Published online May 28, 2011. doi: 10.3748/wjg.v17.i20.2563
Revised: February 15, 2011
Accepted: February 22, 2011
Published online: May 28, 2011
AIM: To evaluate the effect of silencing Livin gene expression with siRNA to apoptosis and proliferation in a colon cancer cell line.
METHODS: To investigate the anticancer effect of silencing Livin gene expression, we established an siRNA transfected cell line using the HCT116 colon cancer cell line. After confirming the successful transfection, MTT assay, flow cytometry and annexin V staining were employed to evaluate the antiapoptotic effect. To confirm the in vivo effect of Livin-siRNA, different doses of Livin-siRNA were injected into xenografted tumors in BALB/c nude mice model.
RESULTS: Livin expression was dramatically decreased after siRNA transfection, especially at 25 μmol/L of siRNA, but this suppression was not dose-dependent. The cell count at 18 h after transfection was significantly reduced as compared with controls (P < 0.01), but tended not to decrease proportionally depending on transfected dose or time. MTT assay revealed that silencing the Livin gene suppressed cellular proliferation at 18 h after transfection (P = 0.04); however, the inhibitory effect disappeared thereafter. Also, there was no significant difference in cellular proliferation depending on siRNA dose. The rate of apoptosis also increased with silencing of the Livin gene. In vivo, the tumor size significantly decreased after Livin-siRNA injection at 20 μmol/L concentration (P = 0.03). There were no significant body weight changes of mice after siRNA injection. Histologic examination revealed no significant toxic reaction in kidney, liver and brain of mice.
CONCLUSION: siRNA-mediated downregulation of Livin expression can induce apoptosis in colon cancer in vitro and in vivo, which suggests the possibility of new cancer therapeutics using siRNA.
- Citation: Oh BY, Lee RA, Kim KH. siRNA targeting Livin decreases tumor in a xenograft model for colon cancer. World J Gastroenterol 2011; 17(20): 2563-2571
- URL: https://www.wjgnet.com/1007-9327/full/v17/i20/2563.htm
- DOI: https://dx.doi.org/10.3748/wjg.v17.i20.2563
Colon cancer is one of the most common malignancies worldwide and has shown increased incidence, especially in developing countries[1]. Until recently, a major treatment strategy has been surgical resection that shows good outcome over other treatments. With the activation of a screening program, early cancer, less than stage I, is curable with only surgical treatment. But the treatment results for metastatic disease are still unsatisfactory with surgery alone. In these cases, clinicians consider other treatment options such as chemotherapeutic agents and targeted agents. Recently, development of techniques in manipulation of nucleic acids makes gene therapy possible to adopt in cancer therapy.
RNA interference (RNAi) is a fundamental protective process in eukaryotic cells including invertebrates and vertebrates, which is able to block harmful signal by targeting complementary mRNA and cleaving thereof[2]. Small interfering RNA (siRNA) is fragments from double-stranded RNA by the enzyme Dicer[3-5]. The natural role of RNAi is supposed to be a defense mechanism against some viral infection or deleterious genomic instability. This special mechanism has been of great interest recently as siRNA targeting small genes are easily manufactured and applied to major clinical problems such as cancer, asthma, inflammatory disease and infection[6]. This methodology is becoming a powerful tool for new drug development in such areas and is replacing the techniques of using antisense oligonucleotide and ribozymes[7,8].
Livin, a 280 amino acid protein and a member of the mammalian type of inhibitor of apoptosis (IAP), are well-conserved proteins across the species. It has a single baculovirus IAP repeat (BIR) domain and a COOH-terminal RING domain and is localized predominantly in the nucleus and a filamentous pattern of cytoplasm[9]. The Livin gene spans 46 kb and is located on chromosome 20 at band q13[10]. The major function of Livin is an inhibition of apoptosis by binding to caspase 3, caspase 7 and caspase 9 and also in inhibition of proteolytic processes of caspase 9[9,11]. Its overexpression protects cells from various proapoptotic stimuli.
The expression of Livin, similar to expression of Survivin, is rarely detected in normal adult tissues but exists abundantly in cancerous tissues and transformed cells[9]. The overexpression of Livin protein has been reported in colorectal cancer, leukemia, hepatocellular carcinoma, and melanoma[12]. Recently, major attention has been focused on the IAP family, especially Survivin and Livin, as they are easy to handle in the genetic field due to their small size. Moreover, the lack of expression in normal adult tissues of Livin is a very attractive characteristic in cancer therapy[9].
In this study, we used siRNA targeting to Livin transfection into the HCT116 colon cancer cell line to confirm the antitumor effect and blockade of Livin gene and performed an additional in vivo study using BALB/c nude mice xenografts to determine the direct tumor regression effect of Livin silencing with siRNA. These results suggested that Livin is an effective target for colorectal cancer treatment using siRNA technique.
Cell culture-The HCT116 colon cancer cell line, purchased from Korean cell line bank, was grown in RPMI1640 medium (Life Technologies, Inc., Grand Island, NY) supplemented with 10% fetal bovine serum (FBS, Life Technologies, Inc.), penicillin (100 U/mL) and streptomycin (100 g/mL). Cells were maintained at 37°C in a humidified atmosphere of 50 mL/L CO2.
Total RNA was extracted using the Trizol reagent (Invitrogen, Carlsbad, CA). The method for extracting total RNA was according to the manufacturer’s protocol. RNA samples were quantified at 260 nm using spectrophotometry. One μg of total RNA was reacted for 15 min at 42°C using the Promega’s Reverse Transcription System (Promega Corp., Madison, WI), reacted for 5 min at 95°C, and cDNA was obtained. For polymerase chain reaction (PCR), cDNA (3 μL) was obtained by reverse transcription, 0.1 μmol primers, 1.25 μL GoTaq DNA polymerase, 0.2 mmol/L deoxynucleotide triphosphate (dNTP), 1 Go Taq reaction buffer and each primer was added, and DNA was amplified by performing PCR with primer of Livin; forward, 5-GTCAGTTCCTGCTCCGGTCAA-3; reverse, 5-GGGCACTTTCAGACTGGACCTC-3. Electrophoresis of PCR products was performed on 1.5% agarose gel and examined by staining with ethidium bromide. By measuring the brightness of bands using a densitometer, their amount was quantified using the value of β-actin as the standard value.
Synthetic 21-nt RNAs were purchased from Dharmacon Research (Lafayette, CO; in deprotected, desalted, and annealed form.). We used 4 primers for silencing the Livin gene to improve the blocking efficiency. The target sequences of Livin for production of siRNA were 5'-GGAGAGAGGTCCAGTCTGA-3', 5'-GGAAGAACCGGAAGACGCA-3', 5'-GCTCTGAGGAGTTGCGTCT-3' and 5'-GCTCTGAGGAGTTGCGTCTTT-3'. The nonspecific control siRNA duplex was also purchased from Dharmacon Research.
Briefly, centrifuge tubes containing siRNA to ensure that the siRNA pellet was collected at the bottom of the tube. Resuspended siRNA to a convenient stock concentration with 1X siRNA buffer (Dharmacon, Inc., Lafayette, CO). After placing the solution on a shaker for 30 min at room temperature, the concentration of siRNA was verified using UV spectrophotometry at 260 nm.
HCT116 cells were diluted in fresh media without antibiotics and transferred to six-well plates (1 × 105 cells/well) 24 h before transfection and maintained at 37°C in a humidified atmosphere of 50 mL/L. HCT116 cells grown to a confluence of 40%-50% were transfected with siRNA using Lipofectamine 2000 (Life Technologies, Inc., Grand Island, NY) according to the manufacturer’s recommendations. To detect the transfection efficiency, the cells were analyzed by FACSan (Becton Dickson, San Jose, CA) 6 h after transfection with FITC-labeled siRNA. After transfection, the cells were incubated and then used to various analyses.
Cell viability was examined by routine 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. HCT116 cells (1 × 106) were planted in 96-well plates with RPMI1640 in a final volume of 500 μL. On the following day, cells were treated with increasing concentration of siRNA and cultured for 48 h, then cell proliferation was assessed by MTT assay. Following incubation at 37°C for 3 h, the reaction was stopped by the addition of 150 μL DMSO. After the crystal dissolved, the absorbency of the samples was determined at 492 nm.
HCT116 cells were planted in 24-well plates with RPMI 1640. Cells were treated with increasing concentration of siRNA and cultured for 48 h, then cells were collected by trypsinization, mixed in PBS and trypan blue added. Cells were counted using a microscope counting chamber.
At 24 h after transfection, HCT116 cells on 60-mm tissue culture plate were treated with 1 × trypsin-EDTA (Invitrogen, Carlsbad, CA) for 5 min at 37°C; cells were then removed from the plate by gentle scraping, dispersed into cold PBS and washed twice. The cells were resuspended in 1 × binding buffer at a concentration of approximately 1 × 106 cells/mL. Five μL of Annexin V- FITC and 10 μL of PI were added to each cell suspension and incubated at room temperature for 10 min in the dark. The cells were subjected to fluorescence estimation using a FACS Calibur flow cytometer. Uninoculated cells stained with Annexin V- FITC and PI were used to determine background levels of apoptosis.
All animal experiments in this report were performed with approval of the Institutional Animal Care & Use Committee in Ewha Womans University, School of Medicine. The 4-wk-old male BALB/c nude mice were grown in a sanitary room and adjusted for 1 wk before the start of the experiments. Viable HCT116 colon cancer cells (2.0 × 106 in PBS/100 μL) were injected subcutaneously into the right flank of the nude mice. Three weeks after tumor cell inoculation with confirmation of successful maturation of tumors, mice were divided randomly into four groups (ten mice per group) and were treated weekly for 4 wk by way of center-intratumoral direct injection at different doses (10, 20, 50 μmol/L) of siRNA manufactured with atelocollagen to achieve effective delivery. To prepare the siRNA/atelocollagen complex, equal volumes of atelocollagen (AteloGene® Systemic Use; Koken, Tokyo, Japan. 0.1% in PBS at pH 7.4) and siRNA solution were combined and mixed by rotation for 20 min at 4°C. In the control group, pure PBS was injected using the same method. The tumors were monitored with a caliper every week over a 4 wk period after siRNA transfection. Tumor volume for each mouse was determined (in cubic millimeter) by measuring in two directions and was calculated as tumor volume = length × (width)2/2. Also, body weight of each mouse was monitored and recorded. Five weeks after the last siRNA injection, all mice were sacrificed according to the animal experimental guidelines and the xenografted tumors were excised and prepared with a routine pathological procedure. Tumor sections were deparaffinized and subjected to Hematoxylin and Eosin staining in the usual manner.
Each experiment was repeated three times or more. Bands from RT-PCR were quantified with Quantity One software (Bio-Rad, Hercules, CA). mRNA levels were calculated by referring them to the amount of b-actin. The difference between means was performed with analysis of variance. All statistical analyses were performed using SPSS 11.0 software (SPSS, Inc., Chicago, IL). Statistical significance was considered when the P value was less than 0.05.
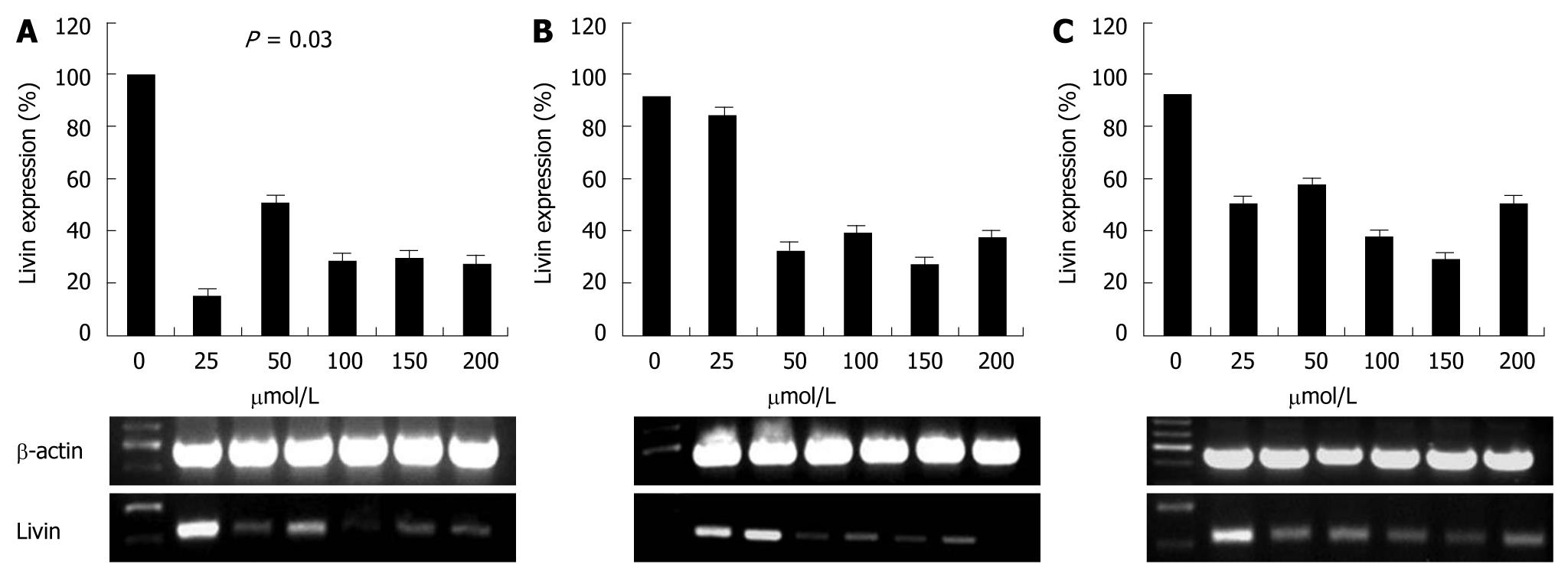
To identify whether Livin expression was affected by siRNA, we transfected siRNA at different concentrations (25, 50, 100, 150 and 200 μmol/L) to HCT 116 cell line, and then detected Livin density at 18, 24 and 30 h after transfection by RT-PCR.
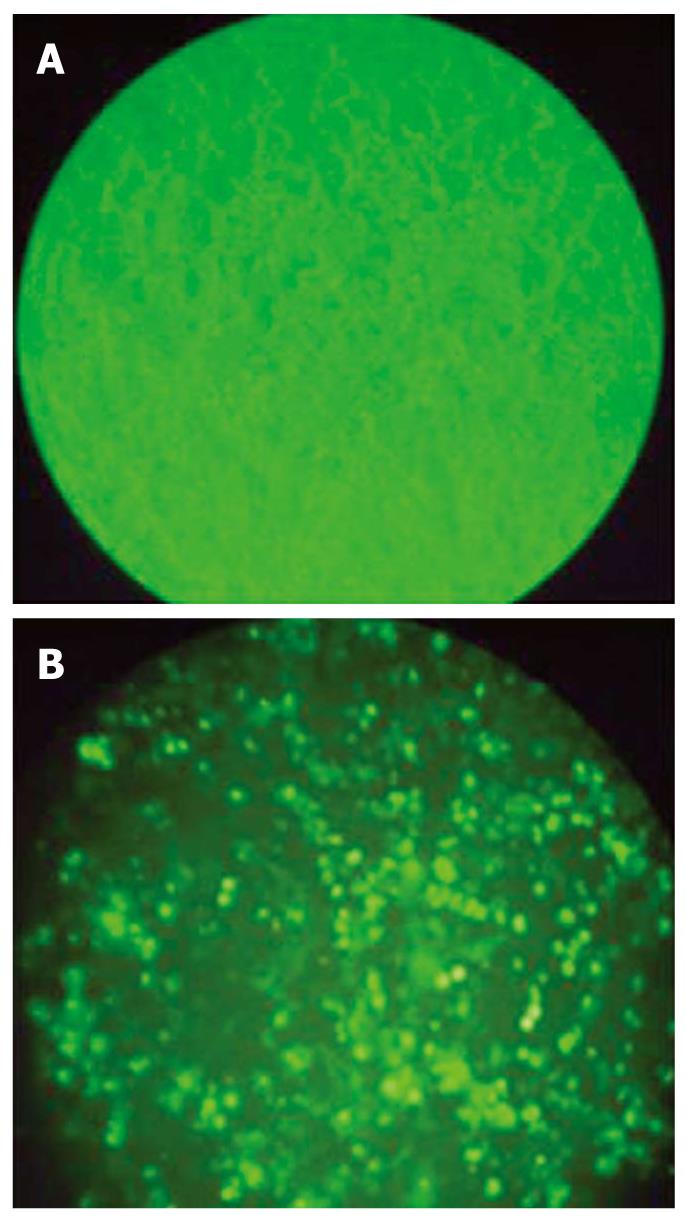
Successful transfection of siRNA was detected with fluorescence staining (Figure 1) and expression of Livin gene was compared to β-actin. Control HCT 116 cells expressed Livin very well and this expression was dramatically decreased after siRNA transfection. But this effect was attenuated with time and Livin expression was retrieved 30 h after transfection. Livin expression was most effectively suppressed at 25 μmol/L of siRNA, but this suppression was also not dose-dependent (Figure 2).
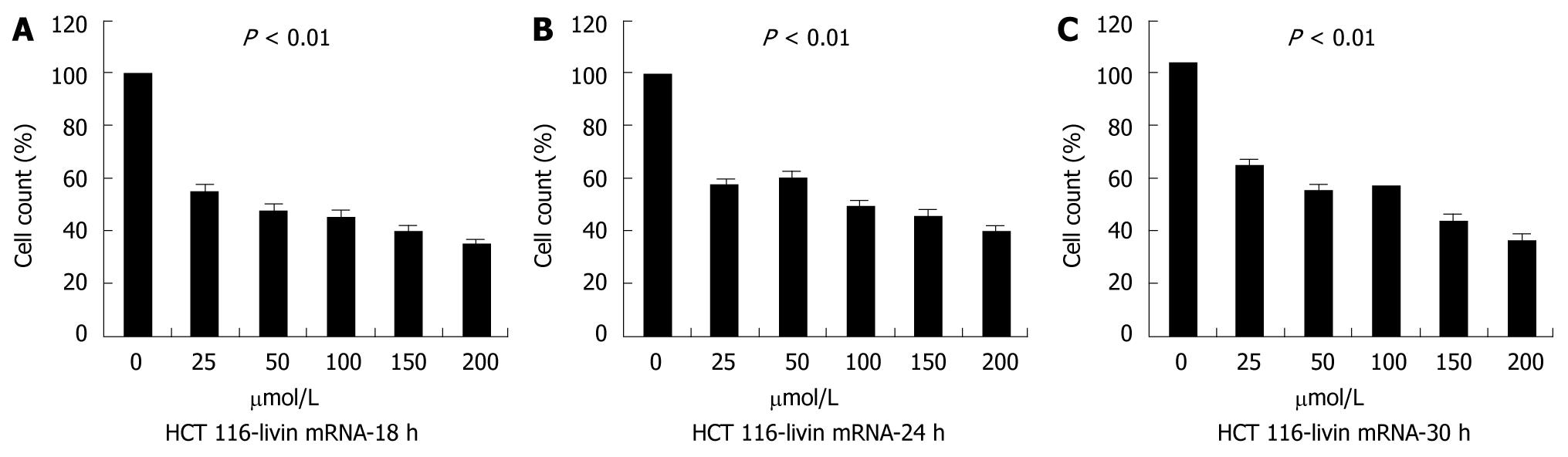
First, we estimated cell counts of each group to evaluate tumor cell growth after Livin silencing. We analyzed cultured cells grown in three dishes per group at one time, and repeated this study three times. As seen in Figure 3, the cell count at 18 h after transfection was significantly reduced as compared with control (P < 0.01), but tended not to decrease proportionally depending on transfected dose or relapsing time.
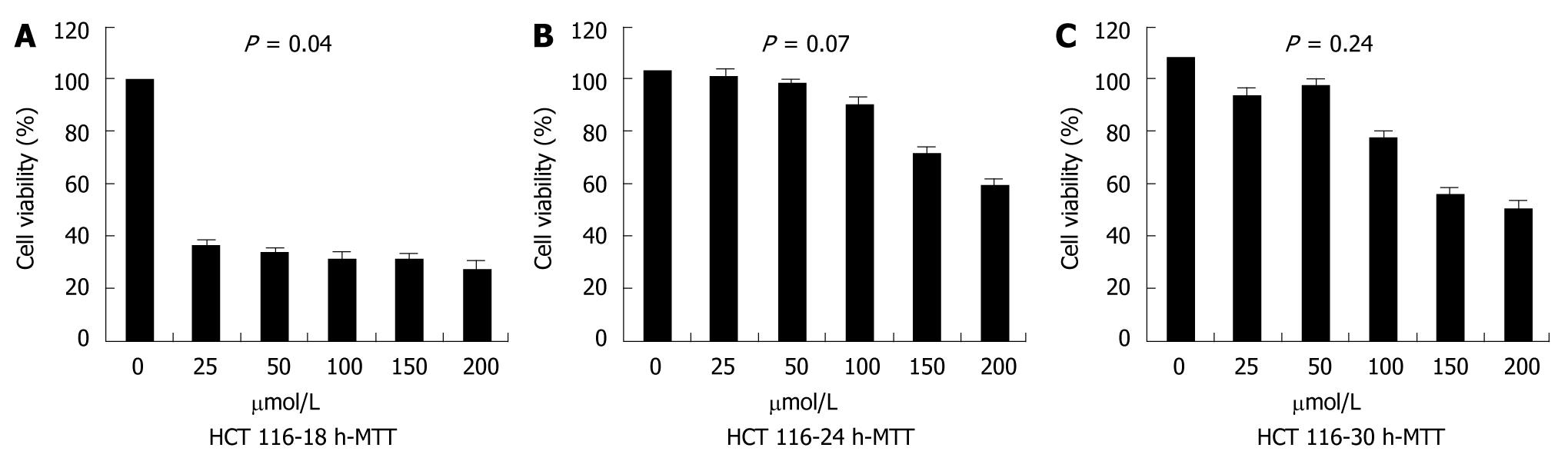
To quantify the cellular viability, MTT assay was performed. The MTT assay showed that, compared with controls, the proliferation of cells transfected with siRNA was remarkably inhibited at 18 h after transfection (P = 0.04), but the inhibitory effect disappeared thereafter, which indicated that the number of viable cells began to increase. There was no significant difference in cellular proliferation depending on siRNA dose (Figure 4).
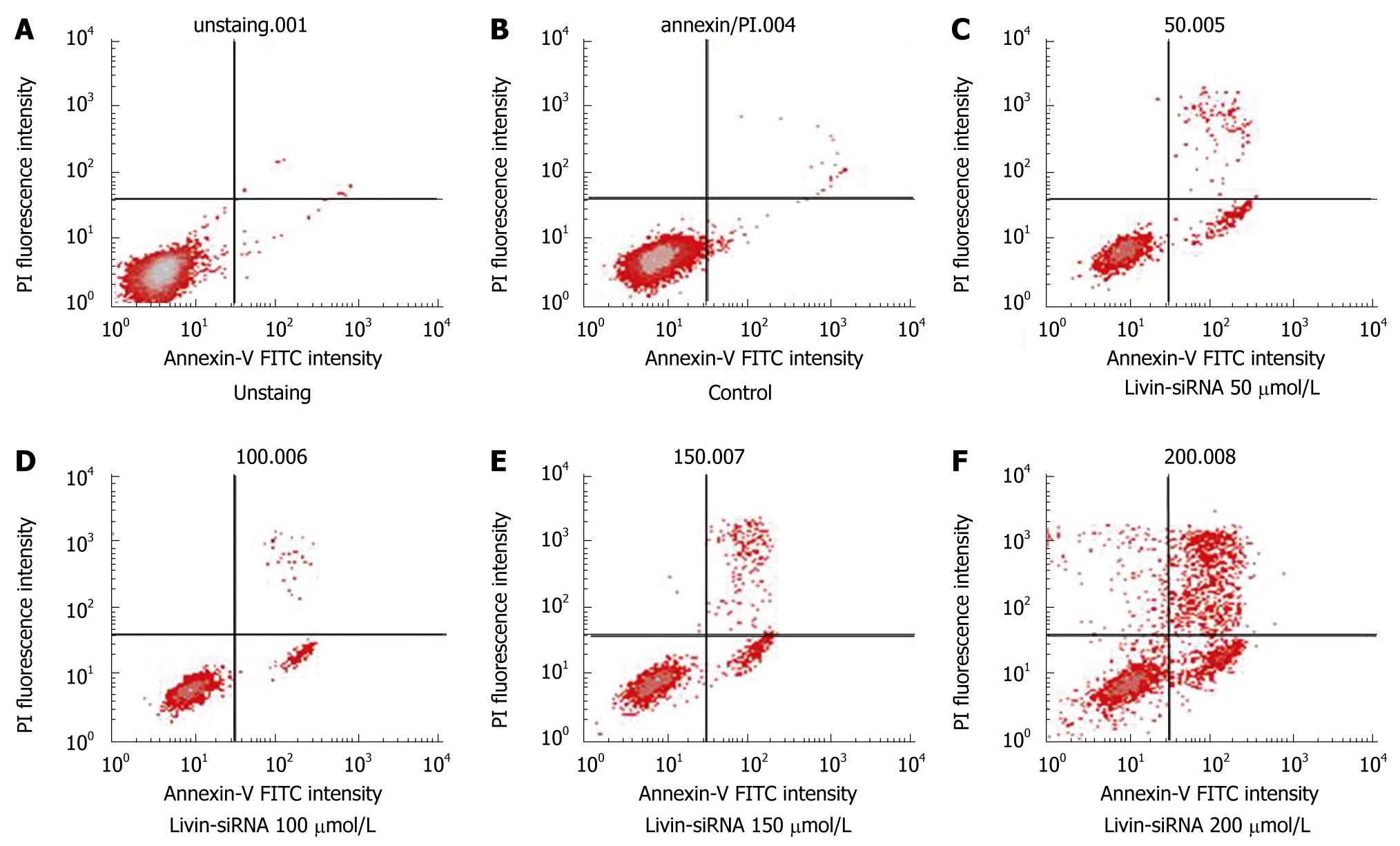
Flow cytometric analysis after Annexin V staining was performed to investigate whether Livin-silencing induces apoptosis resulting in reduction of cell count or suppression of cellular proliferation as per the above findings. As a result, the apoptotic rate was increased continuously since 50 μmol/L of siRNA was treated. The apoptotic rate in control was 3.91%, the apoptotic rate of cells transfected with siRNA 50 μmol/L was 30.32%. The apoptotic rates of cells transfected with siRNA 100 μmol/L, 150 μmol/L and 200 μmol/L increased to 32.88%, 36.91% and 45.08%, respectively (Figure 5). The apoptotic rate was increased dose-dependently, but the necrotic portion was also increased. So, it was thought low dose siRNA was more effective in the clinical setting.
Next, we designed a xenograft model to identify the potential in vivo effects of siRNA on inhibition of the proliferation of colon cancer cells. Viable HCT 116 colon cancer cells were injected subcutaneously into the right flank of the nude mice.
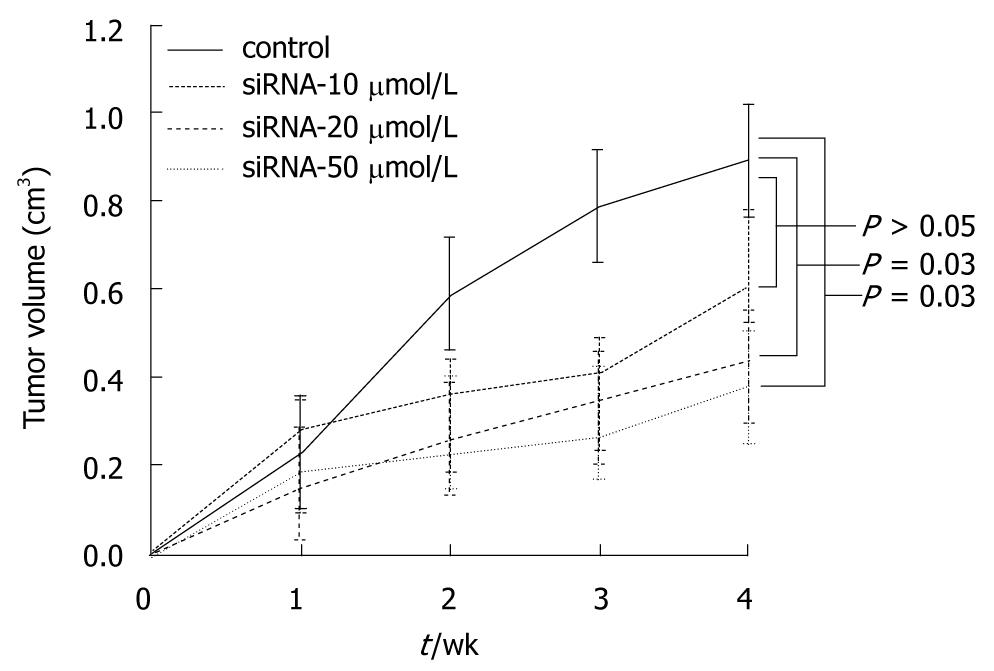
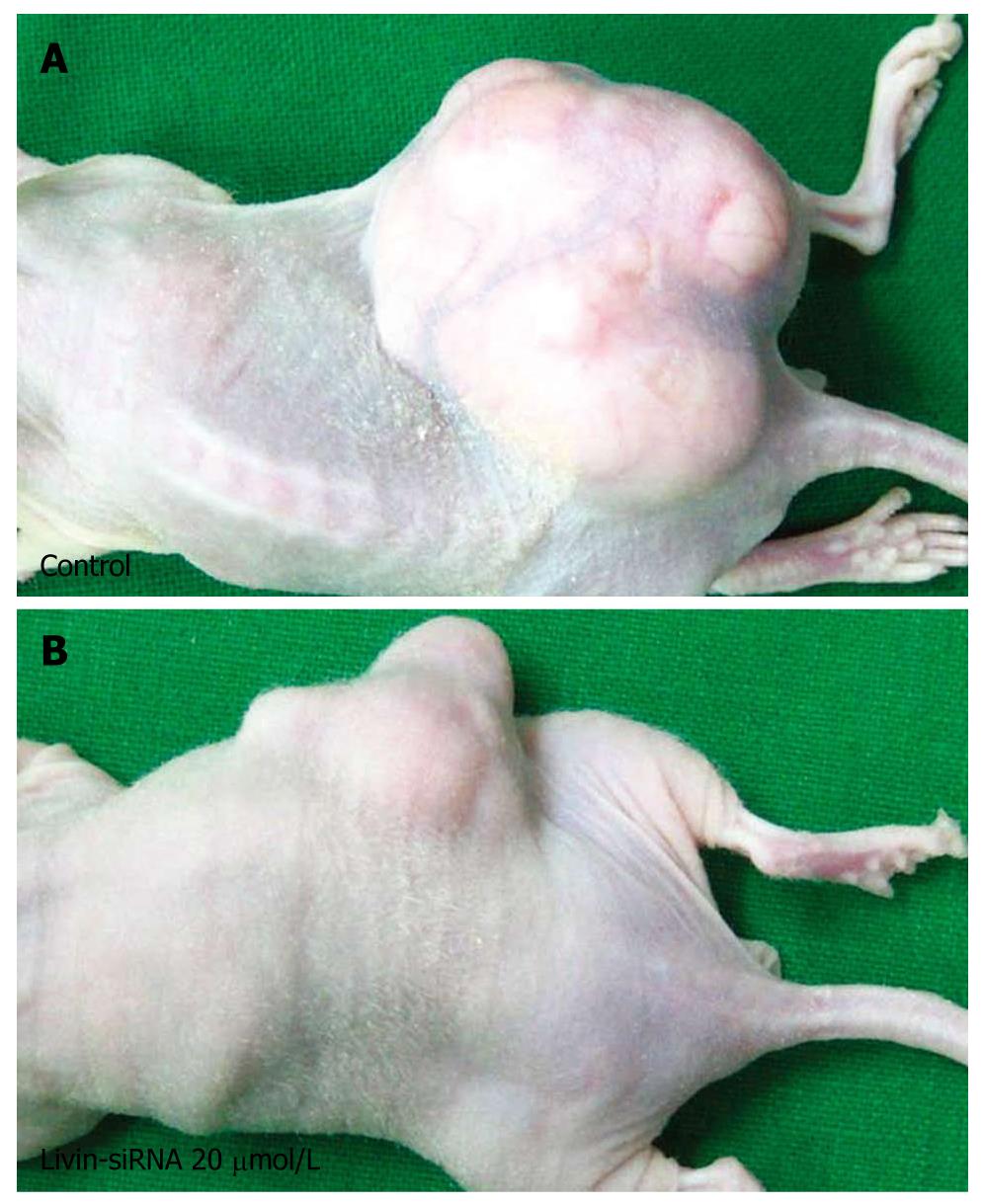
In the first series of experiments, the inoculated mice were divided randomly into four groups of ten mice each and were treated with different doses of siRNA (control, 10, 20 and 50 μmol/L) weekly, for 4 wk. Over a 4 wk period after siRNA injection, the tumor volumes were estimated every week. As seen in Figure 6, the rates of tumor growth significantly decreased in higher than 20 μmol/L transfected groups with siRNA as compared with the control group 2 wk following treatment (P = 0.03). However, there was no difference in antitumor effect between 20 μmol/L and 50 μmol/L in the transfected group.
In the first study series, we detected that siRNA 20 μmol/L was the most effective dose for reduction of tumor growth in xenograft models. Thereafter, in the second series of experiments, we divided the inoculated mice randomly into two groups (control group and siRNA 20 μmol/L treated group) of ten mice per group. The mice of the siRNA 20 μmol/L treated group were injected with siRNA daily for 4 wk, and then the tumor volumes were measured every week during that period. The result was similar with the previous experiment; the mean volume of tumors decreased in transfected groups with siRNA 20 μmol/L as compared with control groups (Figure 7).
To evaluate the toxic effect of siRNA in vivo, the body weights of mice were taken during the experimental period. The body weight of mice in all groups slightly increased over the entire experimental period; there were no significant body weight changes after siRNA injection compared to the control group. Histologic examination was performed with tissues taken from sacrificed mice after experiment completion. It revealed no significant toxic reaction in kidney, liver and brain of siRNA treated mice compared with the control group (data not shown).
Eight human IAP molecules have been reported to date; NAIP (BIRC1), c-IAP1 (BIRC2), c-IAP2 (BIRC3), X-linked IAP (XIAP, BIRC4), Survivin (BIRC5), Apollon (BRUCE, BIRC6), Livin/ML-IAP (BIRC7), and IAP-like protein 2 (BIRC8)[12]. They are categorized as IAP by the presence of a baculovirus IAP repeat (BIR) protein domain, which forms a zinc-fold, a critical motif for regulating apoptosis. Many in this family also harbor a COOH-terminal RING finger domain. The BIR domain constitutes the zinc-fold, which is the important motif in delivering the anti-apoptotic signals[9]. It has been proposed that endogenous Livin has a minor direct effect on caspase activity whereas its anti-apoptotic effect could be ascribed to its antagonizing activity on the XIAP-Smac/DIABLO interaction and has been regarded as counteraction molecules against apoptosis through the blocking the terminal effectors such as caspases, thus far[12]. That the IAPs are highly conserved from yeast to mammals suggests the importance of this family in maintain life[11]. There are numerous reports on overexpression of the IAP family in cancer tissues to normal counterparts. The role of Survivin and Livin, in particular, in carcinogenesis has been well confirmed to date.
Livin was reported in 2001 as a novel IAP family protein[9]. The overactivity of Livin was regarded as a predictor of poor prognosis in cancer patients and a resistance factor to chemotherapeutic agents or radioactive drugs in several cancers[13,14]. The overexpression of Livin mRNA has been found in some tumors including melanoma, breast, cervical, colon and prostate cancers, as well in leukemia, in lymphoma and in hepatoma cell lines[12]. Moreover, researchers have shown that only overexpression of Livin-α isoform is correlated with high risk of relapse in bladder cancer[10].
In the previous study, Livin gene was expressed well in the stools of colon cancer patients as opposed to the healthy control group. This finding suggests that the exfoliated cancer cells from the gastrointestinal tumors were contained in the patients’ stool and overexpressed tumor related genes such as Livin were detected from the stool. Based on this result, we chose Livin gene as a treatment target in this experiments.
In recent years, numerous genetic engineering techniques were introduced in drug development markets worldwide such as antisense oligonucleotides, aptamer (single stranded nucleic acids), use of ribozymes, dominant negative mutant construction and siRNA. We focused on siRNA technology because of its target sequence-specific degradation ability and relatively simple application procedure.
In higher organisms, defense mechanisms are more complex but long dsRNAs should not be used as an experimental tool to trigger RNAi in mammalian cells. Long dsRNAs are processed by the endonuclease Dicer into short 21 bp duplexes with siRNA instead of direct suppression[15]. RNA interference is a post-transcriptional regulation process which exploits a complex pathway that regulates gene expression and includes machinery for sequence specific mRNA degradation[8,16].
But there are still some limitations to overcome in applying siRNA in clinical fields. The greatest interference is the short half-life of the delivered siRNA in vivo due to renal clearance. Another is a normally existing RNase activity that can degrade siRNA administered artificially. The next key to successful in vivo application of siRNA is the delivery system. Transfering the siRNA into target tissues and into the cell cytoplasm is not easily achieved and a most delicate process is required to get the therapeutic potential. Usual methods to achieve maximum therapeutic effects are lipid or protein carriers, liposomes, antibody promoter fusions, cyclodextrin nanoparticles, fusogenic peptides, aptamers, biodegradable polylactide copolymers and polymers[17].
There are several reports on siRNA targeting Livin treatments for cancer in the literature. Crnkovic-Mertens et al[18] first reported the induction effects of apoptosis with Livin targeted-siRNA in a HeLa cell line. Their results suggested that the possibility of intracellular interference with Livin gene expression sensitized human tumor cells to apoptosis. Recently, Wang et al[19] investigated the apoptotic susceptibility of the SGC-7901 gastric cancer cell by shRNA-mediated silencing of the Livin gene. When Livin gene was silenced, the reproductive activity of the gastric cancer cells was significantly lower than the control groups (P < 0.05). The study also showed that IC50 of 5-FU and cisplatin on gastric cancer cells treated by shRNA decreased and the cells were more susceptible to proapoptotic stimuli (5-FU and cisplatin) (P < 0.01). In another study[20], it was reported that expression of Livin was downregulated by siRNA in dose- and time-dependent manners, and silencing Livin promoted apoptosis in malignant melanoma LiBr cells. They also demonstrated that silencing Livin by siRNA leads to cell cycle arrest at the G0/G1 phase, reducing the rate of DNA synthesis or resulting in apoptosis. With these promising results, a combination treatment of Livin-targeting siRNA with proapoptotic drugs appears to be more effective in clinical perspectives.
In this study, we confirmed that silencing of Livin gene expression with siRNA was well documented in HCT116 colon cancer cells associated with an increased apoptotic response. In preliminary experiments, we tested the expression of Livin immediately, 2, 6, 12 and 24 h after treatment of siRNA (data not shown). The decrease of Livin expression was notable from 2 h after treatment but the effect was not much different before 24 h, so we extended the time to 30 h after treatment of siRNA. Testing dosage was arranged from 25 to 200 μmol/L, but the silencing effect of siRNA was not proportional in order of concentration. It was possible that a very small amount of siRNA could create the action to achieve the silencing of Livin gene in the cell line. Unfortunately, the silencing effect weakened from 24 h after treatment, which means duration of siRNA to Livin is very short and time limited. Another consideration is the proliferation of the cells not influenced by siRNA affected the total expression of the Livin with time. On the other, the cell count decreased 45% with 25 μmol/L siRNA at 18 h after treatment and lasted for 30 h and cellular proliferation counted by MTT assay revealed the same results with cell counting. Proliferation was more influenced by siRNA treatment in the early phase compared to the decrease of total cell count numbers. With this phenomenon apoptosis increased 24 h after treatment with dose dependent manner. To integrate the full results, silencing of Livin in colon cancer cell was easily achieved with siRNA treatment into the induction of apoptosis from minimal dosage only in in vitro environment.
We used the mouse xenograft model to identify the antitumor effect of siRNA to Livin in vivo. We selected the direct intratumoral injection method to verify the direct effect on tumors. In the clinical setting, it is very difficult to use the infected cells of siRNA before confirmation of drug safety regulation, so direct implication to the tumor itself would be more readily applicable to the human body. Because of the limitation of uptake of siRNA, we adopted atelocollagen in this experiment to obtain maximal therapeutic effects. Atelocollagen is derived from type I collagen of calf dermis by pepsin digestion. A telopeptide that is an amino acid sequence of N- and C- terminal of the collagen contains the most antigenicity. Atelocollagen has limited antigenicity because of a lack of telopeptides, so it is more easily applicable to drug delivery processes. It is well confirmed that atelocollagen-siRNA complexes are taken up into cells to achieve a silencing effects. Also, atelocollagen-siRNA complexes are resistant to nucleases, which is another important advantage in siRNA application in vivo[8]. Several reports were found in the database revealing siRNA effects with atelocollagen complex. Atelocollagen is one of the more useful DDS available in clinical settings. Kawata et al[21] reported PLK-1 (polo-like kinases-1) siRNA with atelocollagen in hepatic metastatic cells from lung cancer. Takeshita et al[22] demonstrated the evidence of tumor targeted delivery of systemically administered siRNA with atelocollagen, injected siRNA intracardiac to luciferase and luciferase-expressing bone-metastatic tumor models. In another study, a therapeutic effect via atelocollagen-mediated systemic administration of siRNA targeting Bcl-xL into pregrown PC-3 xenografts was reported[17].
A number of studies have reported antitumor effects of siRNA in vivo. Verma et al[23] demonstrated that siRNA was effective in reducing growth of HCT116 cells in nude mice. These mice were injected with HCT116 cells intraperitoneally and then were treated with siRNA directed against β-catenin, the regulator of cellular proliferation in colon cancer. There was a significant decrease in the tumor size of cells transfected with siRNA directed against β-catenin[23]. In another study, it was found that transfected HCT116 colon cancer cells with siRNA against Survivin, a type of IAP family such as Livin, significantly decreased in size as compared with controls in xenograft models[24]. In a recent study, it was observed that siRNA against Survivin remarkably induced apoptosis in SW480 cell as a consequence of the inhibition of Survivin and led to significant inhibition of tumor growth in vivo[1].
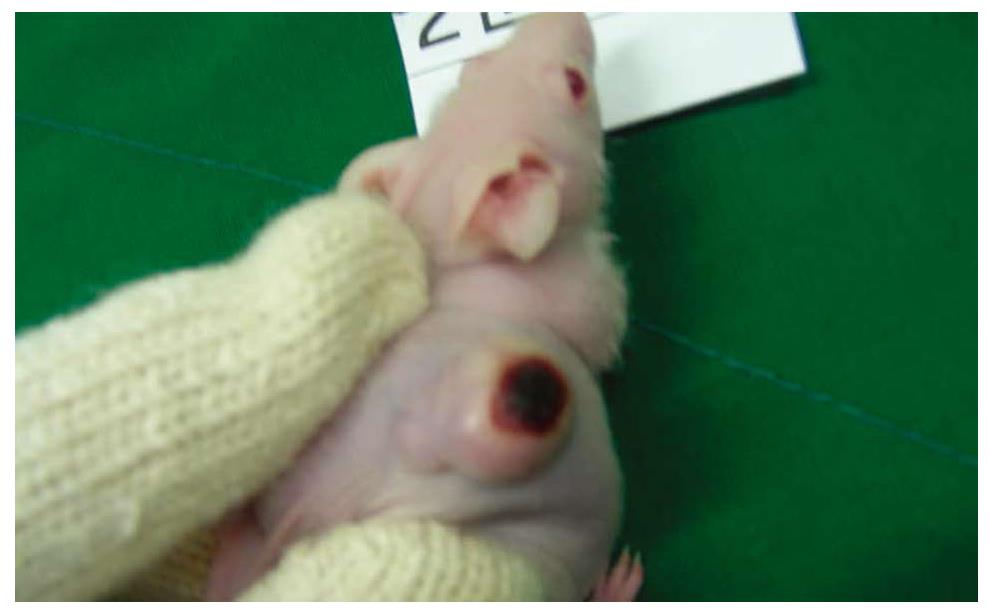
In our xenograft model, the rates of tumor growth were significantly decreased in higher than 20 μmol/L transfected groups with siRNA as compared with control groups. This antitumor effect tended to be greater at a higher dose of siRNA but the differences between each group were not statistically significant. So it would be expected that a low dose of siRNA showed a satisfactory effect on tumor suppression. In addition to the antitumor effect, toxic effects of treatment should be considered. Some therapeutics have been associated with toxic side effects such as weight loss and other organ injury. These systemic toxicities may be related with higher morbidity and lower response rate, resulting in poorer survival. Thus, if there are systemic toxicities caused by drugs, no matter how effective, they should not be used. In this study, measured body weights of mice had no differences, and there were no significant pathologic findings on tissues withdrawn from siRNA-treated group. For these reasons, it could be thought that siRNA targeting Livin can be used in colon cancer therapy without systemic toxic effects in vivo. In our present study, another interesting finding was detected despite an effective reduction of tumor size with siRNA. Local hemorrhagic necrosis was seen in the intratumoral injection site of siRNA with syringe, this finding was not identified in other studies (Figure 8). If there are local side effects such as this, even though there is no evidence of systemic toxicity of siRNA, we should carefully determine administration routes to develop antitumor treatments using siRNA.
In conclusion, siRNA-mediated downregulation of Livin expression can induce apoptosis in colon cancer in vitro and in vivo, which implies new potential cancer therapeutics using siRNA in colon cancer.
Colon cancer is one of the most common malignancies and has increased in incidence in recent years. Several treatment modalities have been used, especially in metastatic diseases. Several studies have been reported to implicate gene therapy for metastatic colon cancer. Livin, a type of inhibitor of apoptosis, protects cells from various proapoptotic stimuli. So Livin is an effective target for colorectal cancer treatment.
The overexpression of Livin has been reported in colorectal cancer. However, the direct tumor regression effect of Livin silencing has not been unequivocally addressed. In this study, the authors investigate the effect of silencing Livin gene expression with siRNA to apoptosis and proliferation in colon cancer cell line.
There are several reports on siRNA targeting Livin treatments for cancer. But these studies were nearly all performed in vitro and there were few studies about colorectal cancer. In this study, the authors confirmed that silencing of Livin gene expression with siRNA was well documented in vitro and in vivo, especially in colorectal cancer.
siRNA-mediated downregulation of Livin expression can induce apoptosis in colon cancer in vitro and in vivo, which may imply new potential cancer therapeutics using siRNA in colorectal cancer.
Livin is a 280 amino acid-protein and a member of the mammalian type of inhibitor of apoptosis. It inhibits apoptosis by binding to caspase 3, caspase 7 and caspase 9. Its overexpression protects cells from various proapoptotic stimuli, and is shown in numerous cancers. RNA interference is a fundamental protective process in eukaryotic cells, which is able to block harmful signals by targeting complementary mRNA and cleaving it. The role of RNAi is supposed to be a defense mechanism against deleterious genomic instability.
The experiments are properly performed, and findings support their conclusion.
Peer reviewer: Jian Wu, Associate Professor of Medicine, Internal Medicine/Transplant Research Program, University of California, Davis Medical Center, 4635 2nd Ave. Suite 1001, Sacramento CA 95817, United States
S- Editor Tian L L- Editor O’Neill M E- Editor Ma WH
| 1. | Shen W, Wang CY, Wang XH, Fu ZX. Oncolytic adenovirus mediated Survivin knockdown by RNA interference suppresses human colorectal carcinoma growth in vitro and in vivo. J Exp Clin Cancer Res. 2009;28:81. |
| 2. | Zhang L, Fogg DK, Waisman DM. RNA interference-mediated silencing of the S100A10 gene attenuates plasmin generation and invasiveness of Colo 222 colorectal cancer cells. J Biol Chem. 2004;279:2053-2062. |
| 3. | Lv W, Zhang C, Hao J. RNAi technology: a revolutionary tool for the colorectal cancer therapeutics. World J Gastroenterol. 2006;12:4636-4639. |
| 4. | Yang L, Kang WK. The effect of HIF-1alpha siRNA on growth and chemosensitivity of MIA-paca cell line. Yonsei Med J. 2008;49:295-300. |
| 5. | Li TJ, Song JN, Kang K, Tong SS, Hu ZL, He TC, Zhang BQ, Zhang CQ. RNA interference-mediated gene silencing of vascular endothelial growth factor in colon cancer cells. World J Gastroenterol. 2007;13:5312-5316. |
| 6. | Ryther RC, Flynt AS, Phillips JA 3rd, Patton JG. siRNA therapeutics: big potential from small RNAs. Gene Ther. 2005;12:5-11. |
| 7. | Zhang YA, Nemunaitis J, Samuel SK, Chen P, Shen Y, Tong AW. Antitumor activity of an oncolytic adenovirus-delivered oncogene small interfering RNA. Cancer Res. 2006;66:9736-9743. |
| 8. | Takeshita F, Ochiya T. Therapeutic potential of RNA interference against cancer. Cancer Sci. 2006;97:689-696. |
| 9. | Kasof GM, Gomes BC. Livin, a novel inhibitor of apoptosis protein family member. J Biol Chem. 2001;276:3238-3246. |
| 10. | Liu B, Han M, Wen JK, Wang L. Livin/ML-IAP as a new target for cancer treatment. Cancer Lett. 2007;250:168-176. |
| 11. | Wang L, Zhang Q, Liu B, Han M, Shan B. Challenge and promise: roles for Livin in progression and therapy of cancer. Mol Cancer Ther. 2008;7:3661-3669. |
| 12. | Augello C, Caruso L, Maggioni M, Donadon M, Montorsi M, Santambrogio R, Torzilli G, Vaira V, Pellegrini C, Roncalli M. Inhibitors of apoptosis proteins (IAPs) expression and their prognostic significance in hepatocellular carcinoma. BMC Cancer. 2009;9:125. |
| 13. | Wang R, Lin F, Wang X, Gao P, Dong K, Zou AM, Cheng SY, Wei SH, Zhang HZ. Silencing Livin gene expression to inhibit proliferation and enhance chemosensitivity in tumor cells. Cancer Gene Ther. 2008;15:402-412. |
| 14. | Crnković-Mertens I, Muley T, Meister M, Hartenstein B, Semzow J, Butz K, Hoppe-Seyler F. The anti-apoptotic livin gene is an important determinant for the apoptotic resistance of non-small cell lung cancer cells. Lung Cancer. 2006;54:135-142. |
| 16. | Verdel A, Vavasseur A, Le Gorrec M, Touat-Todeschini L. Common themes in siRNA-mediated epigenetic silencing pathways. Int J Dev Biol. 2009;53:245-257. |
| 17. | Mu P, Nagahara S, Makita N, Tarumi Y, Kadomatsu K, Takei Y. Systemic delivery of siRNA specific to tumor mediated by atelocollagen: combined therapy using siRNA targeting Bcl-xL and cisplatin against prostate cancer. Int J Cancer. 2009;125:2978-2990. |
| 18. | Crnkovic-Mertens I, Hoppe-Seyler F, Butz K. Induction of apoptosis in tumor cells by siRNA-mediated silencing of the livin/ML-IAP/KIAP gene. Oncogene. 2003;22:8330-8336. |
| 19. | Wang TS, Ding QQ, Guo RH, Shen H, Sun J, Lu KH, You SH, Ge HM, Shu YQ, Liu P. Expression of livin in gastric cancer and induction of apoptosis in SGC-7901 cells by shRNA-mediated silencing of livin gene. Biomed Pharmacother. 2010;64:333-338. |
| 20. | Pirollo KF, Chang EH. Targeted delivery of small interfering RNA: approaching effective cancer therapies. Cancer Res. 2008;68:1247-1250. |
| 21. | Kawata E, Ashihara E, Kimura S, Takenaka K, Sato K, Tanaka R, Yokota A, Kamitsuji Y, Takeuchi M, Kuroda J. Administration of PLK-1 small interfering RNA with atelocollagen prevents the growth of liver metastases of lung cancer. Mol Cancer Ther. 2008;7:2904-2912. |
| 22. | Takeshita F, Minakuchi Y, Nagahara S, Honma K, Sasaki H, Hirai K, Teratani T, Namatame N, Yamamoto Y, Hanai K. Efficient delivery of small interfering RNA to bone-metastatic tumors by using atelocollagen in vivo. Proc Natl Acad Sci USA. 2005;102:12177-12182. |
| 23. | Verma UN, Surabhi RM, Schmaltieg A, Becerra C, Gaynor RB. Small interfering RNAs directed against beta-catenin inhibit the in vitro and in vivo growth of colon cancer cells. Clin Cancer Res. 2003;9:1291-1300. |
| 24. | Williams NS, Gaynor RB, Scoggin S, Verma U, Gokaslan T, Simmang C, Fleming J, Tavana D, Frenkel E, Becerra C. Identification and validation of genes involved in the pathogenesis of colorectal cancer using cDNA microarrays and RNA interference. Clin Cancer Res. 2003;9:931-946. |