Published online Jan 21, 2012. doi: 10.3748/wjg.v18.i3.285
Revised: June 13, 2011
Accepted: June 20, 2011
Published online: January 21, 2012
AIM: To study the metabolic profiling of serum samples from compensated and decompensated cirrhosis patients.
METHODS: A pilot metabolic profiling study was conducted using three groups: compensated cirrhosis patients (n = 30), decompensated cirrhosis patients (n = 30) and healthy controls (n = 30). A 1H nuclear magnetic resonance (NMR)-based metabonomics approach was used to obtain the serum metabolic profiles of the samples. The acquired data were processed by multivariate principal component analysis and orthogonal partial least-squares discriminant analysis (OPLS-DA).
RESULTS: The OPLS-DA model was capable of distinguishing between decompensated and compensated cirrhosis patients, with an R2Y of 0.784 and a Q2Y of 0.598. Twelve metabolites, such as pyruvate, phenylalanine and succinate, were identified as the most influential factors for the difference between the two groups. The validation of the diagnosis prediction showed that the accuracy of the OPLS-DA model was 85% (17/20).
CONCLUSION: 1H NMR spectra combined with pattern recognition analysis techniques offer a new way to diagnose compensated and decompensated cirrhosis in the future.
- Citation: Qi SW, Tu ZG, Peng WJ, Wang LX, Ou-Yang X, Cai AJ, Dai Y. 1H NMR-based serum metabolic profiling in compensated and decompensated cirrhosis. World J Gastroenterol 2012; 18(3): 285-290
- URL: https://www.wjgnet.com/1007-9327/full/v18/i3/285.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i3.285
Liver cirrhosis (LC) and its associated complications are a major cause of morbidity and mortality worldwide[1,2]. During early cirrhosis, the liver is able to compensate the changes resulting from bridging fibrosis, and most patients do not show specific symptoms until they enter the stage of decompensated cirrhosis[3]. Patients often miss the best opportunity for therapy because the hepatic reserve in the decompensated stage is unable to compensate for hepatocyte loss and structural distortions.
The diagnostic confirmation of cirrhosis is based on a histological examination or the combined results of clinical and imaging examinations[1]. However, the proposed methods cannot be satisfactorily applied to a clinical diagnosis. Firstly, histological examination requires the puncturing of liver tissue and causes substantial pain. Secondly, the clinical results derived from a series of laboratory assays are costly and require long periods of time. Finally, imaging studies cannot provide sensitive diagnoses and are vulnerable to the influence of subjective factors. Therefore, establishing a simple and specific strategy to diagnose cirrhosis and distinguish decompensated LC from compensated LC is important for patient care and treatment decisions.
Metabonomics is a comprehensive and fully quantitative analysis of low molecular weight molecules within a particular biological sample. 1H nuclear magnetic resonance (NMR) spectra combined with principal component analysis (PCA) and orthogonal partial least-squares discriminant analysis (OPLS-DA) are frequently conducted in metabolic profiling studies[4-6].
The incidence of acute and chronic viral hepatitis in China is very high and hepatitis B virus (HBV) contributes to 80% of liver cirrhosis[7]. Thus, in the present study, we employed quantitative 1H NMR to analyze the serum metabolic profiling in HBV-infected cirrhosis patients. Our purpose was to establish a diagnostic method for decompensated and compensated cirrhosis and to discover metabolomic biomarkers related to the cirrhosis conditions.
This study enrolled 60 patients (30 compensated and 30 decompensated cirrhosis patients) from the Department of Gastroenterology and Infectious Diseases of Shenzhen People’s Hospital from November 2009 to July 2010. We excluded patients with past or current hepatocellular carcinoma, alcoholic cirrhosis, diabetes, cardiovascular and cerebrovascular disease, kidney disease and any other viral co-infection, including human immunodeficiency virus, hepatitis delta or hepatitis C virus. Thirty healthy volunteers served as controls. The baseline clinical characteristics of the cirrhotic patients and controls are summarized in Table 1. This study was performed according to the guidelines of Chongqing Medical University, which abides by the Declaration of Helsinki on ethical principles for medical research involving human subjects.
| Liver cirrhosis | |||
| Parameter | Control (n = 30) | Compensated LC (n = 30) | Decompensated LC (n = 30) |
| Age (yr) | 48.8 ± 10.5 | 56.3 ± 12.9 | 58.7 ± 14.5 |
| Gender (M:F) | 12:18 | 14:16 | 15:15 |
| ALT (U/L) | 20.4 ± 10.2 | 110.4 ± 20.2a | 85.4 ± 32.6a |
| AST (U/L) | 15.3 ± 11.6 | 187.5 ± 100.5a | 142.4 ± 52.9a |
| TP (g/L) | 65.4 ± 5.6 | 61.5 ± 10.2 | 60.4 ± 5.7a |
| ALB (g/L) | 50.4 ± 10.2 | 48.6 ± 6.9 | 35.8 ± 11.2ab |
| T-BIL (μmol/L) | 11.4 ± 3.6 | 35.8 ± 30.9a | 45.6 ± 20.2a |
| D-BIL (μmol/L) | 5.4 ± 1.2 | 15.4 ± 8.3a | 23.4 ± 10.2ab |
| GLU (mmol/L) | 5.31 ± 1.6 | 5.48 ± 1.23 | 5.47 ± 0.98 |
| CRE (μmol/L) | 113.6 ± 56.3 | 140.3 ± 50.8 | 152.6 ± 34.2a |
| INR | 1.0 ± 0.05 | 1.1 ± 0.06 | 1.2 ± 0.12 |
| MELD score | - | 12.7 ± 3.5 | 14.2 ± 6.3 |
All patients were positive for hepatitis B surface antigen for at least 1 year before screening. Cirrhosis patients were diagnosed according to the results of histological examination or the combined results of clinical and imaging examinations. Decompensated cirrhosis was defined as the presence of at least two of the following five criteria: ascites, hyperbilirubinemia, peripheral edema of noncardiac or renal origin, hypoalbuminemia and an INR (clotting times, as reflected by the international normalized ratio) > 1.3[8]. The severity of liver disease was calculated according to the model for end-stage liver disease[9].
Approximately 5 mL of peripheral venous blood from fasted healthy volunteers and LC patients was collected. The blood was allowed to clot for 30 min at room temperature before being centrifuged at 2000 g for 10 min at 25 °C; the serum was separated and stored at -80 °C. Prior to NMR analysis, serum samples were thawed and 400 μL aliquots were mixed with 150 μL of deuterium oxide. The serum samples were centrifuged at 12 000 g for 10 min at 4 °C, and 500 μL aliquots of the resulting supernatants were placed into 5 mm NMR tubes. All NMR spectra were recorded at 25 °C on a Varian Unity INOVA 600 NMR spectrometer. One-dimensional spectra were recorded using the Carr-Purcell-Meiboom-Gill sequence[10,11] with a spin-spin relaxation delay of 120 ms and a spectral width of 8000 Hz. All spectra were carefully phase- and baseline-corrected and referenced to the internal lactic acid CH3 resonance at 1.33 ppm. Spectra were segmented into 0.005-ppm chemical shift “bins” between 0.5 and 9.0 ppm, and the spectral area within each bin was integrated. Bins between 4.7 and 5.2 ppm containing residual water were removed. The free induction decay was zero-filled to 64 K and multiplied by an exponential line-broadening function of 0.3 Hz prior to Fourier transformation.
Multivariate statistics, including unsupervised PCA and supervised OPLS-DA, were performed using SIMCA-P 11.0 software (Umetrics, Umea, Sweden). Analysis of the metabolite signals in the 1H NMR serum profiles was first performed using unsupervised PCA, which displays the internal structure of datasets in an unbiased way and decreases the dimensionality of data[12-14]. After an initial overview of the PCA analysis, we obtained a more sophisticated OPLS-DA model with the specific discriminant information between the different groups[15,16]. The differences in the metabolites between groups were shown as coefficient of variation plots. Using a significance level of 0.05, we employed a correlation coefficient of ± 0.355 as the threshold to choose the variables that best correlated with the OPLS-DA discriminative scores. To further test whether the metabolic profiling can effectively distinguish decompensated cirrhosis patients from compensated cirrhosis ones, we randomly selected 20 cirrhotic patients (10 compensated and 10 decompensated cirrhosis patients) to validate the discriminatory power of the OPLS-DA model.
Statistical analyses were performed using SPSS 11.5 software (SPSS Company, Chicago, IL, United States). The threshold P value was set at 0.05 throughout the study.
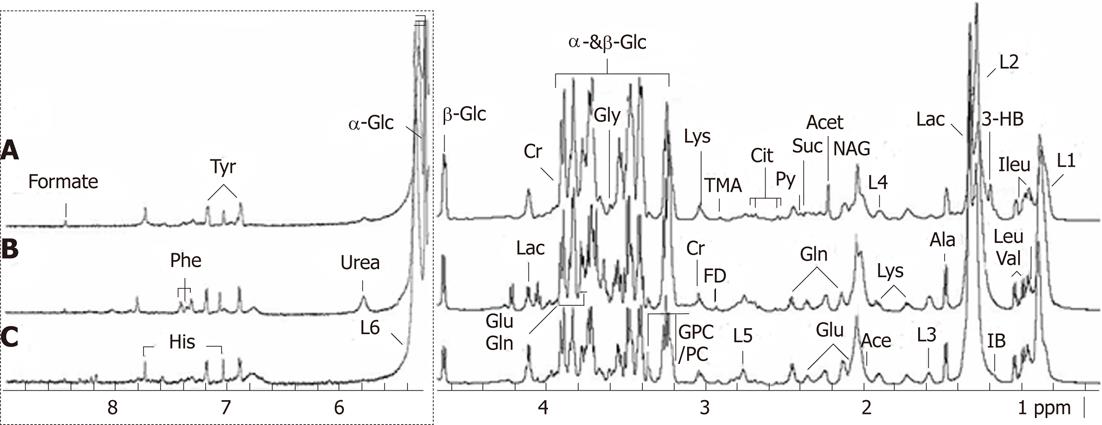
The 1H-NMR signals of all common metabolites, such as amino-acids, organic acids and carbohydrates, were assigned according to previous publications[10]. Examples of typical serum spectra from control, compensated and decompensated cirrhosis groups are shown in Figure 1. The 600 MHz 1H NMR spectra demonstrated resonances arising from glucose, glutamine, acetate, leucine, glycerophosphocholine, histidine, isoleucine, citrate, etc. The useful extracted information was subsequently analyzed using multivariate statistics including PCA and OPLS-DA.
As a proof of principle, we first evaluated whether a metabonomics approach would be capable of distinguishing cirrhotic patients (included compensated and decompensated cirrhosis) from healthy controls.
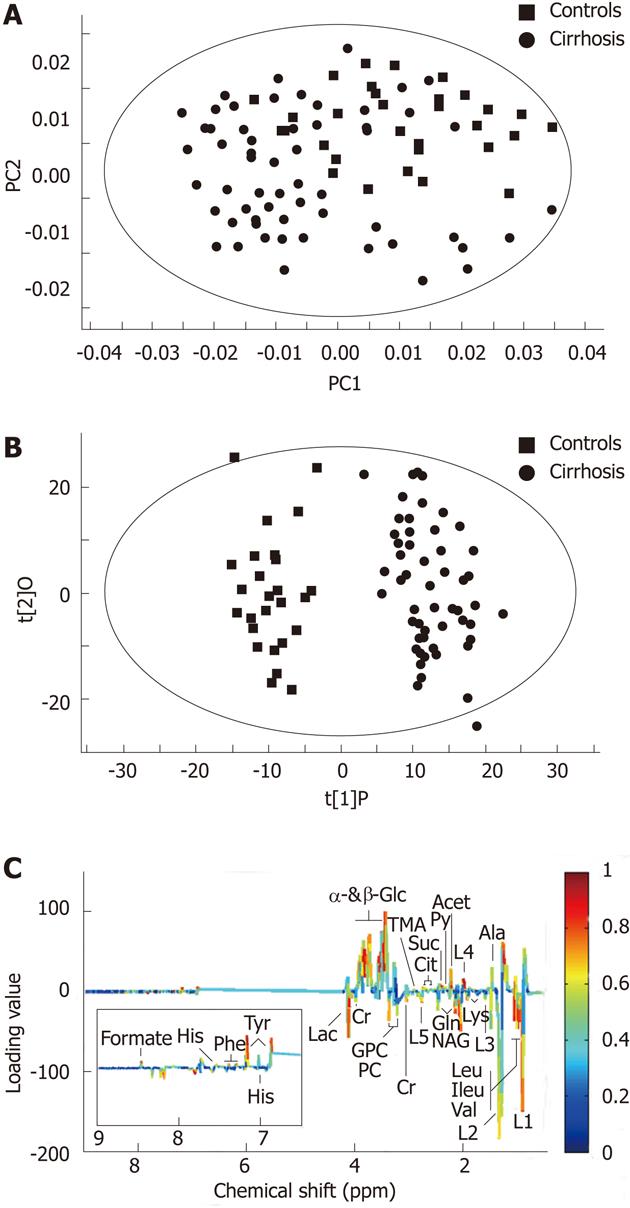
At first, we analyzed the serum metabolic profiles using unsupervised PCA. Figure 2A displays the PCA score plots of cirrhotic patients and healthy controls, with an R2X of 0.812 and a Q2Y of 786. The PCA was followed by OPLS-DA, which is more focused on discriminatory variations. Excellent separation with negligible overlapping was observed in OPLS-DA score plots between controls and cirrhosis patients. This model showed very good fit and predictability values with an R2Y of 0.941 and a Q2Y of 0.836 (Figure 2B). The loading plots (Figure 2C) revealed that the responsible variables were those corresponding to β-glucose, α-glucose, low-density lipoproteins (LDL), very low-density lipoproteins (VLDL), valine, tyrosine, succinate, lipid, isobutyrate, glutamine, glutamate, etc.
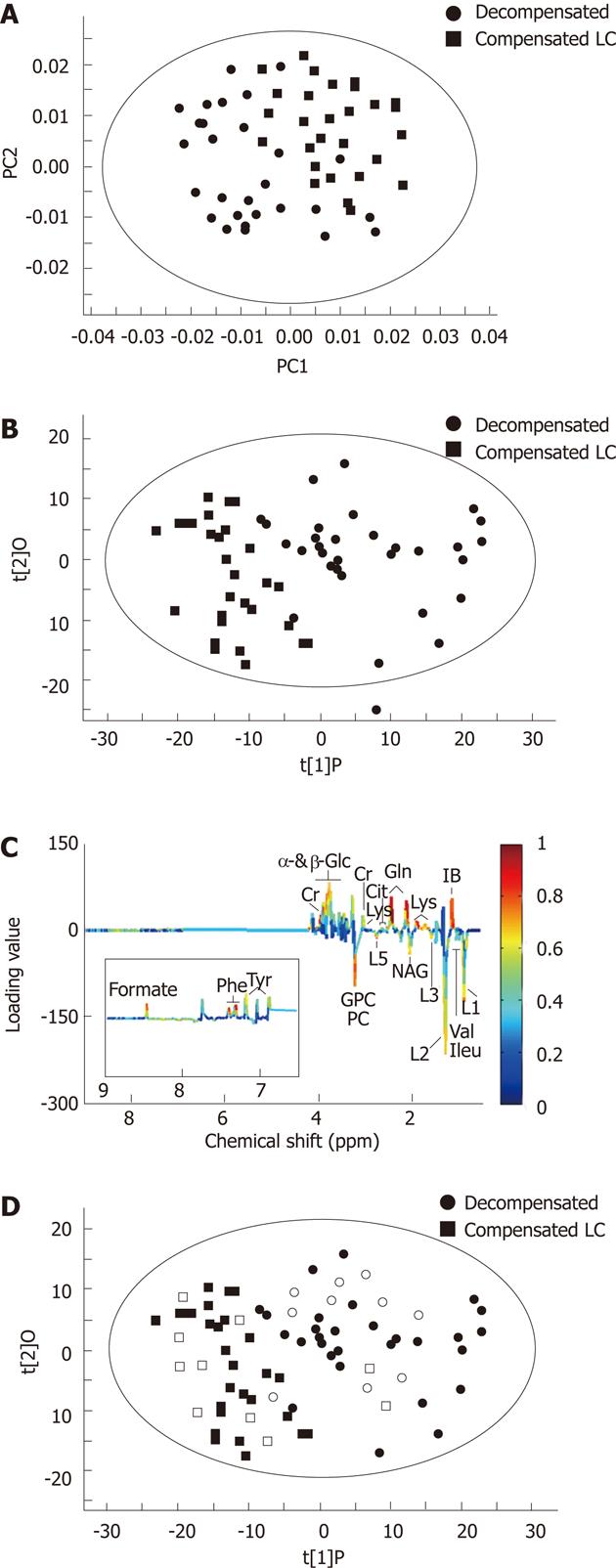
A similar procedure was performed to assess the utility of metabonomics to distinguish decompensated LC from compensated LC patients. A PCA score plot (Figure 3A) could not reveal an obvious separation between compensated and decompensated LC, with an R2X of 0.795 and a Q2Y of 0.736. The overlap in PCA score plots suggested that there was some metabolic variation between the two classes not related to the disease state. OPLS-DA revealed a clear separation between decompensated and compensated LC patients (Figure 3B), with an R2Y of 0.784 and a Q2Y of 0.598. The majority of the disease-related variance could be explained by the first component of the model. The OPLS-DA loading plot (Figure 3C) revealed that the significant variables were those corresponding to pyruvate, phenylalanine, succinate, lysine, etc. The variables meeting the cutoff value (i.e., |correlation coefficient| > 0.355) are summarized in Table 2.
| Metabolites | δ1H | Correlation coefficients | |
| Compensated LC | Decompensated LC | ||
| β-Glucose | 3.24, 4.65 | 0.654 | 0.722 |
| α-Glucose | 3.425.24 | 0.602 | 0.806 |
| CH3-(CH2)n-(LDL&VLDL) | 1.28 | 0.748 | -0.693 |
| Valine | 0.99, 1.04 | -0.903 | -0.910 |
| Tyrosine | 6.89, 7.19 | 0.675 | 0.805 |
| Succinate | 2.40 | - | 0.603 |
| Pyruvate | 2.37 | - | 0.585 |
| Phenylalanine | 7.32, 7.41 | - | 0.749 |
| NAG:N-acetyl glycoprotein | 2.04 | -0.583 | -0.774 |
| Lysine | 1.72, 3.01, 3.78 | -0.602 | - |
| Lipid, H3-(CH2)n-(LDL&VLDL) | 0.86 | 0.805 | -0.793 |
| Leucine | 0.93 | -0.876 | -0.727 |
| Lactate | 1.33, 4.12 | -0.671 | -0.769 |
| Isoleucine | 0.96, 1.01 | -0.916 | -0.859 |
| Histidine | 7.06, 7.75 | - | 0.616 |
| GPC/PC | 3.21, 3.35 | -0.732 | -0.712 |
| Glutamine | 2.14, 2.45, 3.78 | -0.659 | 0.706 |
| Glutamate | 2.03, 2.35, 3.78 | - | 0.554 |
| Creatine | 3.04, 3.93 | -0.709 | 0.555 |
| Citrate | 2.53, 2.67 | 0.657 | 0.604 |
| Alanine | 1.48 | - | 0.571 |
| Acetone | 2.23 | 0.729 | - |
The serum metabolic profiles of 20 cirrhotic patients were overlaid in the OPLS-DA model to evaluate the diagnostic performance (Figure 3D). Of these, eight compensated cirrhosis patients were clustered together in the left section of the OPLS-DA model (compensated group), nine decompensated cirrhosis patients were clustered together in the right section of the OPLS-DA model (decompensated group), and the three remaining cirrhosis patients were situated in an intermediate area between the decompensated and compensated cirrhosis patients.
Previous studies have shown that the transition from compensated to decompensated cirrhosis is so insidious that many patients miss the best opportunity for treatment[8,17]. In the present study, we applied NMR-based metabolic profiling to examine the serum from decompensated and compensated LC patients and identified low molecular weight biomarkers for the diseases.
The findings of this pilot study showed that the 1H NMR serum metabolic profiling of cirrhotic patients and control subjects was clearly separated. Compared with healthy controls, the serum metabolic profiling of cirrhotic patients showed increased levels of glucose and lactate and decreased levels of lipids and choline. In this context, our results are consistent with the studies of Gao et al[18], which showed that 1H NMR metabonomics analysis of serum is helpful for differentiating cirrhotic patients from healthy subjects.
However, little work has been done to discriminate patients with compensated and compensated cirrhosis through metabonomics. Corbin et al[8] used phosphorus-31 magnetic resonance spectroscopy to document the differences in the hepatic metabolite concentrations among patients with compensated and decompensated cirrhosis. Although this study provided a useful and noninvasive diagnostic tool, the researchers only studied five metabolites and did not investigate all metabolites related to cirrhosis.
In contrast, NMR has the advantage of full quantitative analysis and minimal requirements for sample preparation[5]. In the present study, we obtained a ‘metabolic fingerprint’ from the pathological process of cirrhosis. Compared with compensated LC patients, the decompensated LC patients displayed higher levels of pyruvate, phenylalanine, succinate, lysine, histidine, alanine, glutamate, glutamine, creatine and lower levels of LDL, VLDL and acetone.
The increased concentration of pyruvate and succinate in decompensated LC serum is possibly due to the reduced utilization of pyruvate and succinate into the tricarboxylic acid cycle[19]. Phenylalanine belongs to the aromatic group of amino acids, and it is converted to tyrosine by the catalysis of phenylalanine hydroxylase, which is a liver-specific enzyme[20]. The increase in phenylalanine suggests that the BCAA/AAA ratio has changed in patients with decompensated LC. Glutamate is at the very center of hepatic amino acid metabolism. Glutamine, histidine, arginine, ornithine, proline, and glutamate comprise the ‘glutamate family’ of amino acids[21]. The elevated levels of glutamate, glutamine and histidine reflect the abnormal metabolism of amino acids with cirrhosis progression.
Our study showed that the levels of VLDL and LDL were reduced in the decompensated LC group compared to the compensated LC group. Because liver tissue loses some degree of its lipid synthesizing ability in the later stage of cirrhosis, the blood lipid level decreases in the serum of decompensated LC patients[22].
The remaining metabolite changes include increased lysine and alanine and decreased acetone; altered levels of these metabolites in decompensated LC patients may be the result of the impairment of hepatocytes, but may also be the result of liver perfusion.
Taken together, these results imply that hypermetabolism of the liver was prevalent when compensated cirrhosis developed into the decompensated stage. Hence, these metabolites may serve as biomarkers that can be used to monitor the changes in LC patients.
Regarding predictive tests of the discriminatory power, the results showed that the accuracy of the OPLS-DA was 85% (17/20). Three patients located in the middle area of the OPLS-DA model were observed carefully in the following two months. One compensated LC patient developed decompensated cirrhosis, and the remaining two LC patients did not exhibit distinct changes. It is conceivable that the metabolic changes revealed by the NMR spectra could be more sensitive than other clinical symptom and lab tests.
Serum metabolic analysis bears the potential to be a useful and convenient method to diagnose LC patients. However, there are some potential limitations of the present study that require consideration. Firstly, a limited number of samples prevented us from drawing a more reliable conclusion about the predictive power of this model. Larger numbers of patients and controls will be crucial to validate this model. Secondly, the specificity of the biomarkers indentified by OPLS-DA requires a longitudinal study to determine their validity.
In summary, the metabolic profiling obtained from 1H NMR-based metabonomics analysis of serum may be a simple and reliable way to diagnose compensated and decompensated LC. The metabolic differences between the two groups also facilitate a better understanding of the metabolic changes associated with cirrhosis progression.
The authors would like to thank the patients and healthy volunteers who participated in this study.
The diagnostic confirmation of cirrhosis is based on histological examination or the combined results of clinical and imaging examinations. However, the proposed methods cannot be satisfactorily applied in clinical diagnosis.
1H nuclear magnetic resonance (NMR) is an ideal instrumental platform for metabolic analysis of biofluids, and NMR analysis has been reported as successfully applied in the diagnosis and prognosis of some human diseases.
The authors compared the serum metabolic profiling of compensated and decompensated cirrhosis patients. The orthogonal partial least-squares discriminant analysis (OPLS-DA) model was successful in distinguishing decompensated cirrhosis from compensated cirrhosis. Twelve metabolites were identified as the most influential biomarkers for the difference between the two groups.
Serum metabolic analysis bears the potential to be a useful and convenient method to diagnose compensated and decompensated cirrhosis.
Principal component analysis (PCA): PCA is a mathematical procedure that uses an orthogonal transformation to convert a set of observations of possibly correlated variables into a set of values of uncorrelated variables called principal components; orthogonal partial least-squares discriminant analysis (OPLS-DA): OPLS-DA is a method of discriminating between two or more groups. The variables responsible for the differences may be identified.
In this manuscript, the authors employed metabolomics to compare serum metabolic profiles among control, compensated and decompensated cirrhosis patients. Rationale and results are sound.
Peer reviewer: Dr. Hui-kang Liu, PhD, Assistant Research Fellow, National Research Institute of Chinese Medicine, 155-1, Li-nung street section 2, Taipei 112, Taiwan, China
S- Editor Tian L L- Editor Logan S E- Editor Xiong L
| 1. | Schuppan D, Afdhal NH. Liver cirrhosis. Lancet. 2008;371:838-851. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1686] [Cited by in RCA: 1565] [Article Influence: 92.1] [Reference Citation Analysis (0)] |
| 2. | Lu Y, Liu J, Lin C, Wang H, Jiang Y, Wang J, Yang P, He F. Peroxiredoxin 2: a potential biomarker for early diagnosis of hepatitis B virus related liver fibrosis identified by proteomic analysis of the plasma. BMC Gastroenterol. 2010;10:115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 37] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 3. | Kim MY, Baik SK. [Hyperdynamic circulation in patients with liver cirrhosis and portal hypertension]. Korean J Gastroenterol. 2009;54:143-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 4. | Jiménez B, Montoliu C, MacIntyre DA, Serra MA, Wassel A, Jover M, Romero-Gomez M, Rodrigo JM, Pineda-Lucena A, Felipo V. Serum metabolic signature of minimal hepatic encephalopathy by (1)H-nuclear magnetic resonance. J Proteome Res. 2010;9:5180-5187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 49] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 5. | Wang T, Shao K, Chu Q, Ren Y, Mu Y, Qu L, He J, Jin C, Xia B. Automics: an integrated platform for NMR-based metabonomics spectral processing and data analysis. BMC Bioinformatics. 2009;10:83. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 53] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 6. | Schicho R, Nazyrova A, Shaykhutdinov R, Duggan G, Vogel HJ, Storr M. Quantitative metabolomic profiling of serum and urine in DSS-induced ulcerative colitis of mice by (1)H NMR spectroscopy. J Proteome Res. 2010;9:6265-6273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 81] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 7. | Meng QH, Wang JH, Yu HW, Li J, Feng YM, Hou W, Zhang J, Zhang Q, Wang X, Wang X. Resting energy expenditure and substrate metabolism in Chinese patients with acute or chronic hepatitis B or liver cirrhosis. Intern Med. 2010;49:2085-2091. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 8. | Corbin IR, Ryner LN, Singh H, Minuk GY. Quantitative hepatic phosphorus-31 magnetic resonance spectroscopy in compensated and decompensated cirrhosis. Am J Physiol Gastrointest Liver Physiol. 2004;287:G379-G384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 9. | Ahmad J, Downey KK, Akoad M, Cacciarelli TV. Impact of the MELD score on waiting time and disease severity in liver transplantation in United States veterans. Liver Transpl. 2007;13:1564-1569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 10. | Nicholson JK, Foxall PJ, Spraul M, Farrant RD, Lindon JC. 750 MHz 1H and 1H-13C NMR spectroscopy of human blood plasma. Anal Chem. 1995;67:793-811. [PubMed] |
| 11. | Viant MR. Improved methods for the acquisition and interpretation of NMR metabolomic data. Biochem Biophys Res Commun. 2003;310:943-948. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 12. | De Meyer T, Sinnaeve D, Van Gasse B, Rietzschel ER, De Buyzere ML, Langlois MR, Bekaert S, Martins JC, Van Criekinge W. Evaluation of standard and advanced preprocessing methods for the univariate analysis of blood serum 1H-NMR spectra. Anal Bioanal Chem. 2010;398:1781-1790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 31] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 13. | Trygg J, Holmes E, Lundstedt T. Chemometrics in metabonomics. J Proteome Res. 2007;6:469-479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 987] [Cited by in RCA: 898] [Article Influence: 49.9] [Reference Citation Analysis (0)] |
| 14. | Kumar A, Bala L, Kalita J, Misra UK, Singh RL, Khetrapal CL, Babu GN. Metabolomic analysis of serum by (1) H NMR spectroscopy in amyotrophic lateral sclerosis. Clin Chim Acta. 2010;411:563-567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 81] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 15. | Ni Y, Su M, Lin J, Wang X, Qiu Y, Zhao A, Chen T, Jia W. Metabolic profiling reveals disorder of amino acid metabolism in four brain regions from a rat model of chronic unpredictable mild stress. FEBS Lett. 2008;582:2627-2636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 144] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 16. | Ayshamgul H, Batur M, Ilyar S. [1H-MRS metabonomic analysis of plasma samples of esophageal cancer patients based on different pattern recognition]. Zhonghua Zhong Liu Za Zhi. 2010;32:681-684. [PubMed] |
| 17. | Rainey PM. Clinical problem-solving: the landlady confirms the diagnosis. N Engl J Med. 1992;327:895-896. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 318] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 18. | Gao H, Lu Q, Liu X, Cong H, Zhao L, Wang H, Lin D. Application of 1H NMR-based metabonomics in the study of metabolic profiling of human hepatocellular carcinoma and liver cirrhosis. Cancer Sci. 2009;100:782-785. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 152] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 19. | Bubber P, Hartounian V, Gibson GE, Blass JP. Abnormalities in the tricarboxylic acid (TCA) cycle in the brains of schizophrenia patients. Eur Neuropsychopharmacol. 2011;21:254-260. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 73] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 20. | Dejong CH, van de Poll MC, Soeters PB, Jalan R, Olde Damink SW. Aromatic amino acid metabolism during liver failure. J Nutr. 2007;137:1579S-1585S; discussion 1597S-1598S. [PubMed] |
| 21. | Brosnan ME, Brosnan JT. Hepatic glutamate metabolism: a tale of 2 hepatocytes. Am J Clin Nutr. 2009;90:857S-861S. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 147] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 22. | Closs SJ. Postoperative pain at night. Nurs Times. 2010;87:40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 26] [Cited by in RCA: 33] [Article Influence: 2.2] [Reference Citation Analysis (1)] |