Published online Feb 14, 2012. doi: 10.3748/wjg.v18.i6.570
Revised: July 17, 2011
Accepted: August 15, 2011
Published online: February 14, 2012
AIM: To screen the differential expressed genes in colorectal cancer and polyp tissue samples.
METHODS: Tissue specimens containing 16 cases of colorectal adenocarcinoma and colorectal polyp vs normal mucosae were collected and subjected to cDNA microarray and bioinformatical analyses. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was used to confirm some of the cDNA microarray data.
RESULTS: The experimental data showed that eight genes were differentially expressed, most of which were upregulated in adenomatous polyp lesions. Forty-six genes expressions were altered in colorectal cancers, of which 29 were upregulated and 17 downregulated, as compared to the normal mucosae. In addition, 18 genes were similarly altered in both adenomatous polyps and colorectal cancer. qRT-PCR analyses confirmed the cDNA microarray data for four of those 18 genes: MTA1, PDCD4, TSC1 and PDGFRA.
CONCLUSION: These differentially expressed genes likely represent biomarkers for early detection of colorectal cancer and may be potential therapeutic targets after confirmed by further studies.
- Citation: Dai YC, Zhu XS, Nan QZ, Chen ZX, Xie JP, Fu YK, Lin YY, Lian QN, Sang QF, Zhan XJ. Identification of differential gene expressions in colorectal cancer and polyp by cDNA microarray. World J Gastroenterol 2012; 18(6): 570-575
- URL: https://www.wjgnet.com/1007-9327/full/v18/i6/570.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i6.570
Colorectal polyp (CRP) is considered as a premalignant lesion for development of colorectal cancer (CRC)[1]. Although the mechanism underlying colorectal cancer development remains to be defined, a series of genetic and epigenetic events are thought to play important roles in colorectal carcinogenesis, including oncogene activation and tumor suppressor gene inactivation[2,3].
By attaining a detailed understanding of the altered gene expression profile of colorectal cancer novel strategies may be developed for earlier detection and more effective prevention and treatment, thereby reducing colorectal cancer incidence and increasing survival rates.
In this study, we performed a cDNA microarray analysis to profile differential gene expressions in tissue specimens of polyps and colorectal carcinoma and compared the expression profiles to that in corresponding normal tissues. We chose the genes with marked differential expressions for verification by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). These data provide insightful information into the genetic mechanisms of colorectal cancer and identify genes that may be useful as biomarkers for early disease detection.
This study was carried out in accordance with the Declaration of Helsinki (2000) of the World Medical Association and approved by the Medical Ethics Committee of Fujian Province, China. All patients read and signed an informed consent form prior to surgery and sample collection.
A total of 16 patients with colorectal adenocarcinoma and adenomatous polyp lesions were collected from The 174th Hospital of the Chinese PLA between May 2006 and December 2010. Diagnosis of these patients was confirmed by surgical pathology. None of the patients received any pre-surgical chemo- or radiation-therapy. All tissue specimens were immediately taken from the operation room upon excision from the patient, snap frozen in liquid nitrogen, and stored at -80 °C until use. The tissue specimens from these patients were divided into two groups: adenomatous polyp lesions vs proximal non-cancerous colorectal mucosae (Group A) or colorectal cancer vs proximal non-cancerous colorectal mucosae (Group B). The clinicopathological characteristics of these patients are summarized Table 1.
| Case | Sex | Age | Size in cm/n | Differentiation | Depth | Dukes | |
| CRC | CRP | CRC | Polyps | ||||
| 1 | F | 41 | 1.7/2 | 0.6/2 | High | S | B1 |
| 2 | M | 37 | 0.9/1 | 1.2/3 | Poor | Ss | C1 |
| 3 | M | 68 | 2.9/2 | 0.7/1 | Poor | Sm | D1 |
| 4 | F | 27 | 2.8/1 | 1.4/1 | Poor | Ss | C2 |
| 5 | M | 59 | 1.8/1 | 0.5/3 | Moderate | S | C2 |
| 6 | F | 52 | 2.9/1 | 1.9/4 | Poor | Ss | B2 |
| 7 | M | 71 | 3.3/2 | 0.4/1 | High | Sm | B1 |
| 8 | M | 46 | 2.7/1 | 3.1/1 | Moderate | Ss | C1 |
| 9 | M | 28 | 0.9/1 | 2.8/2 | Poor | Sm | B1 |
| 10 | F | 36 | 2.6/1 | 0.4/1 | Poor | Ss | C2 |
| 11 | F | 30 | 2.4/1 | 2.8/2 | Moderate | S | C1 |
| 12 | M | 46 | 3.1/1 | 1.3/2 | Poor | Sm | B1 |
| 13 | M | 62 | 1.5/1 | 2.0/3 | Poor | Ss | C2 |
| 14 | F | 24 | 0.8/1 | 2.4/2 | Moderate | Sm | C2 |
| 15 | M | 70 | 2.5/1 | 4.3/1 | Poor | S | B2 |
| 16 | M | 43 | 3.2/2 | 3.7/1 | High | Ss | B2 |
Total cellular RNA was extracted from the tissue samples by using the Trizol reagent (Sigma-Aldrich Inc., Germany)[2,4]. mRNA isolation was then carried out with Qiagen Oligotex beads, (Valencia, CA, United States) according to the manufacturer’s instructions. The final concentration of mRNA was measured by spectrophotometer.
Next, the mRNAs from colorectal cancer or adenomatous polyp lesions were reverse transcribed into cDNA by means of Cy5-dUTP labeling, while the mRNAs from the normal mucosae were processed with Cy3-dUTP labeling by following the manufacturer’s protocols (NEN Company, Boston, MA, United States). The labeled probes were then hybridized to the cDNA microarray (Chipscreen, Shenzhen, China), which contained 8064 human genes.
Hybridized cDNA microarrays were scanned using a Gene PIX 4000 microarray fluorescence scanner (Axon Instruments, Foster City, CA, United States). Accompanying bioinformatical software was used convert the output images to data form and perform analysis. Ratios of Cy5: Cy3 were normalized to the median ratio value of all the microarray spots detected. Spots with intensities in both channels that were 0.5 to 2.0-fold higher than the local background were excluded from further analysis. SPSS v13.0 statistical software (Chicago, IL, United States) was used to carry out Student’s t-test statistical analysis to determine significant intergroup differences of gene expression. P-values < 0.05 were considered statistically significant.
Differentially expressed genes between the adenomatous polyp lesions and colorectal cancer samples detected by the cDNA microarray, as compared to non-cancerous tissues, were verified by using qRT-PCR. The primers for these mRNAs used for qRT-PCR is listed in Table 2.
| Gene tag | Sequences | Tm (°C) | Cycle | Product (bp) |
| MTA1 | 5’-AGCCGTGCTTCGGTATCTT-3’ | 57 | 30 | 580 |
| 5’-CCCGTTGTGCTGCTCGTA-3’ | ||||
| PDCD4 | 5′-GCTGAATTCGGATGGATGTAGAAAATGAGCAGA-3’ | 54 | 27 | 470 |
| 5’-CTGCTCGAGTCAGTAGCTCTCTGGTTTAAGA-3’ | ||||
| TSC1 | 5′-ATCGCCTTTATGGAATGT-3’ | 49 | 29 | 510 |
| 5’-GCTTGTGGTGGTTCAGTT-3’ | ||||
| PDGFRA | 5’-ACCATAAGGCTCTTACTCT-3’ | 45 | 31 | 490 |
| 5’-TTCTGGCACTTACCTACA-3’ |
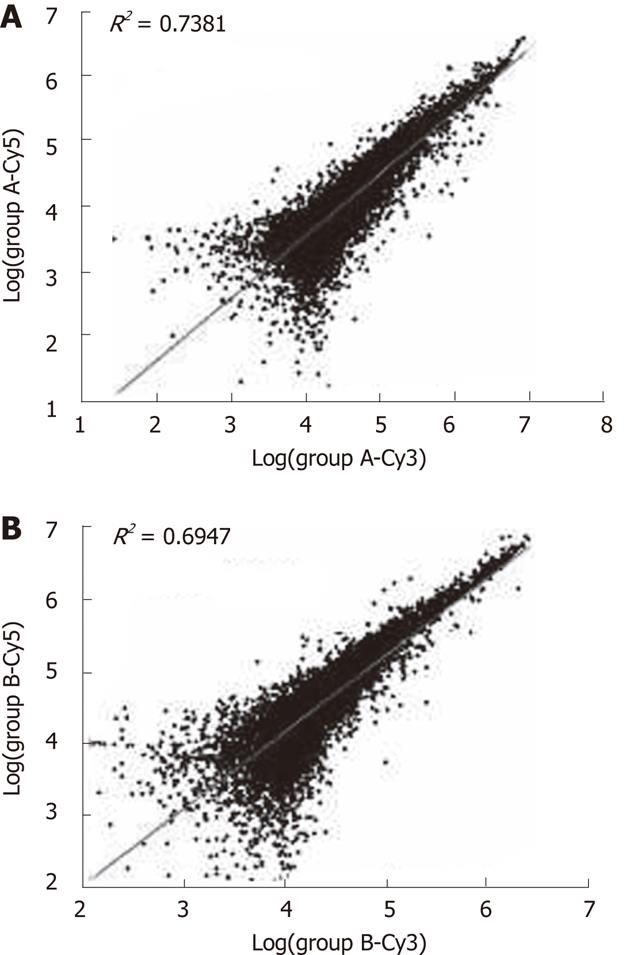
Quality of the isolated total RNA and mRNA from the tissue samples was found to have high correlation coefficients (Figure 1). The fluorescence signal was consistent with the expectations and standards (Figure 2); for example, the good 28S to 18S RNA subunit ratio in these samples indicated that there was no significant degradation (data not shown).
cDNA microarray analysis of these tissue mRNAs revealed that eight genes were differentially expressed between adenomatous polyp lesions and the normal mucosae, and most of these were upregulated in the polyps (Table 3, P < 0.05). Meanwhile, 46 genes were differentially expressed between colorectal cancer and normal tissues, of which 29 were up-regulated in the cancer samples (Table 3, P < 0.05). A total of 18 genes were found to be similarly altered in both adenomatous polyp lesions and colorectal cancer samples (Table 4). qRT-PCR confirmed the observed patter for four of those 18 genes: MTA1, PDCD4, TSC1 and PDGFRA (Table 5).
| Accession | Gene function | Gene tag | Ratio |
| AY421086 | Programmed cell death 4 | PDCD4 | 6.41 ± 0.10 |
| AA630800 | Interferon, γ-inducible protein 30 | IFI30 | 1.31 ± 0.18 |
| AA400973 | Lipocalin 2 (oncogene 24p3) | LCN2 | 3.16 ± 0.22 |
| AI817942 | Zeta-chain associated protein kinase (70 kD) | ZAP70 | 3.29 ± 0.31 |
| AA447515 | Mad4 homolog | MAD4 | 2.35 ± 0.20 |
| W47350 | Retinoic acid receptor responder 3 | RARRES3 | 0.18 ± 0.13 |
| AA436401 | TU3A protein | TU3A | 5.40 ± 0.27 |
| AI650283 | Serum/glucocorticoid regulated kinase 2 | SGK2 | 1.29 ± 0.13 |
| NM003542 | H4 histone family, member G | H4FG | 7.04 ± 0.17 |
| NM205510 | Fibroblast growth factor receptor 1 | FGFR1 | 2.94 ± 0.21 |
| NM204434 | Cyclin-dependent kinase inhibitor 2A | CDKN2A | 3.16 ± 0.28 |
| NM005438 | FOS-like antigen-1 | FOSL1 | 1.93 ± 0.25 |
| NM005439 | Myeloid leukemia factor 2 | MLF2 | 2.17 ± 0.24 |
| BC08072 | v-raf murine sarcoma 3611 viral oncogene homolog 1 | ARAF1 | 0.98 ± 0.15 |
| NM020531 | Chromosome 20open reading frame 3 | C20ORF3 | 2.23 ± 0.21 |
| NM033158 | Hyaluronoglucosaminidase 2 | HYAL2 | 4.16 ± 0.28 |
| NM001950 | E2F transcription factor 4,p107/p130-binding | E2F4 | 5.26 ± 0.28 |
| BC059522 | Ribosomal protein S30 | FAU | 2.58 ± 0.13 |
| NM008583 | Multiple endocrine neoplasia I | MEN1 | 3.89 ± 0.11 |
| NM011492 | Serine/threonine kinase 11 | STK11 | 5.12 ± 0.27 |
| NM133862 | Fibrinogen, gamma polypeptide | FGG | 3.0 4 ± 0.28 |
| BC162533 | GRO2 oncogene | GRO2 | 1.39 ± 0.30 |
| NM000612 | Insulin-like growth factor 2 receptor | IGF2 | 5.06 ± 0.29 |
| NM005343 | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | HRAS | 4.74 ± 0.27 |
| NM002634 | Prohibitin | PHB | 2.98 ± 0.15 |
| BC046375 | p53-induced protein | PIG11 | 6.67 ± 0.29 |
| NM004448 | v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 | ERBB2 | 3.15 ± 0.17 |
| NM010658 | v-maf musculoaponeurotic fibrosarcoma oncogene family | MAFG | 5.61 ± 0.04 |
| NM022012 | Mitogen-activated protein 3 kinase 11 | MAP3K11 | 3.32 ± 0.06 |
| NM023983 | Melanoma adhesion molecule | MCAM | 7.38 ± 0.14 |
| NM014567 | Breast cancer anti-estrogen resistance 1 | BCAR1 | 5.12 ± 0.23 |
| NM000535 | Postmeiotic segregation increased 2 | PMS2 | 5.17 ± 0.25 |
| NM183243 | Inosine monophosphate dehydrogenase 1 | IMPDH1 | 7.14 ± 0.10 |
| NM005380 | Neuroblastoma, suppression of tumorigenicity 1 | NBL1 | 5.62 ± 0.16 |
| NM002429 | Matrix metalloproteinase 19 | MMP19 | 3.90 ± 0.22 |
| NM002466 | v-myb avian myeloblastosis viral oncogene homolog-like 2 | MYBL2 | 9.70 ± 0.21 |
| NM022588 | Metastasis associated 1 | MTA1 | 10.41 ± 0.37 |
| NM017045 | Retinoblastoma 1 | RB1 | 2.81 ± 0.14 |
| NC006104 | SET translocation | SET | 2.69 ± 0.11 |
| NM002439 | Phosphatase and tensin homolog | PTEN | 3.94 ± 0.21 |
| NM053455 | Fibrinogen-like 2 GTPase activating | FGL2 | 3.40 ± 0.27 |
| NM005638 | ADP-ribosylation factor protein 1 | ARFGAP | 5.14 ± 0.25 |
| NM032415 | Mucosa associated lymphoid tissue lymphoma translocation gene 1 | MALT1 | 4.84 ± 0.21 |
| NM006283 | Transforming acidic coiled-coil containing protein 1 | TACC1 | 3.41 ± 0.13 |
| NM00288 | v-ral simian leukemia viral oncogene homolog B | RALB | 4.12 ± 0.18 |
| NM003766 | Myosin-like BCL2-interacting protein | BECN1 | 5.05 ± 0.14 |
| NG027821 | TRK-fused gene | TFG | 4.33 ± 0.28 |
| NM005805 | Cadherin 1,type 1,E-cadherin (epithelial) | CDH1 | 4.11 ± 0.26 |
| NM001982 | v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 | ERBB3 | 5.82 ± 0.15 |
| NM133250 | MutS (Escherichia coli) homolog 2 | MSH2 | 2.13 ± 0.21 |
| AY805747 | Ras homolog gene family, member E | ARHE | 3.61 ± 0.22 |
| NM002884 | RAP1A, member of RAS oncogene family | RAP1A | 5.18 ± 0.27 |
| NM000368 | Tuberous sclerosis 1 | TSC1 | 6.02 ± 0.14 |
| L36953 | Mothers against decapentaplegic homolog 4 | MADH4 | 4.93 ± 0.26 |
| Accession | Gene function | Gene tag | Ratio | |
| A | B | |||
| AA191692 | Stratifin | SFN | 2.36 ± 0.25 | 3.86 ± 0.11a |
| H15456 | Calpain 1, (mu/I) large subunit | CAPN1 | 8.17 ± 0.20 | 10.25 ± 0.24a |
| AA043501 | v-maf musculoaponeurotic fibrosarcoma (avian) oncogene homolog | MAF | 1.36 ± 0.08 | 4.85 ± 0.27a |
| AA495936 | Microsomal glutathione S-transferase 1 | MGST1 | -2.14 ± 0.23 | 2.90 ± 0.16a |
| H23235 | Platelet-derived growth factor receptor | PDGFRA | -0.15 ± 0.31 | 4.81 ± 0.14a |
| AA430032 | Pituitary tumor-transforming 1 | PTTG | 4.91 ± 0.23 | 11.46 ± 0.18a |
| N94468 | Jun B proto-oncogene | JUNB | 1.27 ± 0.20 | 11.09 ± 0.14a |
| T61948 | FBJ murine osteosarcoma viral oncogene B homolog | FOSB | 1.37 ± 0.31 | 10.21 ± 0.20a |
| AA460168 | Growth arrest and DNA damage inducible 34 | GADD34 | 1.07 ± 0.15 | 4.15 ± 0.20a |
| L36870 | MAP kinase kinase 4 | MKK4 | 4.01 ± 0.23 | 6.93 ± 0.24a |
| AA426216 | Malignant cell expression-enhanced gene | LENG4 | 7.03 ± 0.16 | 8.81 ± 0.23a |
| AA486219 | SRp25 nuclear protein | LOC51329 | 2.09 ± 0.12 | 6.18 ± 0.23a |
| AA457705 | Immediate early response 3 | IER3 | 1.94 ± 0.25 | 9.17 ± 0.20a |
| AA485377 | v-fos FBJ murine osteosarcoma viral oncogene homolog | FOS | 4.03 ± 0.27 | 7.21 ± 0.05a |
| AA463204 | Pleiomorphic adenoma gene-like 1 | PLAGL1 | 7.11 ± 0.24 | -4.16 ± 0.06a |
| AA434373 | E74-like factor 3 (epithelial-specific ) | ELF3 | 1.09 ± 0.29 | 7.03 ± 0.15a |
| AI677994 | Fms-associated tyrosine kinase 3 ligand | FLT3LG | 1.94 ± 0.32 | 4.35 ± 0.02a |
| AA464600 | v-myc avian myelocytomatosis viral oncogene homolog | MYC | 3.27 ± 0.17 | 8.01 ± 0.13a |
Colorectal carcinoma is a major cause of cancer-related deaths in China[5,6]. Unfortunately, little is known about the gene expression profiles between colorectal cancer and adenomatous polyp lesions. Genes that are differentially expressed in adenomatous polyp lesions may represent useful biomarkers for risk of colorectal cancer development, while altered genes in cancerous tissues may be used as therapeutic targets for colorectal cancer treatment[7].
Adenomatous polyp is the premalignant lesion of colorectal cancer[8-12]. Therefore, we conducted the current study to profile the differential gene expressions between normal mucosae and adenomatous polyp lesions, between normal mucosae and colorectal cancer, and between adenomatous polyp lesions and colorectal cancer. We found that eight genes were differentially expressed in adenomatous polyp lesions, as compared to normal mucosae. In addition, 46 genes were differentially expressed in colorectal cancer, as compared to the normal mucosae; twenty-nine of which were up-regulated. A total of 18 genes were significantly upregulated in both colorectal cancer and adenomatous polyp lesions, further indicating that adenomatous polyp is a precancerous lesion. However, some genes were downregulated in adenomatous polyp lesions (such as MGST1 and PDGFRA) or upregulated in colorectal cancers only (PLAGL1). These genes encode proteins that are known to be involved in cell growth, apoptosis, and metastasis and are likely to contribute to colorectal carcinogenesis, as purported by previous studies[13,14].
The MGST1 gene is located in 12p13.1-13.2 and was previously considered to be a “housekeeping” gene[15]. However, it has been frequently observed as upregulated in various cancers. A recent in vitro study has implicated the role of MGST1 in development of multiple drug resistance during breast cancer chemotherapy with several cytostatic drugs (such as cisplatin)[16]. Polymorphisms in MGST1 have also been associated with colorectal cancer risk in Chinese[17]. In this study, we found that MGST1 mRNA levels were upregulated in colorectal cancer, as compared to those detected in normal mucosae (2.90 ± 0.16). Intriguingly, MGST1 was down-regulated in adenomatous polyps, as compared to the normal mucosae (2.14 ± 0.23), but further study is necessary to fully understand the implications of this finding.
PDGFRA gene mutation is commonly observed in tissues of gastrointestinal stromal tumors[18]. Mutated PDGFRA proteins demonstrate constitutively elevated tyrosine kinase activity and possess transforming ability, which can be reversed through PDGFR blockade[19]. Thus, mutants of PDGFRA protein behave as oncogenes, as has been demonstrated in glioma samples[20]. Here, we observed high expression of PDGFRA in colorectal cancers, as compared to that in normal tissues (4.81 ± 0.14). This observation suggests that PDGFRA may contribute to cancer development or maintenance of the tumor phenotype, possibly by supporting properties of tumor cell growth and invasiveness. However, to the reason why PDGFRA was down-regulated in adenomatous polyps remains unclear.
Finally, PLAGL1, a tumor suppressor gene, is localized on the chromosome 6q24-25 and is the target of several types of chromosomal rearrangement, including one identified in pleomorphic adenomas and malignant tumors. PLAGL1 is ubiquitously expressed in many human tissues where it regulates normal physiological functions; however, it has also been demonstrated to functionally contribute to complex pathologies such as cancer[21-23]. Our current study showed that PLAGL1 mRNA was down-regulated in colorectal cancer, as compared to adenomatous polyps, suggesting that PLAGL1 protein may also play a role in suppressing colorectal cancer development.
The functional roles for each of these genes in colorectal tumorigenesis remain to be verified. Nonetheless, our data provide insightful information into their potential roles in this complex and diverse disease[24]. Future studies will aim to verify these differentially expressed genes as biomarkers for early detection and/or therapeutic targets for treatment of colorectal cancers.
The mechanism of colorectal carcinogenesis remains to be defined and this study aims to obtain the gene expression profiles between colorectal cancer and adenomatous polyp lesions.
Many researchs on genetic and epigenetic events which are thought to play important roles in colorectal carcinogenesis, such as oncogene activation and tumor suppressor gene inactivation.
The study firstly screened the differential expressed genes in colorectal adenocarcinoma and colorectal polyp vs normal mucosae.
These differentially expressed genes maybe as biomarkers for early detection and/or therapeutic targets for treatment of colorectal cancers.
Study was well designed and performed methodologically.
Peer reviewers: Jean Wang, Professor, Washington University, 660 S. Euclid Ave, Campus Box 8124, St. Louis, United States; Zoran Krivokapic, Professor, MD, FRCS, Institute for Digestive Disease, First Surgical Clinic, Clinical Center of Serbia, 6, Dr Koste Todorovica, Belgrade, 11 000, Serbia
S- Editor Lv S L- Editor Webster JR E- Editor Xiong L
| 1. | Gopalappa C, Aydogan-Cremaschi S, Das TK, Orcun S. Probability model for estimating colorectal polyp progression rates. Health Care Manag Sci. 2011;14:1-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 2. | Klementa I, Skalický P, Vyslouzil K, Starý L, Zboril P, Vomáckova K, Klementa B, Konecný M. [Risk of colorectal carcinoma in a patient with ulcerative colitis]. Rozhl Chir. 2010;89:754-759. [PubMed] |
| 3. | Sobala A, Herbst F, Novacek G, Vogelsang H. Colorectal carcinoma and preceding fistula in Crohn's disease. J Crohns Colitis. 2010;4:189-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 4. | Meng L, Feldman L. A rapid TRIzol-based two-step method for DNA-free RNA extraction from Arabidopsis siliques and dry seeds. Biotechnol J. 2010;5:183-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 100] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 5. | Cacheux W, Le Tourneau C, Baranger B, Mignot L, Mariani P. Targeted biotherapy in metastatic colorectal carcinoma: Current practice. J Visc Surg. 2011;148:12-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 6. | Nabi U, Nagi AH, Riaz S, Sami W. Morphological evaluation of colorectal carcinoma with grading staging and histological types. J Pak Med Assoc. 2010;60:998-1001. [PubMed] |
| 7. | Zhao R, Li J. Perspectives on the treatment of colorectal carcinoma. World J Gastrointest Oncol. 2010;2:229-234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 3] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 8. | Park SY, Kim BC, Shin SJ, Lee SK, Kim TI, Kim WH. Proximal shift in the distribution of adenomatous polyps in Korea over the past ten years. Hepatogastroenterology. 2009;56:677-681. [PubMed] |
| 9. | Valls Bautista C, Piñol Felis C, Reñé Espinet JM, Buenestado García J, Viñas Salas J. Telomerase activity and telomere length in the colorectal polyp-carcinoma sequence. Rev Esp Enferm Dig. 2009;101:179-186. [PubMed] |
| 10. | Jass JR, Baker K, Zlobec I, Higuchi T, Barker M, Buchanan D, Young J. Advanced colorectal polyps with the molecular and morphological features of serrated polyps and adenomas: concept of a 'fusion' pathway to colorectal cancer. Histopathology. 2006;49:121-131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 187] [Cited by in RCA: 187] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 11. | Sohns C, Heuser M, Sossalla S, Wolff H, Obenauer S. Current role and future potential of computed tomographic colonography for colorectal polyp detection and colon cancer screening-incidental findings. Clin Imaging. 2008;32:280-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 12. | Zheng J. [Colorectal polyps and colorectal cancer]. Zhonghua Bing Li Xue Za Zhi. 2005;34:4-5. [PubMed] |
| 13. | Bliek J, Verde G, Callaway J, Maas SM, De Crescenzo A, Sparago A, Cerrato F, Russo S, Ferraiuolo S, Rinaldi MM. Hypomethylation at multiple maternally methylated imprinted regions including PLAGL1 and GNAS loci in Beckwith-Wiedemann syndrome. Eur J Hum Genet. 2009;17:611-619. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 173] [Cited by in RCA: 156] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 14. | Braggio E, Braggio Dde A, Small IA, Lopes LF, Valadão M, Gouveia ME, Moreira Ados S, Linhares E, Romano S, Bacchi CE. Prognostic relevance of KIT and PDGFRA mutations in gastrointestinal stromal tumors. Anticancer Res. 2010;30:2407-2414. [PubMed] |
| 15. | Schneider A, Tögel S, Barmada MM, Whitcomb DC. Genetic analysis of the glutathione s-transferase genes MGST1, GSTM3, GSTT1, and GSTM1 in patients with hereditary pancreatitis. J Gastroenterol. 2004;39:783-787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 16. | Johansson K, Ahlen K, Rinaldi R, Sahlander K, Siritantikorn A, Morgenstern R. Microsomal glutathione transferase 1 in anticancer drug resistance. Carcinogenesis. 2007;28:465-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 42] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 17. | Zhang H, Liao LH, Liu SM, Lau KW, Lai AK, Zhang JH, Wang Q, Chen XQ, Wei W, Liu H. Microsomal glutathione S-transferase gene polymorphisms and colorectal cancer risk in a Han Chinese population. Int J Colorectal Dis. 2007;22:1185-1194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 18. | Du CY, Shi YQ, Zhou Y, Fu H, Zhao GF. [Status and clinical analysis of c-kit and PDGFRA mutations in the gastrointestinal stromal tumors]. Zhonghua Wei Chang Wai Ke Za Zhi. 2008;11:371-375. [PubMed] |
| 19. | Ozawa T, Brennan CW, Wang L, Squatrito M, Sasayama T, Nakada M, Huse JT, Pedraza A, Utsuki S, Yasui Y. PDGFRA gene rearrangements are frequent genetic events in PDGFRA-amplified glioblastomas. Genes Dev. 2010;24:2205-2218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 170] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 20. | Barreca A, Fornari A, Bonello L, Tondat F, Chiusa L, Lista P, Pich A. KIT and PDGFRA mutations and PDGFRA immunostaining in gastrointestinal stromal tumors. Mol Med Report. 2011;4:3-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 21. | Goumidi L, Spengler D, Cottel D, Wagner A, Ducimetière P, Ruidavets JB, Legry V, Arveiler D, Bingham A, Ferrières J. Study of the genetic variability of ZAC1 (PLAGL1) in French population-based samples. J Hypertens. 2009;27:314-321. [PubMed] |
| 22. | Valleley EM, Cordery SF, Bonthron DT. Tissue-specific imprinting of the ZAC/PLAGL1 tumour suppressor gene results from variable utilization of monoallelic and biallelic promoters. Hum Mol Genet. 2007;16:972-981. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 47] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 23. | Abdollahi A. LOT1 (ZAC1/PLAGL1) and its family members: mechanisms and functions. J Cell Physiol. 2007;210:16-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 71] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 24. | Liu X, Lazenby AJ, Siegal GP. Signal transduction cross-talk during colorectal tumorigenesis. Adv Anat Pathol. 2006;13:270-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 346] [Cited by in RCA: 255] [Article Influence: 12.8] [Reference Citation Analysis (0)] |