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 17. | Fang W, Piao Z, Buyse IM, Simon D, Sheu JC, Perucho M, Huang S. Preferential loss of a polymorphic RIZ allele in human hepatocellular carcinoma. Br J Cancer. 2001;84:743-747. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 18. | Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, Stockert E, Day RS, Johnson BE, Skolnick MH. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436-440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1955] [Cited by in RCA: 1887] [Article Influence: 60.9] [Reference Citation Analysis (0)] |
| 19. | Dong SW, Ma L, Xu N, Yan HQ, Liu HY, Li YW, Zhang P. Research on the reactivation of Syk expression caused by the inhibition of DNA promoter methylation in the lung cancer. Neoplasma. 2011;58:89-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 20. | Ma L, Dong S, Zhang P, Xu N, Yan H, Liu H, Li Y, Zhou Q. The relationship between methylation of the Syk gene in the promoter region and the genesis of lung cancer. Clin Lab. 2010;56:407-416. [PubMed] |
| 21. | Du Y, Carling T, Fang W, Piao Z, Sheu JC, Huang S. Hypermethylation in human cancers of the RIZ1 tumor suppressor gene, a member of a histone/protein methyltransferase superfamily. Cancer Res. 2001;61:8094-8099. [PubMed] |
| 22. | Oshimo Y, Oue N, Mitani Y, Nakayama H, Kitadai Y, Yoshida K, Chayama K, Yasui W. Frequent epigenetic inactivation of RIZ1 by promoter hypermethylation in human gastric carcinoma. Int J Cancer. 2004;110:212-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 46] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 23. | Fang W, Piao Z, Simon D, Sheu JC, Huang S. Mapping of a minimal deleted region in human hepatocellular carcinoma to 1p36.13-p36.23 and mutational analysis of the RIZ (PRDM2) gene localized to the region. Genes Chromosomes Cancer. 2000;28:269-275. [PubMed] |