INTRODUCTION
Hepatitis C virus (HCV) is an enveloped, positive-stranded RNA virus classified in the hepacivirus genus of the Flaviviridae family[1]. HCV infection is a major cause of chronic hepatitis, liver cirrhosis and hepatocellular carcinoma[2]. Interferon (IFN) and the nucleotide analogue ribavirin were used as the basic treatment, and sustained virologic response (SVR) rate was achieved in only 38%-63% of the treated patients although substantial improvements have been made[3]. The response to antiviral therapy depends highly on the HCV genotype. HCV is classified into six major genotypes and 30 subtypes, of which, 1a and 1b are distributed most frequently and studied intensively[4,5]. Patients infected with HCV genotype 1 showed a response rate of 38%-52% to IFN therapy, whereas patients infected with genotypes 2 or 3, achieved a SVR in up to 90% of treated patients[6-8]. The HCV genome is about 9600 nucleotides in length, encoding a single polyprotein that is processed by host and viral proteases into at least 10 distinct structural and nonstructural proteins in the order of C-E1-E2-P7-NS2-NS3-NS4A-NS4B-NS5A-NS5B[9]. The major structural proteins (Core, E1 and E2), together with a host derived lipid bilayer and the viral RNA, comprise the viron. p7 and NS2 are located within the polyprotein between these structural and nonstructural proteins. p7 is a small membrane protein with ion channel activity, NS2 and NS3 contain proteases that cleave the junction between NS2 and NS3. The nonstructural proteins NS3-NS5B are considered to assemble into membrane-associated HCV replication complex.
In the last decade, studies of HCV replication and translation have been conducted using subgenomic or full-length genomic replicon culture system[10-12]. Until recently, studies about the entire life cycle of HCV, especially the processes involved in virus entry, assembly and release, have been advanced with the development of genotype 2a JFH1 based cell culture systems, which could produce infectious HCV particles[13-15]. However, robust cell culture systems for HCV genotypes 1a and 1b, the most prevalent genotypes in the world, have not yet been developed successfully, with the exception for strains H77 and H77-S of genotype 1a[16,17], although several groups have constructed intragenotypic and intergenotypic chimeras[18-21]. What renders the JFH1 genome so special, and which element affects viral particle production and release? Wakita’s work suggested that NS5B, 3’UTR and NS3 helicase activity of JFH-1 contribute to the efficient replication of JFH-1[13]. Since the NS5A protein has been the subject of intensive research over the last decade, in this paper, we focused on the NS5A protein and tried to elucidate its functions in the production of infectious virus[22].
NS5A is part of a multi-protein, membrane-bound replication complex, and plays a key role in both viral RNA replication and modulation of the physiology of the host cell, and in the establishment and maintenance of persistent infection as well[23-25]. NS5A has been shown to interact with a number of proteins which contribute to the replication of HCV, including grb2, PI3 kinase, hVAP, FBL2, and FKBP8[26-29]. NS5A is a phosphoprotein that can be found in basally phosphorylated (56 kDa) and hyperphosphorylated forms (58 kDa)[30,31]. As observed in HCV genotype 1a and 1b, the adaptive mutations of NS5A region related to phosphorylation form could efficiently promote the replication of HCV, whereas, in the JFH1 isolate, the robust replication does not need the adaptive mutation of NS5A. In order to clarify the function of NS5A domain, especially that of JFH1 isolate in the replication and infection of HCV, a chimera containing genotype 1b NS5A domain on the background of FL-J6JFH1 was constructed to study the effects of gene substitution. The results indicate that the intergenotypic chimera FL-J6JFH/J4NS5A did replicate and produce infectious viral particles in vitro, however, with much lower titers than the prototype FL-J6JFH, which suggested that JFH1 NS5A plays an important role in the replication and cell culture-produced HCV (HCVcc) production.
MATERIALS AND METHODS
Construction of chimera
pFL-J6JFH1 and pFL-J6JFH1 (GND) were generous gifts from Rice CM (Rockefeller University, USA). Full-length clone FL-J6JFH/J4NS5A chimera was constructed as follows. First, HCV 1b NS5A gene fragment was amplified from pHCV-J4[32] with the primers FJ45A-JFH (5'-GCCCCATCCCATGCTCCGGCTCGTGGCT-3') and RJ45A-JFH (5'-GAGTATGACATGGAGCAGCAGACGACATC-3'). Then, overlapping polymerase chain reaction (PCR) was carried out to amplify the close sequences in the following steps. The junction of J6JFH1 NS4B with J4-NS5A was amplified with the primers F-BamJFH (5'-CTACTGCCTGGGATCCTGTCTC-3') and RJFH4B (5'-GCATGGGATGGGGCAGTCC-3'). The two PCR products were gel-purified and extended using the primers F-BamJFH and RJFH4B to get J6JFH1 NS4B-J4NS5A. JFHNS5B was amplified with the primers FJFH5B (5'-TCCATGTCATACTCCTGGAC-3') and RJFH-Bsp (5'-GATGTTGTACAGT ACACCTTG-3'). The fragment of JFHNS4B-J4NS5A-JFHNS5B was amplified using JFHNS4B-J4NS5A and JFHNS5B as templates with the primers F-BamJFH and RJFH-Bsp, and the PCR product was cloned into pMD18-T vector (TaKaRa, Dalian, China) and digested with Mfe I + Kpn I. The vector was gel purified and ligated with 4960bp fragment from pFL-J6JFH/ Mfe I + Kpn I to get subclone pT-I. Finally, the full-length pJ6JFH/J4-NS5A was constructed by ligating the 6506bp fragment from pT-I/Kpn I + Bsp1407 I with 5866bp fragment from pFL-J6JFH/Kpn I + Bsp1407 I. All cloned PCR products were verified by DNA sequencing.
Cells
Huh7.5 cells (a generous gift from Rice CM) were cultured in complete Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum, 10 mmol/L Hepes, 100 units/mL penicillin, 100 mg/mL streptomycin, 2 mmol/L L-glutamine (Invitrogen) at 37°C in a 5% CO2 incubator. Cells were split every 3 to 4 d at a ratio of 1:3 to 1:4.
In vitro transcription and transfection
Plasmid DNA was linearized with Xba I (TaKaRa, Dalian, China), and transcribed using the RiboMAXTM Large Scale RNA Production System (Promega, CA, USA) according to the manufacturer’s instructions. The RNA transcripts were treated with RNase-free DNase I (TaKaRa, Dalian, China) and purified, and the RNA concentrations were checked by spectrophotometry. Twenty-four hours prior to transfection, the Huh-7.5 cells were plated at 3 × 105 per well to a 12-well plate with fresh medium without antibiotics. For the transfection, a total of 5 μg RNA transcripts were mixed with Lipofectamine TM 2000 (Invitrogen, CA, USA), and the transfection was carried out according to the manufacturer’s instructions.
Quantitation of HCV RNA
At different time points after transfection, cells were collected and total RNAs were extracted with RNAex reagent (Watson, Shanghai, China) according to the manufacturer’s instructions. The RNAs were reversely transcribed into cDNA with random primer and then quantitated by real-time PCR using SYBR Premix Ex Taq (TaKaRa, Dalian, China). In brief, 1 μg RNA was reversely transcribed in a 25 μL reaction mixture at 37°C for 30 min, followed by inactivation of the reverse transcriptase at 95°C for 15 min. Products were then amplified by PCR for 35 cycles each, at 95°C for 10 s, 60°C for 15 s and 72°C for 5 s. Primer sets targeting the 5’-non coding regions of J6JFH1 included forward primer F-HCVN: 5'-GCGTTAGTATGAGTGTCGTG-3', and reverse primer R-HCVN: 5'-TCGCAAGCACCCTACAG-3'. Reverse transcription PCR (RT-PCR) amplification of cellular glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in the parallel control with the primer sets (forward primer, 5'-TGGGCTACACTGAGCACCAG-3'; reverse primer, 5'-AAGTGGTCGTTGAGGGCAAT-3')[33]. RNA levels of HCV and GAPDH were determined according to a standard curve consisting of serial dilutions of the plasmid containing the full-length HCV cDNA or human GAPDH PCR products.
Immunofluorescence assay for HCV-specific proteins
Cells were fixed for 20 min with methanol (-20°C), and then permeabilized with 0.1% tritonX-100(v/v) for 30 min at room temperature. After washing with PBS, cells were blocked for 1 h with normal sera. HCV-specific proteins were checked by incubation with sera from hepatitis C patients (HCV-positive sera collected from Changhai Hospital, Shanghai, China) as primary antibody followed by incubation with a 1:1000 dilution of FITC-conjugated goat anti-human IgG (Jackson Immunology Institute, USA) for 1 h at room temperature. The percentage of HCV-positive cells was evaluated under microscopy as 0% (no cell infected), 5%, 10%-90%, 95% and 100% (all cells infected).
Titration of HCVcc infection
At indicated time points after transfection, cell culture supernatants were harvested and cleared with low-speed centrifugation and 0.45 μm filter. Cell supernatants were serially diluted at 10 folds with complete DMEM and used to infect 104 naïve Huh-7.5 cells per well in 96-well plates. The inoculums were removed after 1 h incubation at 37°C, and cells were supplemented with fresh complete DMEM. The level of HCV infection was determined 3 d after infection by immunofluorescence assay (IFA) for HCV proteins. The viral titer was calculated at focus forming units per milliliter of supernatant (ffu/mL), determined by the average number of positive foci at the highest dilutions.
Sequence analysis of HCVcc NS5A
HCV RNA was isolated from cell supernatants harvested at day 13, 34 and 59 after transfection using RNAex LS reagent (Watson, Shanghai, China) and then used as template to generate cDNA in a reverse transcription reaction. The primer sets (forward primer: 5′-CGTCGACAAATGTCCGGCTCGTGGCTAAG-3′; reverse primer: 5′-GGTACCGCTCGAGTGCAGCAGACGACATCC-3′) for full-length HC-J4 NS5A gene were used for PCR with PrimeSTARTM HS DNA polymerase (TaKaRa, Dalian, China). Nucleotide sequencing of the products was performed by Invitrogen Biotechnology Corporation (Shanghai, China).
Statistical analysis
All results were expressed as the mean ± SD. Statistical comparison was made using Student’s t test after analysis of variance. Differences were considered statistically significant at P < 0.05.
RESULTS
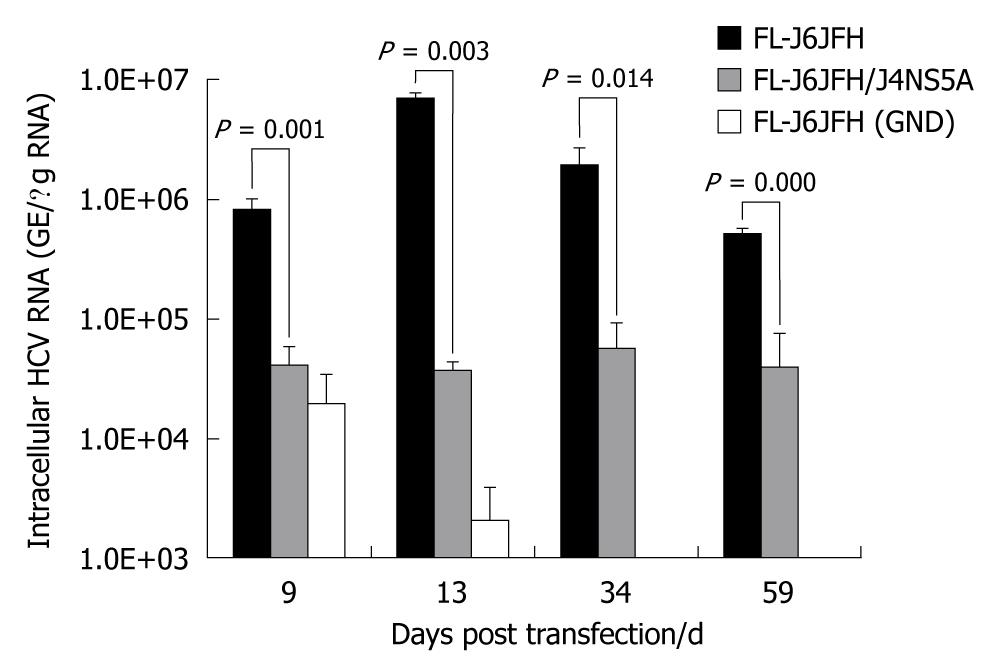
The intergenotypic chimera FL-J6JFH/J4NS5A was generated and confirmed by DNA sequencing and restriction enzyme reaction (Figure 1). Cells transfected with genomic RNA transcripts of FL-J6JFH1, FL-J6JFH/J4NS5A and FL-J6JFH1 (GND) were cultured and passaged for more than 59 d. At the indicated time points, total RNA was isolated from the transfected Huh-7.5 cells, and the levels of HCV RNA were determined by HCV-specific fluorescent quantitive RT-PCR. In the prototype FL-J6JFH1 transfected cells, the minimum level of cellular RNA, 8.2 ± 1.5 × 105 copies/μg HCV RNA, was detected at day 9 after transfection (Figure 2). Thereafter, the intracellular HCV RNA levels began to increase, reaching a maximal level of 7.1 × 106± 7.2 × 105 GE/μg RNA by day 13 after transfection, and similar levels were maintained until the experiment was terminated on day 59. While in the chimera FL-J6JFH/J4NS5A RNA transfected cells, the RNA level was significantly lower than that of prototype at any indicated time point (2.58 ± 5.97 × 106vs 4.27 ± 1.72 × 104, P = 0.032). The maximal level of HCV RNA in chimera was 5.6 ± 1.8 × 104 GE/μg RNA at day 34 after transfection, while the prototype reached a peak by 126 folds at day 13 (70.65 ± 14.11 × 105vs 0.56 ± 0.90 × 105, P = 0.028). As a negative control, FL-J6JFH1 (GND) showed a rapid disappearance of HCV intracellular RNA after transfection (Figure 2).
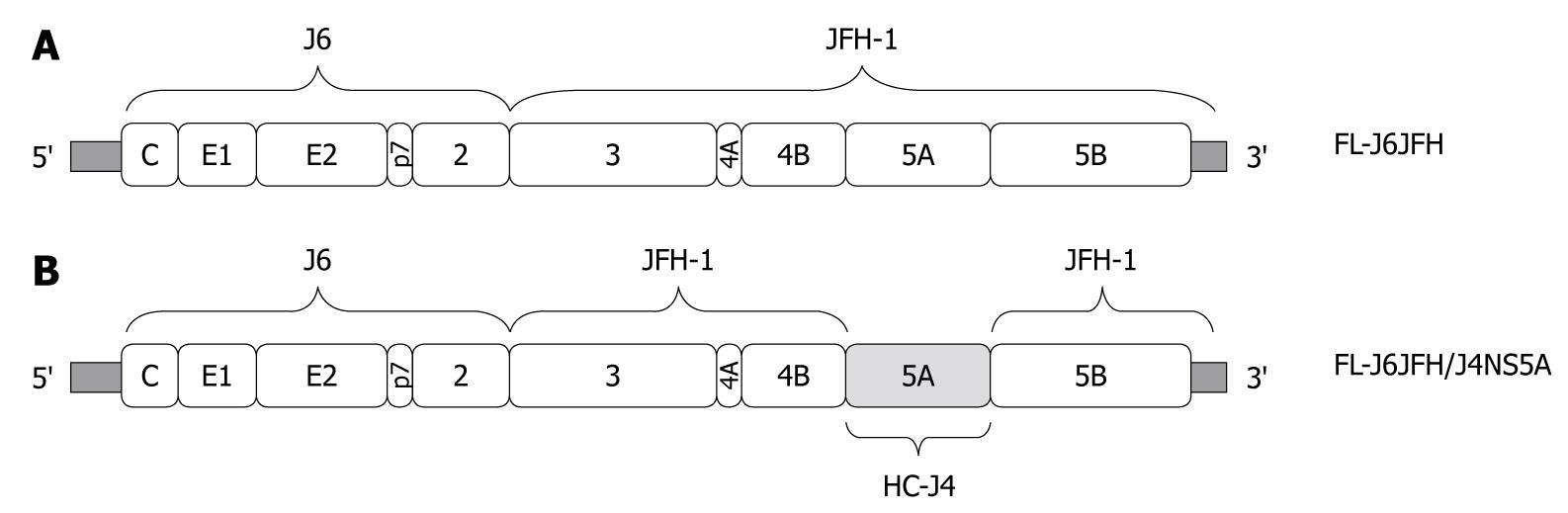
Figure 1 Construction of chimeric FL-J6JFH/J4NS5A genome containing genotype 1b (HC-J4) NS5A domain.
FL-J6JFH/J4NS5A was constructed by inserting full length gene of HC-J4 NS5A into FL-J6JFH1 using overlapping polymerase chain reaction combined with enzyme restriction reaction. A: Prototype FL-J6JFH1; B: Chimera FL-J6JFH/J4NS5A. Vertical bars: HC-J4 NS5A.
Figure 2 Fluorescence quantitative reverse transcription polymerase chain reaction assay for hepatitis C virus RNAs in cell lysis of RNA transcripts transfected Huh-7.
5 cells. Five micrograms in vitro transcribed RNAs were electroporated into 3 × 105 Huh-7.5 cells. Transfected cells were harvested at the indicated time points after transfection. Intracellular hepatitis C virus (HCV) RNA was analyzed by fluorescence quantitative reverse transcription polymerase chain reaction and displayed as genome equivalents per microgram total RNA (GE/μg RNA).
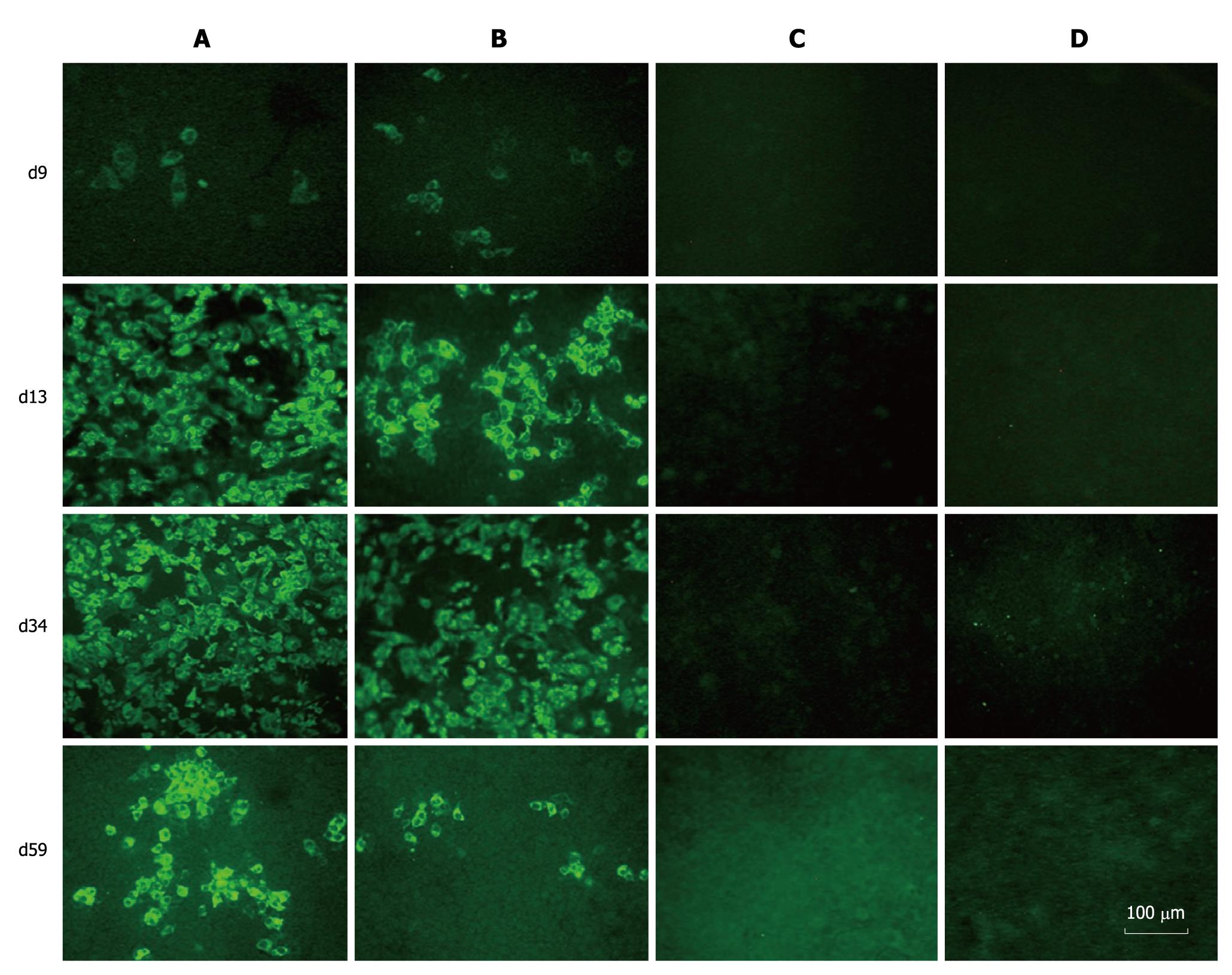
When the transfected cells were passaged, parts of them were collected, and the expression of HCV proteins was examined by IFA with sera from hepatitis C patients. HCV-specific proteins were detectable in both prototype and chimera RNA transfected cells. The Huh7.5 cells transfected with FL-J6JFH1 (GND) and FL-J6JFH1 incubated with normal sera were negative for HCV proteins (Figure 3). The IFA results indicated that the percentage of HCV protein-positive cells in the prototype-transfected cell cultures increased from 5% on day 9 to almost 100% on days 13 and 34. On day 59, it declined to about 50% (Figure 3). On the other hand, the proportion of positive cells in the chimera-transfected cells increased from 5% on day 9 to 50% on day 13, and then to nearly 70% on day 34. About 10% of the cells were still positive for HCV protein staining on day 59 when the experiment was terminated (Figure 3).
Figure 3 Immunofluorescence assay of hepatitis C virus proteins in RNAs transfected Huh-7.
5 cells. Huh-7.5 cells were passaged at day 9, 13, 34 and 59 after transfection, and cultured in 96-well plates for the detection of hepatitis C virus (HCV) proteins using sera from HCV patients as primary antibody. A: FL-J6JFH1-transfected cells; B: FL-J6JFH/J4NS5A-transfected cells; C: FL-J6JFH1 (GND)-transfected cells; D: FL-J6JFH1-transfected cells incubated with normal sera as primary antibody.
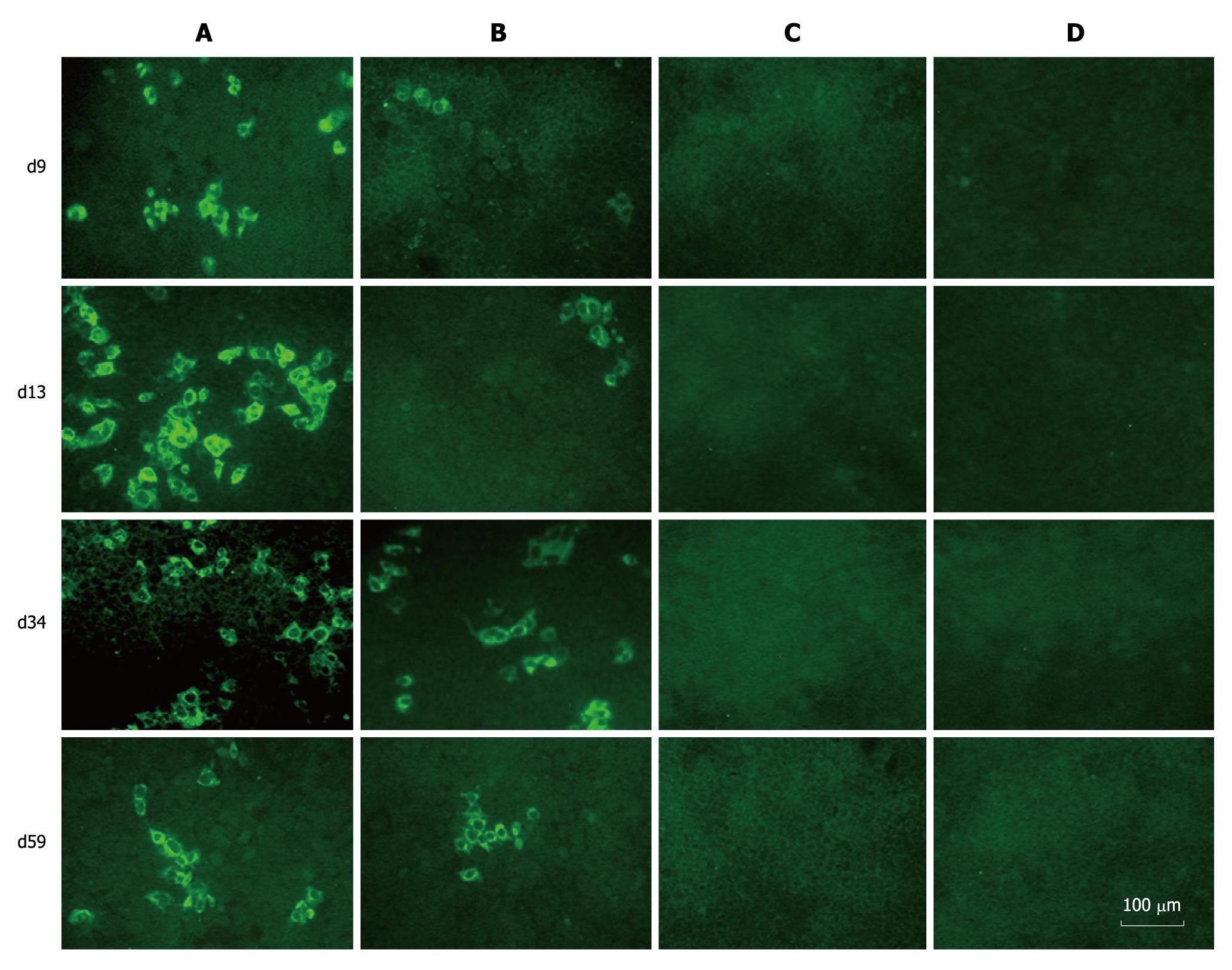
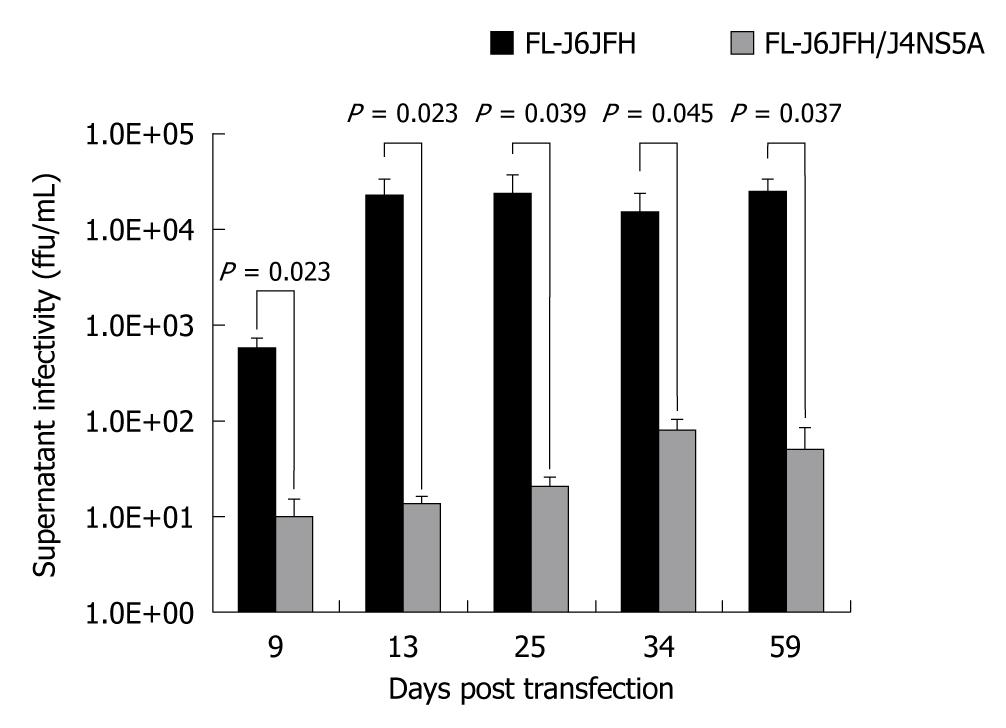
To determine whether FL-J6JFH/J4NS5A produced HCVcc in the transfected Huh-7.5 cells, naïve Huh-7.5 cells were inoculated with cell supernatants harvested from cultured Huh-7.5 cells at days 9, 13, 34 and 59 after transfection. The IFA results revealed that HCV protein-positive cells were detectable at day 3 after inoculation (Figure 4). To further assess the infectivity titers of HCVcc collected at different time points, the supernatants were serially diluted, and the discrete foci of HCV protein-positive cells were counted (Figure 5). HCVcc of FL-J6JFH transfected cells was first detectable with 5.8 ± 1.5 × 102 ffu/mL at day 9 after transfection, and then reached a maximum of 2.4 ± 1.3 × 104 ffu/mL by day 25 after transfection, which is consistent with the levels of intracellular J6JFH1 RNA (Figure 2). The chimera FL-J6JFH/J4NS5A was replicated with slower kinetics than the prototype (Figure 2 and 5). The HCVcc produced by chimera was lower than that of the prototype at any time point (1.96 ± 1.52 × 106vs 5.56 ± 8.99 × 104, P = 0.014), the peak titer of 78.3 ± 23.6 ffu/mL appeared to be delayed and was 300 folds lower than the prototype(2.39 ± 0.18 × 104vs 64.17 ± 39.26, P < 0.001). Collectively, the substitution of JFH1 NS5A gene with that of HC-J4 disrupted the high efficient production of HCV virions of FL-J6JFH1.
Figure 4 Immunofluorescence assay of hepatitis C virus proteins in naïve Huh-7.
5 cells infected with cell supernatants from prototype and chimera transfected Huh-7.5 cells. The naïve Huh-7.5 cells were inoculated with supernatants collected at different time points during the experiment. Three days after transfection, the cells were subjected to immunofluorescence assay using sera from hepatitis C virus patients as primary antibody. A: Cells inoculated with supernatants from FL-J6JFH1-transfected cells; B: Cells inoculated with supernatants from FL-J6JFH/J4NS5A-transfected cells; C: Cells inoculated with supernatants from FL-J6JFH1 (GND)-transfected cells; D: Cells inoculated with supernatants from FL-J6JFH1-transfected cells incubated with normal sera as primary antibody.
Figure 5 Comparison of cell culture-produced hepatitis C virus produced by chimera and prototype.
The full-length RNA transcripts of the prototype and chimera were transfected into Huh-7.5 cells. The supernatants were harvested at the indicated time points after transfection. The infectivity titers of the supernatants were determined, and the values were presented as focus forming unit per milliliter (ffu/mL).
Since NS5A gene of HCV genotype 1a and 1b is easy to introduce compensatory mutations during the evolution with the hosts, the variation of NS5A gene in the nascent recombinant viruses was analyzed by sequencing to investigate if any mutation occurred in the HC-J4 NS5A genes in the backbone of FL-J6JFH1. The viral RNAs were extracted from the cell culture supernatants of days 13, 34 and 59 after transfection and were subsequently used to amplify the whole gene of NS5A by RT-PCR. At the same time, the NS5A gene of prototype was also checked for the mutation. Nucleotide sequencing of the products did not reveal any mutation in the sequence of NS5A region in both the chimera and prototype during the culture (data not shown).
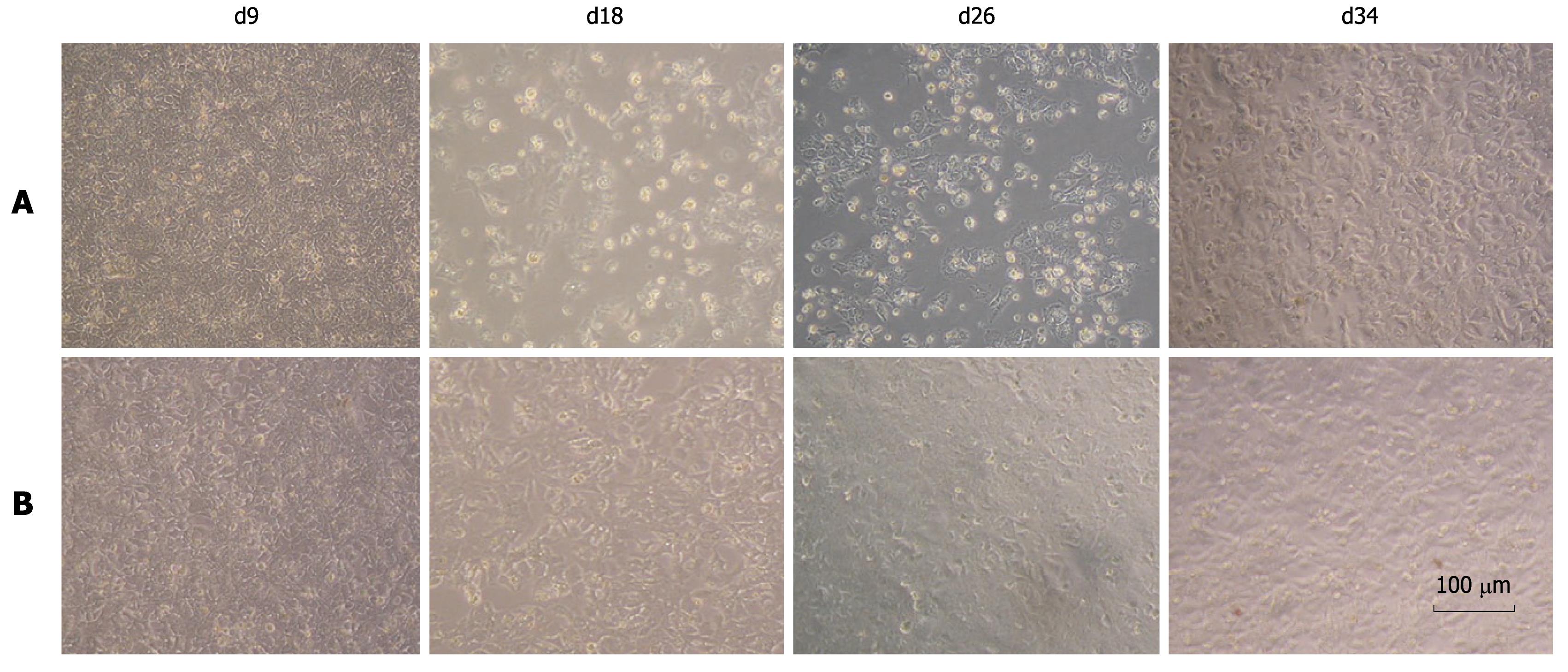
We observed that FL-J6JFH/J4NS5A and FL-J6JFH1 had different cell cytopathogenicities. At day 13 after transfection, a high number of non-adherent cells were observed in the cell culture transfected with the prototype, possibly due to a high titer of HCVcc produced by FL-J6JFH1 (Figure 6). Thirteen days after transfection, the levels of HCV RNA and HCVcc produced by prototype reached a maximum (Figures 2 and 5), and cell growth recovered to normal around day 30 after transfection. In contrast, the chimera RNA transfected cells showed no cytopathic effect (CPE) until the experiment was terminated at day 59, which also indicated that virus production was much less than the prototype.
Figure 6 Cell morphology of chimera and prototype genome RNA transfected cells.
The cytopathic effect was observed in prototype RNA tansfected Huh-7.5 cells under phase-contrast microscopy from day 9 to day 34 after transfection. No cytopathic effect was observed in the chimeric RNA transfected cells. A: FL-J6JFH1-transfected cells; B: FL-J6JFH/J4NS5A-transfected cells.
DISCUSSION
The discovery of high efficiently replicative JFH1 strain enabled us to develop an authentic in vitro cell culture system of HCV, however, other natural HCV strains have not yet been shown to replicate efficiently and demonstrate robust infectivity in cell culture without adaptive mutations. Since JFH1 isolate is unique among the HCV strains and not necessarily representative of HCV biology, there is an urgent demand for establishing cell culture systems for all the HCV genotypes, especially for genotype 1b, the major genotype of human infections in most Asian countries[34-36].
A growing body of evidences showed that NS5A protein possesses multiple and diverse functions in RNA replication, interferon resistance, and viral pathogenesis of HCV. In this study, the intergenotypic chimera constructed with gene substitution was studied to elucidate the roles of NS5A in both HCV replication and virus particle production. In vitro transcripts of FL-J6JFH/J4NS5A were prepared and transfected into Huh-7.5 cells, the influence of substitution of JFH1 NS5A with J4 NS5A on the replication and infection of HCV was observed. In a long culture period, the chimeric genome did replicate and produce infectious virus in vitro, but the levels of RNA replication, protein expression and HCVcc production decreased significantly compared with the prototype. The above results suggested that the NS5A domain of JFH1 isolate played an important role in the replication of J6JFH1 and the release of infectious particles, and may be one of the determinants to establish robust in vitro infection system based on the JFH1 consensus clone. The growth kinetics of the cells transfected with both chimera and prototype were also observed. At day 13 after transfection, a large number of non-adherent cells were noted continuously in the culture transfected with the prototype, which may be caused by robust replication of J6JFH1. In contrast, the chimeric RNA transfected cells showed no CPE during the whole experiment, which also indicated that virus production was much less than the prototype. Recent studies suggest that NS5A is involved in the assembly and maturation of infectious viral particles. Three studies have confirmed the participation of NS5A in the assembly of HCV particles[37-39].
In addition, a recent study suggested that the interaction between the structural and nonstructural genes of the same genotype may be important for infectious particle formation, and Linderbach[14] generated chimeras comprising the core to NS2 region of either the infectious genotype 2a strain J6 or the genotype 1a H77 and the nonstructural region of JFH-1. Transfection of the J6/JFH1 chimera into Huh-7.5 cells resulted in efficient virus replication and release of viral particles, while the H77/JFH1 chimera was only detectable in virus replication but not in infectious particle formation. Several other studies showed that other proteins such as core, p7 and NS2 proteins were involved in the process of virus particle production either in the early step or in the late step[39,40]. How the related proteins are regulated remains an interesting question to be addressed. And the answers will be greatly help establish cell culture systems for all the other HCV genotypes and search for efficient antiviral therapies for HCV infection.
In conclusion, a chimeric FL-J6JFH/J4NS5A genome by replacement of the entire NS5A gene of genotype 2a with that from genotype 1b was constructed. The replication efficiency and virus production of HCV chimera significantly decreased compared with the prototype. Since the NS5A gene is the only difference between chimera and prototype, JFH1 NS5A plays a critical role in robust replication of JFH1-based isolate or chimeras. Moreover, intergenotypic chimera FL-J6JFH/J4NS5A showed lower replication and infectivity than FL-J6JFH1 without CPE, which might be similar to natural HCV infection, and could be used as a valuable tool for comparative studies on the functions of NS5A in a genuine cell culture system and for the in vitro examination of the effects of potential anti-viral drugs targeting NS5A.
ACKNOWLEDGMENTS
We thank Dr. Rice CM (Rockefeller University, USA) for generously providing pFL-J6JFH1 and pFL-J6JFH1 (GND) plasmids and Huh 7.5 cells.
COMMENTS
Background
Hepatitis C virus (HCV) is a major cause of chronic hepatitis, liver cirrhosis, and even hepatocellular carcinoma. Lack of in vitro cell culture system hindered the development of new preventive and therapeutic methods for HCV. Recently, the establishment of cell culture system for HCV genotype 2a JFH1 and JFH1 based chimera genome, greatly promoted the study of life cycle and antiviral drugs of HCV. However, this model is only limited to HCV genotype 2a JFH1. Why the JFH1 genome is so special, and which protein affects virus production and release? Since HCV Nonstructural 5A (NS5A) is supposed to be pleiotropic protein playing a key role in both viral replication and virus production, the role of NS5A of JFH1 should be further illustrated, which will help establish cell culture models for all the other HCV genotypes.
Research frontiers
The establishment of JFH1-based cell culture models is a milestone of HCV research. Recent studies showed that genotype 1-7 cell culture was developed with intragenotypic and intergenotypic chimeras of JFH1, and these cell culture systems could be used for the selection of antiviral drugs or compounds and the search for new antiviral therapies targeting viral or host factors.
Innovations and breakthroughs
Recent reports have used the JFH1-based in vitro cell culture model to study the cell cycle and the function of structural and nonstructural proteins of HCV. Since the in vitro cell culture model is only limited to JFH1, it is necessary to explore the important proteins contributed to the production of cell culture-produced HCV (HCVcc). This paper focused on the NS5A protein of JFH1, and FL-J6JFH/J4NS5A chimera was constructed. Since NS5A gene is the only difference between the chimera and the prototype, the data reflecting the role of JFH1 NS5A in the robust replication of JFH1 are required in the future studies.
Applications
In this study, a 2a/1b chimera FL-J6JFH/J4NS5A was established by the method of gene substitution, and the function of JFH1 NS5A in the replication and infection was studied. By using this platform, other key proteins responsible for HCV replication and infection could be studied alone or in combination so as to establish in vitro cell models for other genotypes of HCV.
Terminology
Gene substitution is usually used to construct chimera to study the function of particular genes. The part or whole of the target gene can be replaced by related or unrelated genes.
Peer review
Wang et al investigated the function of NS5A protein of genotype 2a from JFH1 strain in replication and infection of HCV. They demonstrated that NS5A protein from JFH1 strain play an important role in the replication and production of HCVcc. Experimental design and methods seem appropriate and they presented convincing evidence, although some issues need to be addressed.
Peer reviewer: Yoshiaki Iwasaki, MD, PhD, Associate Professor, Health Service Center, Okayama University, 2-1-1, Tsushima-Naka, Kita-ku, Okayama 700-8530, Japan
S- Editor Sun H L- Editor Ma JY E- Editor Zheng XM