Published online Jan 7, 2010. doi: 10.3748/wjg.v16.i1.30
Revised: September 8, 2009
Accepted: September 15, 2009
Published online: January 7, 2010
AIM: To profile protein expression in mucosal biopsies from patients with chronic refractory pouchitis following antibiotic or probiotic treatment, using a comparative proteomic approach.
METHODS: Two-dimensional polyacrylamide gel electrophoresis and matrix-assisted laser desorption/ionization-time of flight mass spectrometry were used to characterize the changes related to antibiotic therapy in the protein expression profiles of biopsy samples from patients with chronic refractory pouchitis. The same proteomic approach was applied to identify differentially expressed proteins in the non-inflamed pouch before and after probiotic administration.
RESULTS: In the first set of 2D gels, 26 different proteins with at least 2-fold changes in their expression levels between the pouchitis condition and antibiotic-induced remission were identified. In the second set of analysis, the comparison between mucosal biopsy proteomes in the normal and probiotic-treated pouch resulted in 17 significantly differently expressed proteins. Of these, 8 exhibited the same pattern of deregulation as in the pouchitis/pouch remission group.
CONCLUSION: For the first time, 2D protein maps of mucosal biopsies from patients with ileal pouch-anal anastomosis were provided, and differentially expressed proteins following antibiotic/probiotic treatment were identified.
- Citation: Turroni S, Vitali B, Candela M, Gionchetti P, Rizzello F, Campieri M, Brigidi P. Antibiotics and probiotics in chronic pouchitis: A comparative proteomic approach. World J Gastroenterol 2010; 16(1): 30-41
- URL: https://www.wjgnet.com/1007-9327/full/v16/i1/30.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i1.30
Total proctocolectomy with ileal J-pouch-anal anastomosis (IPAA) is the surgical treatment of choice for patients with refractory ulcerative colitis (UC) or UC with dysplasia. Although the surgery generally cures UC and has been shown to result in a significant improvement of health-related quality of life, complications can occur after IPAA[1].
The most common long-term complication is pouchitis, an idiopathic inflammatory disease of the ileal reservoir. The reported incidence of pouchitis is variable, largely because of differences in the type and duration of follow-up. However, studies have shown that as many as 15%-46% of patients with UC develop at least 1 episode of pouchitis within 5 years after surgery[2].
Clinically, pouchitis is characterized by variable symptoms, including increased stool frequency and fluidity, abdominal cramping, pelvic discomfort, bleeding, tenesmus, fever and weight loss, and extra-intestinal manifestations in more severe cases[3]. For an unequivocal diagnosis, endoscopic examination and histologic investigation are mandatory[4]. Pouchitis Disease Activity Index (PDAI) is the most commonly used diagnostic instrument and represents an objective and reproducible scoring system for pouchitis[5]. Active pouchitis is defined as a score ≥ 7 and remission is defined as a score < 7.
The etiology and pathophysiology of pouchitis are still poorly understood. However, the fact that pouchitis almost exclusively occurs in patients with underlying UC and that it generally responds to antibacterial therapy suggests a role for the gut microbiota and a genetic predisposition[6].
The disease activity of pouchitis can be defined as remission, mild-moderate or severe based primarily on symptoms. Duration can be classified as acute (< 4 wk) or chronic (≥ 4 wk). Disease pattern can be infrequent (1-2 acute episodes), relapsing (≥ 3 acute episodes) or chronic (a treatment-responsive form requiring maintenance therapy or a treatment-resistant form). Approximately 10%-15% of patients with pouchitis experience a chronic pouchitis, either treatment-responsive or treatment-refractory, and some of them require surgical excision or exclusion of the pouch because of impairment of reservoir function and poor quality of life[7].
Treatment of pouchitis is largely empirical. Broad-spectrum antibiotics have been widely used and represent the mainstay of treatment. Small randomized trials have shown that both metronidazole and ciprofloxacin, alone, sequentially or in combination, are effective in reducing the PDAI score and achieving a significant improvement in clinical symptoms and endoscopic and histologic findings. However, metronidazole is poorly tolerated and treatment with systemically active antibiotics is not ideal from the perspective of the development of antibiotic resistance. In addition, in chronic pouchitis antibiotic-induced remission periods are often short and the condition is complicated by frequent relapses[8].
Recently, several studies have suggested that altering the microbiota in the pouch by administering probiotic bacteria can be effective in maintaining remission and reducing the incidence of flare-ups in chronic pouchitis[9,10]. Moreover, the efficacy of probiotic therapy as prophylaxis to delay the first onset of pouchitis after pouch surgery, has been demonstrated[11,12].
Comparative proteomic analysis represents an effective tool to identify proteins critical for functional pathways in normal cells and phenotype changes that occur during disease development. Since biological and functional output of cells is governed primarily by proteins, the applications of proteomic technologies are beginning to have a profound impact on understanding of the molecular mechanisms underlying several disease processes, which, in turn, will help to reduce disease-related morbidity and mortality. However, despite their extensive use in proteomic profiling of gene expression in various diseases, the applications of such technologies in inflammatory bowel diseases are still in their infancy[13] and, so far, no proteomic study has been reported in IPAA research.
In the present study, we apply 2-dimensional polyacrylamide gel electrophoresis (2D-PAGE) and matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) to define the differential protein displays of mucosal biopsy samples from patients with chronic refractory pouchitis before and after antibiotic treatment. The same proteomic approach has also been applied to identify specific changes in protein expression in the non-inflamed vs probiotic-administered pouch in order to provide a picture of the intestinal mucosa protein modulation by probiotics.
Six patients who underwent restorative proctocolectomy with IPAA were recruited for this study and routinely followed up by the Department of Internal Medicine and Gastroenterology, University of Bologna, Polyclinic S. Orsola. Patients were included if they had a chronic refractory pouchitis, defined as no response to at least 4 wk of standard antibiotic therapies (ciprofloxacin 1 g twice daily (bid) or metronidazole 400 mg 3 times daily). They were divided in 2 groups according to PDAI score at study entry and treatment received. In the first group, 3 patients with PDAI ≥ 7 were orally administered with a combination of metronidazole (500 mg bid) and ciprofloxacin (500 mg bid) for 1 mo. The second group, including the other 3 patients with chronic refractory pouchitis but with a total PDAI < 7 at study entry, received VSL#3 (VSL pharmaceuticals Inc., Ft. Lauderdale, FL, USA) 2 packets bid for 3 mo. VSL#3 contains 450 billion viable lyophilized bacteria per packet, comprised of 4 strains of lactobacilli (Lactobacillus acidophilus, L. casei, L. delbrueckii subsp. bulgaricus and L. plantarum), 3 strains of bifidobacteria (Bifidobacterium breve, B. infantis and B. longum) and one strain of Streptococcus thermophilus. Mucosal biopsies were collected during pouch endoscopy before and after antibiotic/probiotic therapy.
All samples were immediately snap frozen in liquid nitrogen. The institutional ethics committee approved all protocols and all enrolled subjects gave their informed consent.
Frozen mucosal biopsies (about 10-20 mg) were washed in 200 μL of cold low salt washing buffer (3 mmol/L KCl, 1.5 mmol/L KH2PO4, 68 mmol/L NaCl, 9 mmol/L NaH2PO4), with Complete Protease Inhibitor (Roche Molecular Biochemicals, Mannheim, Germany). After centrifugation at 13 000 r/min for 2 min, tissue samples were homogenized in 1 mL of lysis solution (0.11 mol/L DTT, 0.11 mol/L CHAPS, 8 mol/L urea, 2 mol/L thiourea, 35 mmol/L Tris and Complete Protease Inhibitor) using an Ultra-Turrax® homogenizer (IKA Labortechnik, Staufen, Germany). Protein extraction was performed as previously described[14]. Total protein concentration of the cell extract was calculated using the PlusOne 2D Quant Kit™ (GE Healthcare, Uppsala, Sweden). The protein extract preparation was immediately used or aliquoted and frozen at -20°C.
Samples containing 100 μg of protein were diluted to 250 μL with rehydration solution (8 mol/L urea, 2% CHAPS, 10 mmol/L DTT, 2% (v/v) ampholine, pH 3.5-9.5 (GE Healthcare) and trace bromophenol blue) and applied to Immobiline DryStrips (13 cm, pH 3-10, GE Healthcare) for 12 h rehydration at 50 V. Isoelectric focusing was performed using IPGphor apparatus (GE Healthcare) to give a total of 19 kVh. IPG strips were then reduced and alkylated[15] prior to loading onto 15% acrylamide separating gels (20 cm long, 1 mm thickness). Electrophoresis was performed at 250 V for 7 h using Protean II xi Cell (Bio-Rad, Hercules, CA, USA). Protein spots were visualized with a MS-compatible silver-staining procedure[16].
Protein patterns in the gels were recorded as digitalized images using a GS-800 imaging densitometer (Bio-Rad). Spot detection, matching and the examination of differentially expressed proteins were performed by PDQuest v6.2 software (Bio-Rad). Three technical replicates were made per patient and condition and formed 1 replicate group with average normalized spot intensities. The comparison was carried out for each patient before and after antibiotic/probiotic therapy. Proteins that showed at least 2 times enhanced/decreased expression were selected for identification along with a few spots that showed a similar expression pattern in all 2D gels.
Protein spots with conserved expression levels throughout the gels in all patients and conditions were identified. Two identification methods were employed: comparison of our reference proteome map with Swiss-2D PAGE (http://www.expasy.ch/ch2d/) and other published 2D proteome patterns[17-21] obtained under very similar experimental conditions, and MALDI-TOF MS analysis. Since both methods provided the same identification result for each spot, we used the gel matching method to identify the differentially expressed proteins in pouchitis/antibiotic-induced remission and normal pouch/probiotic-treated pouch groups. When gel matching produced an unreliable and doubtful identification, because of excessive deviations in pI and Mr values across gels, MALDI-TOF MS was employed.
Protein spots were manually excised from 2D gels, washed and in-gel digested as previously reported[22]. Crude digests were concentrated and desalted using mC18 ZipTips (Millipore, Bedford, MA, USA). Peptide extracts were mixed on the MALDI-TOF target (Applied Biosystems, Foster City, CA, USA) with an equal matrix volume of 5 mg/mL α-cyano-4-hydroxycinnamic acid (Sigma-Aldrich, St. Louis, MO, USA) saturated with 50% acetonitrile/0.2% trifluoroacetic acid, and analyzed using a Voyager-DE Pro Biospectrometry Workstation (Applied Biosystems). All mass spectra were obtained in a reflectron mode, with an accelerating voltage of 20 kV and a delayed extraction of 40 ns. Internal mass calibration with peptides arising from trypsin autoproteolysis was performed. Peptide masses were searched against Swiss-Prot, TrEMBL and NCBI non-redundant protein databases using ProFound (http://prowl.rockefeller.edu/prowl-cgi/profound.exe) and Aldente (http://expasy.org/tools/aldente) programs. Search parameters were set to allow up to one missed tryptic cleavage and a peptide mass tolerance of 50 ppm. Only protein hits with a significant probability score calculated by software and at least 3 matching peptide masses were considered.
Statistical analysis of protein expression was performed using the Student’s t-test carried out with SigmaStat v3.5 software (Systat Software, Point Richmond, CA, USA). A P value < 0.05 was considered as statistically significant. Bibliometric analysis for co-citation was performed using Bibliosphere Pathway Edition from Genomatix (Genomatix Software, Munich, Germany).
All the enrolled subjects completed the study. In the first group of patients, after 1 mo of antibiotic therapy, clinical and endoscopic remission was achieved with a significant decrease in both PDAI and median stool frequency (data not shown). In the second group, no episodes of active pouchitis were recorded during the probiotic administration. Both treatments were well tolerated and no side effects were recorded.
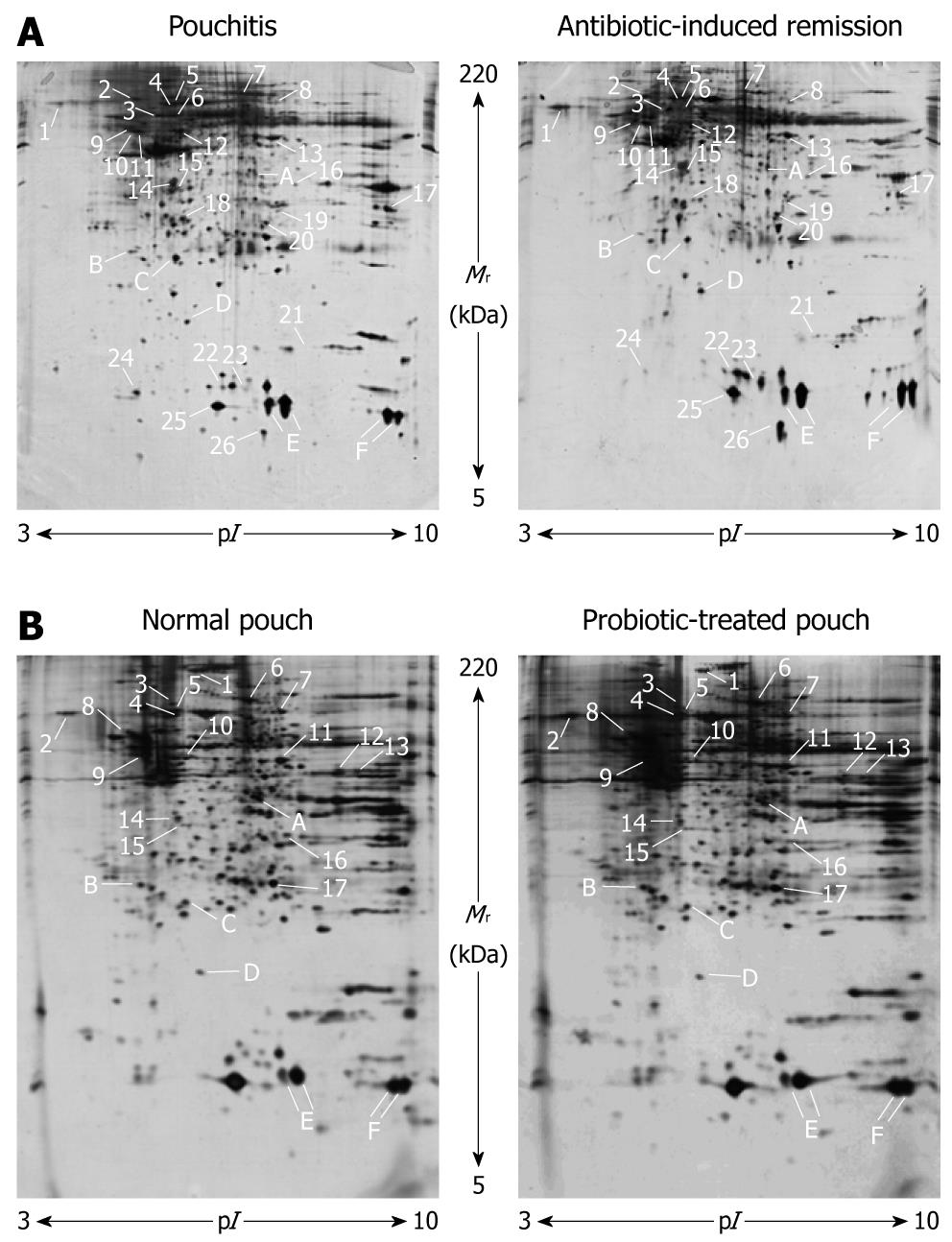
An example of 2D gels obtained from mucosal biopsies in pouchitis and pouch remission is provided in Figure 1A. Approximately 1200 protein spots per gel were detected within a pI range of 3-10 and a Mr range of 5-220 kDa. The resolution of the polypeptides showed better quality in the low molecular mass area and toward the acidic side of the gels whereas increased streaking and precipitation, a well known phenomenon observed in 2D-PAGE, were visible on the basic side.
| Spot ID | Swiss-Prot Acc. No. | Protein name | COG1 | Subcellular location | Theoretical Mr/pI | Experimental Mr/pI | Method of identification2 |
| A | Q15365 | Poly(rC)-binding protein 1 (PCBP1) | A | Nucleus | 37.53/6.66 | 35.99/7.17 | MALDI-TOF MS |
| B | Q6IBM5 | Rho GDP dissociation inhibitor (GDI) α, isoform CRA_a (ARHGDIA) | T | Cytoplasm | 23.21/5.03 | 25.73/4.99 | MALDI-TOF MS |
| C | Q5R8R5 | Glutathione S-transferase P (GSTP1) | O | Cytoplasm | 23.36/5.93 | 24.33/5.80 | MALDI-TOF MS |
| D | P61088 | Ubiquitin-conjugating enzyme E2 N (UBE2N) | O | Nucleus | 17.14/6.13 | 19.00/6.10 | MALDI-TOF MS |
| E | P68871 | Hemoglobin subunit β (HBB) | C | Extracellular | 16.00/6.74 | 13.10/7.70 | MALDI-TOF MS |
| F | Q1HDT5 | Hemoglobin α 1-2 hybrid (HBA1) | C | Extracellular | 15.27/9.04 | 12.50/9.55 | MALDI-TOF MS |
For each patient, 2D patterns of mucosal biopsies collected before and after antibiotic administration were compared by PDQuest. Because of the high intrinsic variability among individuals, a stringent criterion was applied whereby only those proteins with at least 2 times increased or decreased expression and deregulation in the same way in all patients were considered. Out of 40 differentially expressed protein spots, 26 (65%) were identified, of which 15 were upregulated and 11 downregulated in antibiotic-induced remission of pouchitis (Figure 1A and Table 1). In addition, 6 protein spots with a similar expression pattern in all 2D gels were selected and identified (Figure 1 and Table 2).
| Spot ID | Swiss-ProtAcc. No. | Protein name | COG1 | Subcellular location | Theoretical Mr/pI | ExperimentalMr/pI | Method of identification2 | Change in protein expression with AB/PB treatment3 |
| Pouchitis/antibiotic-induced remission | ||||||||
| 1 | P27797 | Calreticulin (CALR) | O | Endoplasmic reticulum | 48.14/4.29 | 68.52/4.35 | GM (Swiss-2D PAGE) | Up |
| 2 | P11021 | 78 kDa glucose-regulated protein (GRP78) | O | Endoplasmic reticulum | 72.33/5.07 | 73.88/4.95 | GM (Swiss-2D PAGE) | Down |
| 3 | P10809 | 60 kDa heat shock protein, mitochondrial precursor (HSP60) | O | Mitochondrial matrix | 61.05/5.70 | 60.20/5.32 | GM (Swiss-2D PAGE) | Up |
| 4 | P11142 | Heat shock cognate 71 kDa protein (HSPA8) | O | Nucleolus | 70.90/5.37 | 69.20/5.18 | GM[20] | Up |
| 5 | P38646 | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP75) | O | Mitochondrion | 73.68/5.87 | 71.41/5.70 | GM (Swiss-2D PAGE) | Down |
| 6 | Q9BU08 | Putative uncharacterized protein, fragment (CCT5) | S | Undefined | 59.47/5.45 | 60.46/5.58 | GM[21] | Up |
| 7 | P02787 | Serotransferrin precursor (TF) | P | Extracellular | 77.05/6.81 | 79.49/7.09 | GM (Swiss-2D PAGE) | Down |
| 8 | Q16822 | Phosphoenolpyruvate carboxykinase (GTP), mitochondrial precursor (PCK2) | C | Mitochondrion | 70.73/7.56 | 71.67/7.62 | GM[19] | Up |
| 9 | P68371 | Tubulin β-2C chain (TUBB) | Z | Cytoplasm | 49.83/4.79 | 52.44/4.79 | MALDI-TOF MS | Up |
| 10 | P06576 | ATP synthase subunit β, mitochondrial precursor (ATP5B) | C | Mitochondrion | 56.56/5.26 | 48.675.01 | MALDI-TOF MS | Up |
| 11 | Q8NBS9 | Thioredoxin domain-containing protein 5, precursor (TXNDC5) | R | Endoplasmic reticulum | 47.63/5.63 | 49.43/5.09 | GM[20] | Down |
| 12 | P35900 | Keratin, type I cytoskeletal 20 (KRT20) | W | Cytoplasm | 48.49/5.52 | 48.15/5.54 | GM[19] | Down |
| 13 | P06733 | α-enolase (ENO1) | G | Cytoplasm | 47.17/7.01 | 46.80/7.57 | MALDI-TOF MS | Down |
| 14 | P11177 | Pyruvate dehydrogenase E1 component subunit β, mitochondrial precursor (PDHB) | C | Mitochondrion | 39.25/6.20 | 32.96/5.64 | MALDI-TOF MS | Down |
| 15 | P17707 | S-adenosylmethionine decarboxylase proenzyme (AMD1) | T | Cytoplasm | 38.34/5.71 | 31.91/5.74 | MALDI-TOF MS | Up |
| 16 | P13804 | Electron transfer flavoprotein subunit α, mitochondrial precursor (ETFA) | C | Mitochondrion | 35.08/8.62 | 34.01/7.91 | GM[20] | Up |
| 17 | P21796 | Voltage-dependent anion-selective channel protein 1 (VDAC1) | P | Mitochondrion | 30.77/8.62 | 30.60/9.20 | GM[19] | Down |
| 18 | P07339 | Cathepsin D, precursor (CTSD) | O | Lysosome | 44.55/6.10 | 28.02/5.70 | GM (Swiss-2D PAGE) | Down |
| 19 | Q99439 | Calponin-2 (CNN2) | Z | Cytoplasm | 33.70/6.94 | 29.64/7.55 | MALDI-TOF MS | Up |
| 20 | P00915 | Carbonic anhydrase I (CA1) | R | Cytoplasm | 28.87/6.59 | 27.52/7.45 | GM[19] | Down |
| 21 | P62937 | Peptidyl-prolyl cis-trans isomerase A (PPIA) | O | Cytoplasm | 18.01/7.68 | 16.42/8.09 | GM[19] | Up |
| 22 | P12104 | Fatty acid-binding protein, intestinal (FABP2) | I | Cytoplasm | 15.21/6.62 | 14.07/6.99 | MALDI-TOF MS | Up |
| 23 | P51161 | Ileal lipid binding protein (FABP6) | I | Cytoplasm | 14.37/6.29 | 13.58/7.22 | MALDI-TOF MS | Up |
| 24 | P09382 | Galectin-1 (LGALS1) | W | Extracellular | 14.72/5.33 | 13.25/5.26 | MALDI-TOF MS | Down |
| 25 | P07148 | Fatty acid-binding protein, liver (FABP1) | I | Cytoplasm | 14.21/6.60 | 12.13/6.80 | MALDI-TOF MS | Up |
| 26 | Q5T1C5 | Protein S100-A10 (S100A10) | R | Plasma membrane | 11.20/6.82 | 10.52/7.25 | MALDI-TOF MS | Up |
| Non-inflamed pouch/probiotic-treated pouch | ||||||||
| 1 | P18206 | Vinculin (VCL) | Z | Cytoplasm | 123.80/5.50 | 114.44/5.81 | GM (Swiss-2D PAGE) | Up |
| 2 | P27797 | CALR | O | Endoplasmic reticulum | 48.14/4.29 | 68.52/4.35 | GM (Swiss-2D PAGE) | Down |
| 3 | P28331 | NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial precursor (NDUFS1) | C | Mitochondrion | 79.47/5.89 | 77.54/5.52 | GM[21] | Down |
| 4 | P11142 | HSPA8 | O | Nucleolus | 70.90/5.37 | 69.20/5.18 | GM[20] | Up |
| 5 | P38646 | GRP75 | O | Mitochondrion | 73.68/5.87 | 71.41/5.70 | GM (Swiss-2D PAGE) | Down |
| 6 | P02787 | TF | P | Extracellular | 77.05/6.81 | 79.49/7.09 | GM (Swiss-2D PAGE) | Down |
| 7 | Q16822 | PCK2 | C | Mitochondrion | 70.73/7.56 | 71.67/7.62 | GM[19] | Up |
| 8 | Q71U36 | Tubulin α-1A chain (TUBA1A) | Z | Cytoplasm | 50.15/4.94 | 56.47/4.82 | GM[17] | Down |
| 9 | Q8NBS9 | TXNDC5 | R | Endoplasmic reticulum | 47.63/5.63 | 49.43/5.09 | GM[20] | Down |
| 10 | P35900 | KRT20 | W | Cytoplasm | 48.49/5.52 | 48.15/5.54 | GM[19] | Down |
| 11 | P06733 | ENO1 | G | Cytoplasm | 47.17/7.01 | 46.80/7.57 | MALDI-TOF MS | Down |
| 12 | P12532 | Creatine kinase, ubiquitous mitochondrial precursor (CKMT1B) | C | Mitochondrion | 47.04/8.60 | 43.16/8.48 | GM[20] | Down |
| 13 | P22695 | Cytochrome b-c1 complex subunit 2, mitochondrial precursor (UQCRC2) | C | Mitochondrion | 48.44/8.74 | 44.10/8.83 | GM[21] | Up |
| 14 | P11177 | PDHB | C | Mitochondrion | 39.25/6.20 | 32.96/5.64 | MALDI-TOF MS | Up |
| 15 | P17707 | AMD1 | T | Cytoplasm | 38.34/5.71 | 31.91/5.74 | MALDI-TOF MS | Up |
| 16 | P00918 | Carbonic anhydrase II (CA2) | R | Cytoplasm | 29.25/6.87 | 30.75/7.69 | MALDI-TOF MS | Down |
| 17 | P60174 | Triosephosphate isomerase (TPI1) | G | Cytoplasm | 26.67/6.45 | 26.14/7.32 | MALDI-TOF MS | Up |
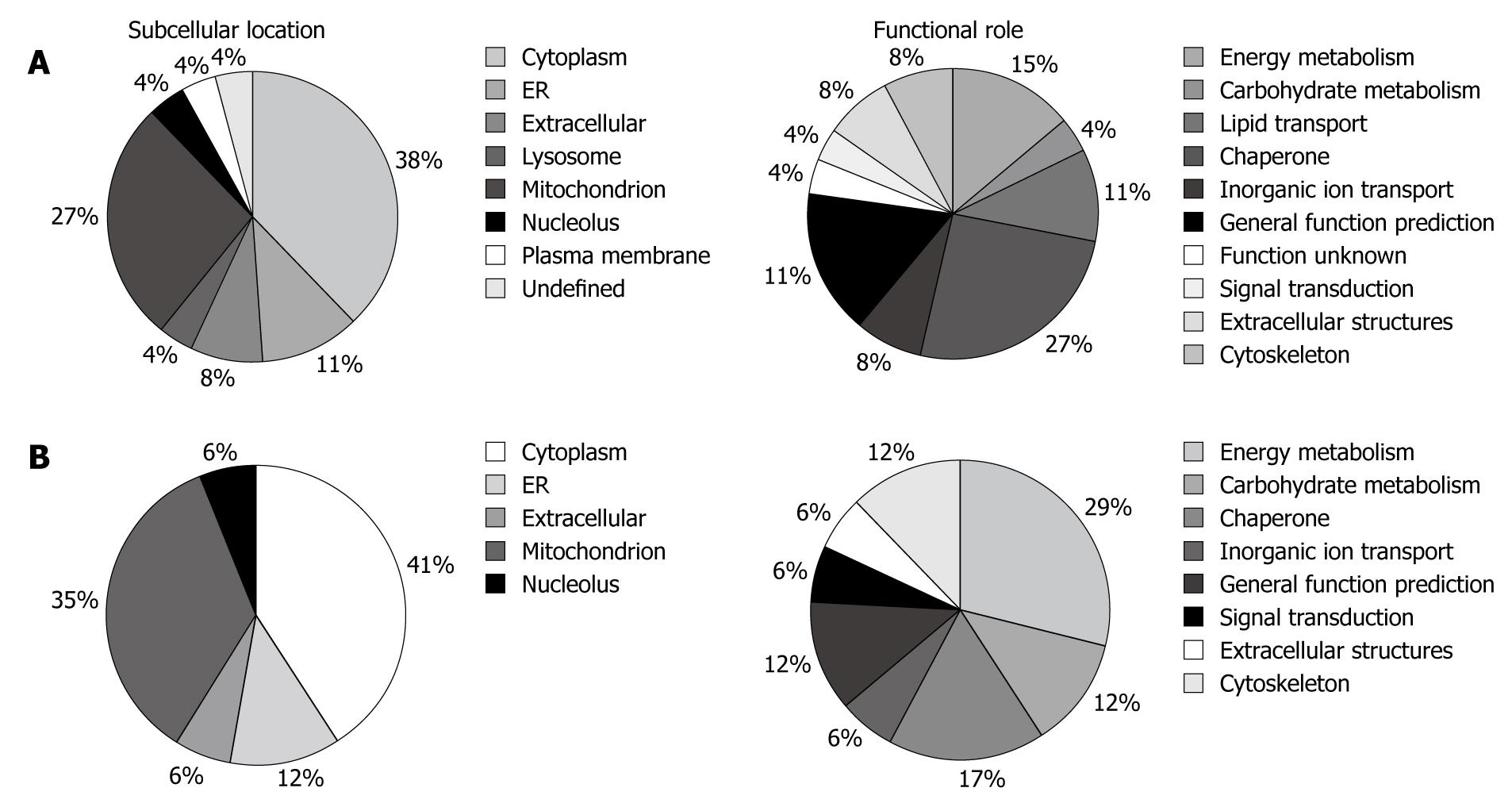
The altered proteins were classified in terms of their subcellular location and biological function by information from Swiss-Prot, HPRD (Human Protein Reference Database, http://www.humanproteinpedia.org), and COGs (Cluster of Orthologous Groups of proteins, http://www.ncbi.nlm.nih.gov/COG/) (Figure 2A). The majority of the identified proteins were located in the cytoplasm (38%), mitochondria (27%) and endoplasmic reticulum (11%). Twenty-seven percent of the altered proteins play a key role in post-translational modifications and protein turnover as chaperones, 15% are involved in energy production and conversion, and 11% are related to lipid transport and metabolism.
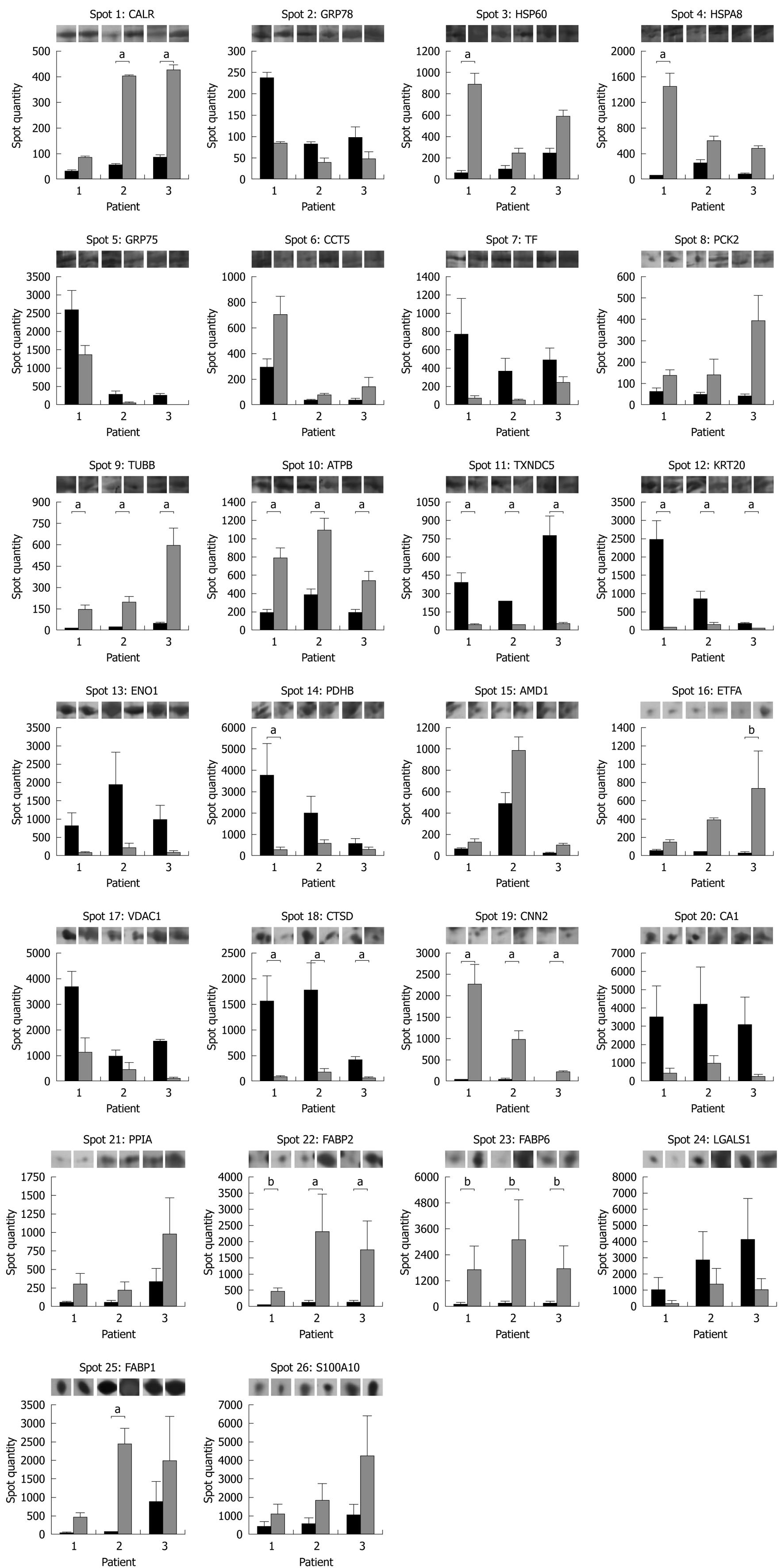
The results of a histogram data analysis carried out on the spot quantity values determined by PDQuest are displayed in Figure 3 together with representative gel images for each protein spot in each patient and clinical condition. A statistically significant increased expression in pouch remission was detected for tubulin β-2C chain (TUBB), ATP synthase subunit β (ATP5B) and calponin-2 (CNN2) in all patients, whereas calreticulin (CALR), 60 kDa heat shock protein (HSP60), heat shock cognate 71 kDa protein (HSPA8), and intestinal (FABP2) and liver fatty acid-binding proteins (FABP1) expression patterns showed an increase with statistical significance in only 1 or 2 out of the 3 patients enrolled. For ileal lipid binding protein (FABP6) and electron transfer flavoprotein subunit α (ETFA), P values of 0.07 and 0.06, respectively, near the threshold of significance were obtained. Among downregulated protein spots after antibiotic treatment, statistical significance was achieved in all patients for thioredoxin domain-containing protein 5 (TXNDC5), type I cytoskeletal keratin 20 (KRT20) and cathepsin D (CTSD). Pyruvate dehydrogenase E1 component subunit β (PDHB) showed a statistically significant decreased expression in only 1 patient.
Representative 2D gels obtained from mucosal biopsies in normal pouch and after probiotic therapy are shown in Figure 1B, confirming the protein maps reported in Figure 1A in terms of number, Mr and pI of the spots.
For each of the 3 subjects enrolled, the comparison of the 2D patterns of non-inflamed mucosal biopsies before and after VSL#3 administration was performed by PDQuest as reported above. Seventeen spots, which represented 75% of total proteins recognized as differentially expressed, were identified, of which 7 were upregulated and 10 were downregulated in the probiotic-treated pouch (Figure 1B and Table 1). In addition, it was possible to identify 6 protein spots that showed a similar expression pattern in all 2D gels (Figure 1 and Table 2).
Pie charts representing the subcellular location and the functional distribution of the probiotic administration-altered proteins are reported in Figure 2B. The majority of the identified proteins were in the cytoplasm (41%), mitochondria (35%) and endoplasmic reticulum (12%). The functional classification indicated that 29% play a key role in energy production and conversion, 17% are related to post-translational modifications and protein turnover as chaperones and 12% are involved in carbohydrate transport and metabolism.
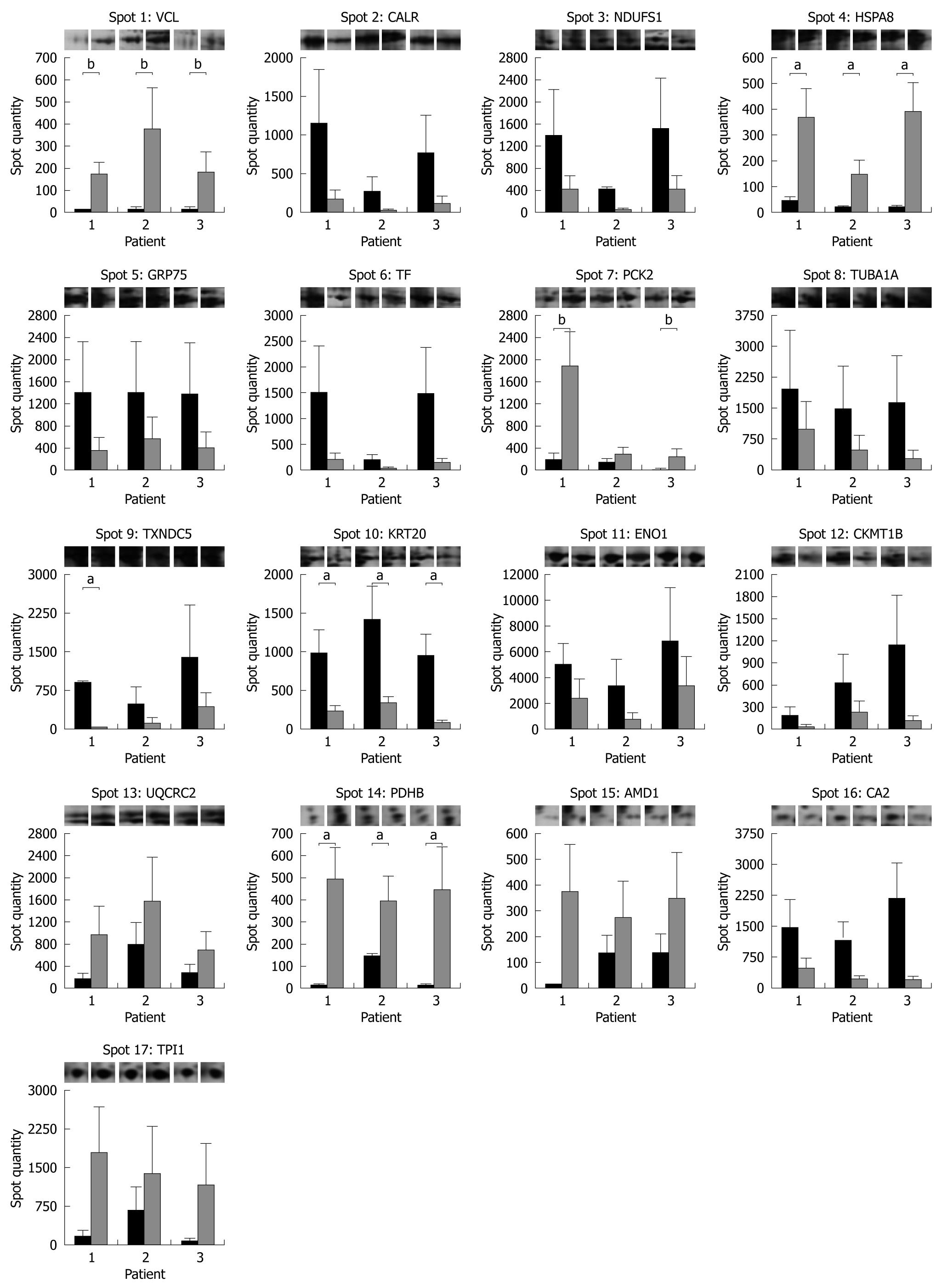
The spot quantity values determined by PDQuest are shown in the form of a histogram in Figure 4 together with representative gel images for each protein spot in each subject and condition. A statistically significant increased expression after 3 mo of probiotic administration was detected for HSPA8 and PDHB in all the subjects enrolled. P values of 0.06, near the threshold of significance, were obtained for vinculin (VCL) and phosphoenolpyruvate carboxykinase (PCK2) in 3 and 2 patients, respectively. Among protein spots with downregulated expression levels after VSL#3 therapy, statistical significance was achieved in all patients for KRT20 and in only 1 for TXNDC5.
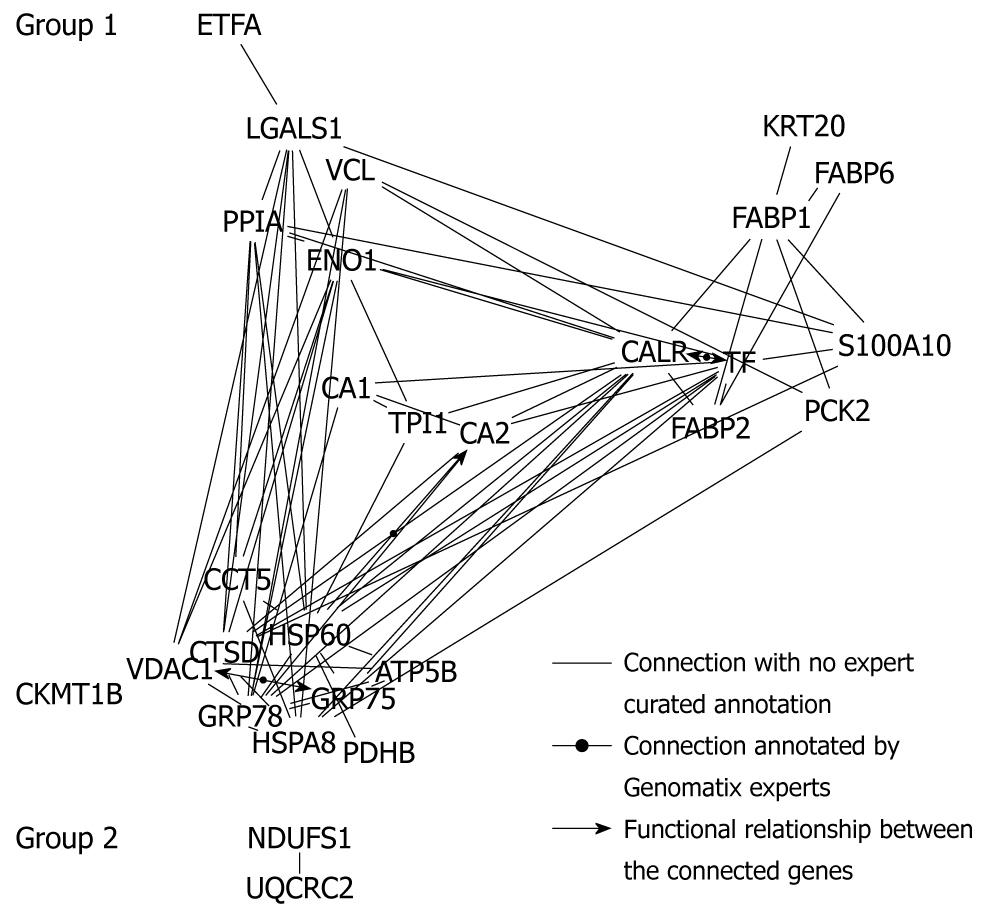
On the basis of literature co-citation from NCBI PubMed, a protein-protein network tree using the data-mining program Bibliosphere software was generated. As shown in Figure 5, the network tree was compiled of 28 different proteins forming 2 network clusters. Group 1 consisted of 26 highly interrelated proteins including ATP5B, carbonic anhydrase I (CA1) and II (CA2), creatine kinase (CKMT1B), α-enolase (ENO1), PCK2, PDHB and triosephosphate isomerase (TPI1), associated with energy, carbohydrate and amino acid metabolism, as well as glycolysis/gluconeogenesis, oxidative phosphorylation and electron transport chain. The second group was formed by 2 linear co-cited proteins, NADH-ubiquinone oxidoreductase 75 kDa subunit (NDUFS1) and cytochrome b-c1 complex subunit 2 (UQCRC2), related to energy production and conversion. The residual 5 detected proteins, S-adenosylmethionine decarboxylase (AMD1), CNN2, tubulin α-1A chain (TUBA1A), TUBB and TXNDC5 were completely disconnected from the network tree.
In this study, we provided for the first time 2D protein maps of mucosal biopsy samples collected during pouch endoscopy in patients who underwent IPAA.
The comparison between mucosal biopsy proteomes in pouchitis and in antibiotic-induced remission enabled the identification of 26 different proteins with at least 2-fold changes in their expression levels. Statistical significance was achieved for ATP5B, CNN2, CTSD, KRT20, TUBB and TXNDC5. In addition, a statistically significant altered expression pattern was obtained for CALR, HSP60, HSPA8, FABP1, FABP2 and PDHB in 1 or 2 of the 3 patients enrolled.
Among the identified mitochondrial proteins, ATP5B, ETFA and PCK2 directly participate in the process of energy production. The decrease of their expression levels in the inflamed pouch suggests the decline of mitochondrial function with pouchitis onset. This assumption is consistent with a previous hypothesis that chronic intestinal inflammation represents an energy-deficiency disease with alterations in the oxidative metabolism of the epithelial cells[23]. Moreover, the low expression of FABP1, FABP2 and FABP6, involved in enhancing the uptake of fatty acids into cells and facilitating their transport to intracellular organelles, could reinforce the speculation that pouchitis-diseased enterocytes do not perform β-oxidation/oxidative phosphorylation owing to a lack of normal supply of fatty acids[24]. Combined with these results, the overexpression of ENO1 found in the inflamed pouch may reflect a shift toward anaerobic glycolysis to overcome the decreased ATP formation by a dysfunctional oxidative phosphorylation[21].
The hypothesis of cellular stress and hypoxic conditions in chronically inflamed tissues is supported by the induction of several chaperone proteins, including 75 (GRP75) and 78 kDa glucose-related proteins (GRP78), TXNDC5, voltage-dependent anion-selective channel protein 1 (VDAC1), CTSD and peptidyl-prolyl cis-trans isomerase A (PPIA)[25-27]. In addition, we detected a statistically significant altered expression pattern for TUBB, KRT20 and CNN2, suggesting changes in cytoskeletal architecture with potential alterations in signal transduction and cellular transcription profiles[27,28].
Next, we compared mucosal biopsy proteomes in the normal pouch before and after probiotic administration and we identified 17 different proteins with significant changes in their expression levels. Interestingly, 8 of the differentially expressed proteins exhibited the same pattern of deregulation as in the pouchitis/pouch remission group. Indeed, both antibiotic and probiotic therapy resulted in downregulation of GRP75, serotransferrin (TF), TXNDC5, KRT20, ENO1 and in upregulation of HSPA8, PCK2 and AMD1, suggesting profound structural and metabolic alterations in enterocytes. In particular, TXNDC5 is a newly identified member of the thioredoxin family of endoplasmic reticulum proteins[29], and it has been proposed as a promising biomarker for cancer diagnosis[30]. Because of its important role in redox regulation[31], the altered expression profile of TXNDC5 in IPAA may be related to the increased oxidative stress with significantly lower plasma concentrations of lipophilic antioxidants and higher free radical activity measured in patients with restorative proctocolectomy compared to normal subjects[32]. Furthermore, for KRT20 and ENO1, widely applied as diagnostic markers for colon adenocarcinomas and many other tumors[33,34], as well as for PCK2 and AMD1 a differential protein profile in inflammatory bowel disease has been already reported[21,35,36].
In addition to these results, in the VSL#3-treated pouch we found a statistically significant upregulation of VCL and an altered expression pattern for TUBA1A, supporting the assumption of a positive modulation exerted by probiotics at cytoskeleton level for cell morphology and integrity[37,38]. In addition, the dysregulated expression levels of NDUFS1, CKMT1B, UQCRC2, PDHB, CA2 and TPI1, directly involved in energy metabolism, strengthen the hypothesis of significant changes in the metabolic profiles of the host associated with probiotic administration[39,40]. Nonetheless, although the manipulation of the ubiquitin/proteasome pathway and the ability to intervene with the complex host system of detoxification of potentially harmful xenobiotics and endobiotic compounds may account for some of the cytoprotective effects of probiotics[37,41,42], we did not find any significant change in glutathione S-transferase P (GSTP1) and ubiquitin-conjugating enzyme E2 N (UBE2N) protein expression levels.
The bibliometric data analysis including all the 33 differentially regulated proteins from the pouchitis/pouch remission and non-inflamed/probiotic-treated pouch group comparison generated a complex network with 26 highly interrelated proteins. As expected, the majority of clustered proteins were associated with glycolysis/gluconeogenesis, oxidative phosphorylation and electron transfer chain pathways.
In conclusion, the identified proteins, both upregulated and downregulated, may be involved in pouchitis pathophysiology and participate in disease onset or in maintenance of the non-inflamed pouch.
Restorative proctocolectomy with ileal pouch-anal anastomosis (IPAA) is the procedure of choice for complicated ulcerative colitis. In the long-term, up to 50% of patients develop pouchitis, an idiopathic inflammatory disease of the ileal reservoir. The management of pouchitis is largely empirical and only few small placebo-controlled clinical trials have been conducted. Although antibiotics represent the mainstay of treatment, probiotics have recently gained more attention as an effective therapeutic option for pouchitis management.
The etiology and pathophysiology of pouchitis are still not entirely clear but the bulk of the evidence points towards an abnormal mucosal immune response to altered microbiota patterns. By investigating the dynamic nature of protein expression, cellular and subcellular distribution, posttranslational modifications and protein-protein interaction networks, proteomic technologies could play a major role in unraveling the mystery of immunopathogenic mechanisms of pouchitis and in discovering novel biomarkers for disease activity, diagnosis and prognosis.
The current study is the first proteomic study to be reported in IPAA research. The authors provided the 2D protein maps of mucosal biopsy samples collected during pouch endoscopy in patients with chronic refractory pouchitis. The changes in the protein expression profiles following antibiotic or probiotic treatment were characterized.
The identified proteins, upregulated or downregulated following antibiotic/probiotic treatment, may be involved in pouchitis pathophysiology and participate in disease onset or in maintenance of the non-inflamed pouch. Future work will be focused in validating the list of proteins identified in larger patient cohorts.
The results are well described and interesting, although the number of patients is a bit on the small side. Even though this manuscript does not give a clear understanding to the mechanistic differences, the results may aid other scientists in making a follow-up study.
Peer reviewer: Dr. Sara K Lindén, Professor, Mucosal Immunobiology and Vaccine Center, Gothenburg University, Box 435, Göteborg, 405 30, Sweden
S- Editor Tian L L- Editor Cant MR E- Editor Zheng XM
| 1. | Baumgart DC, Sandborn WJ. Inflammatory bowel disease: clinical aspects and established and evolving therapies. Lancet. 2007;369:1641-1657. |
| 2. | McLaughlin SD, Clark SK, Tekkis PP, Ciclitira PJ, Nicholls RJ. Review article: restorative proctocolectomy, indications, management of complications and follow-up--a guide for gastroenterologists. Aliment Pharmacol Ther. 2008;27:895-909. |
| 3. | Madden MV, Farthing MJ, Nicholls RJ. Inflammation in ileal reservoirs: 'pouchitis'. Gut. 1990;31:247-249. |
| 4. | Shen B, Achkar JP, Lashner BA, Ormsby AH, Remzi FH, Bevins CL, Brzezinski A, Petras RE, Fazio VW. Endoscopic and histologic evaluation together with symptom assessment are required to diagnose pouchitis. Gastroenterology. 2001;121:261-267. |
| 5. | Sandborn WJ, Tremaine WJ, Batts KP, Pemberton JH, Phillips SF. Pouchitis after ileal pouch-anal anastomosis: a Pouchitis Disease Activity Index. Mayo Clin Proc. 1994;69:409-415. |
| 7. | Sandborn WJ. Pouchitis: definition, risk factors, frequency, natural history, classification, and public perspectives. Trends in Inflammatory Bowel Disease 1996. Lancaster: Kluwer Academic Publishers 1997; 51-63. |
| 8. | Maser EA, Present DH. Pouch-ouch. Curr Opin Gastroenterol. 2008;24:70-74. |
| 9. | Gionchetti P, Rizzello F, Venturi A, Brigidi P, Matteuzzi D, Bazzocchi G, Poggioli G, Miglioli M, Campieri M. Oral bacteriotherapy as maintenance treatment in patients with chronic pouchitis: a double-blind, placebo-controlled trial. Gastroenterology. 2000;119:305-309. |
| 10. | Mimura T, Rizzello F, Helwig U, Poggioli G, Schreiber S, Talbot IC, Nicholls RJ, Gionchetti P, Campieri M, Kamm MA. Once daily high dose probiotic therapy (VSL#3) for maintaining remission in recurrent or refractory pouchitis. Gut. 2004;53:108-114. |
| 11. | Gionchetti P, Rizzello F, Helwig U, Venturi A, Lammers KM, Brigidi P, Vitali B, Poggioli G, Miglioli M, Campieri M. Prophylaxis of pouchitis onset with probiotic therapy: a double-blind, placebo-controlled trial. Gastroenterology. 2003;124:1202-1209. |
| 12. | Gosselink MP, Schouten WR, van Lieshout LM, Hop WC, Laman JD, Ruseler-van Embden JG. Delay of the first onset of pouchitis by oral intake of the probiotic strain Lactobacillus rhamnosus GG. Dis Colon Rectum. 2004;47:876-884. |
| 13. | Alex P, Gucek M, Li X. Applications of proteomics in the study of inflammatory bowel diseases: Current status and future directions with available technologies. Inflamm Bowel Dis. 2009;15:616-629. |
| 14. | Vitali B, Wasinger V, Brigidi P, Guilhaus M. A proteomic view of Bifidobacterium infantis generated by multi-dimensional chromatography coupled with tandem mass spectrometry. Proteomics. 2005;5:1859-1867. |
| 15. | Görg A, Postel W, Günther S. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis. 1988;9:531-546. |
| 16. | Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850-858. |
| 17. | Melis R, White R. Characterization of colonic polyps by two-dimensional gel electrophoresis. Electrophoresis. 1999;20:1055-1064. |
| 18. | Mori Y, Kondo T, Yamada T, Tsuchida A, Aoki T, Hirohashi S. Two-dimensional electrophoresis database of fluorescence-labeled proteins of colon cancer cells. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;823:82-97. |
| 19. | Lenaerts K, Bouwman FG, Lamers WH, Renes J, Mariman EC. Comparative proteomic analysis of cell lines and scrapings of the human intestinal epithelium. BMC Genomics. 2007;8:91. |
| 20. | Li M, Xiao ZQ, Chen ZC, Li JL, Li C, Zhang PF, Li MY. Proteomic analysis of the aging-related proteins in human normal colon epithelial tissue. J Biochem Mol Biol. 2007;40:72-81. |
| 21. | Shkoda A, Werner T, Daniel H, Gunckel M, Rogler G, Haller D. Differential protein expression profile in the intestinal epithelium from patients with inflammatory bowel disease. J Proteome Res. 2007;6:1114-1125. |
| 22. | Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856-2860. |
| 23. | Roediger WE. The colonic epithelium in ulcerative colitis: an energy-deficiency disease? Lancet. 1980;2:712-715. |
| 24. | Esteve M, Navarro E, Klaassen J, Abad-Lacruz A, González-Huix F, Cabré E, Ramos E, Condom E, Fernández-Bañares F, Pastor C. Plasma and mucosal fatty acid pattern in colectomized ulcerative colitis patients. Dig Dis Sci. 1998;43:1071-1078. |
| 25. | Abu-Hamad S, Sivan S, Shoshan-Barmatz V. The expression level of the voltage-dependent anion channel controls life and death of the cell. Proc Natl Acad Sci USA. 2006;103:5787-5792. |
| 26. | Menzel K, Hausmann M, Obermeier F, Schreiter K, Dunger N, Bataille F, Falk W, Scholmerich J, Herfarth H, Rogler G. Cathepsins B, L and D in inflammatory bowel disease macrophages and potential therapeutic effects of cathepsin inhibition in vivo. Clin Exp Immunol. 2006;146:169-180. |
| 27. | Werner T, Haller D. Intestinal epithelial cell signalling and chronic inflammation: From the proteome to specific molecular mechanisms. Mutat Res. 2007;622:42-57. |
| 28. | Keshavarzian A, Banan A, Farhadi A, Komanduri S, Mutlu E, Zhang Y, Fields JZ. Increases in free radicals and cytoskeletal protein oxidation and nitration in the colon of patients with inflammatory bowel disease. Gut. 2003;52:720-728. |
| 29. | Knoblach B, Keller BO, Groenendyk J, Aldred S, Zheng J, Lemire BD, Li L, Michalak M. ERp19 and ERp46, new members of the thioredoxin family of endoplasmic reticulum proteins. Mol Cell Proteomics. 2003;2:1104-1119. |
| 30. | Wang Y, Ma Y, Lü B, Xu E, Huang Q, Lai M. Differential expression of mimecan and thioredoxin domain-containing protein 5 in colorectal adenoma and cancer: a proteomic study. Exp Biol Med (Maywood). 2007;232:1152-1159. |
| 31. | Edman JC, Ellis L, Blacher RW, Roth RA, Rutter WJ. Sequence of protein disulphide isomerase and implications of its relationship to thioredoxin. Nature. 1985;317:267-270. |
| 32. | El Muhtaseb MS, Talwar D, Duncan A, St J O'reilly D, McKee RF, Anderson JH, Foulis A, Finlay IG. Free radical activity and lipid soluble anti-oxidant vitamin status in patients with long-term ileal pouch-anal anastomosis. Colorectal Dis. 2009;11:67-72. |
| 33. | Durany N, Joseph J, Campo E, Molina R, Carreras J. Phosphoglycerate mutase, 2,3-bisphosphoglycerate phosphatase and enolase activity and isoenzymes in lung, colon and liver carcinomas. Br J Cancer. 1997;75:969-977. |
| 34. | Stenling R, Lindberg J, Rutegård J, Palmqvist R. Altered expression of CK7 and CK20 in preneoplastic and neoplastic lesions in ulcerative colitis. APMIS. 2007;115:1219-1226. |
| 35. | Obayashi M, Matsui-Yuasa I, Matsumoto T, Kitano A, Kobayashi K, Otani S. Polyamine metabolism in colonic mucosa from patients with ulcerative colitis. Am J Gastroenterol. 1992;87:736-740. |
| 36. | Vermeulen N, Arijs I, Joossens S, Vermeire S, Clerens S, Van den Bergh K, Michiels G, Arckens L, Schuit F, Van Lommel L. Anti-alpha-enolase antibodies in patients with inflammatory bowel disease. Clin Chem. 2008;54:534-541. |
| 37. | Yang F, Wang J, Li X, Ying T, Qiao S, Li D, Wu G. 2-DE and MS analysis of interactions between Lactobacillus fermentum I5007 and intestinal epithelial cells. Electrophoresis. 2007;28:4330-4339. |
| 38. | Seth A, Yan F, Polk DB, Rao RK. Probiotics ameliorate the hydrogen peroxide-induced epithelial barrier disruption by a PKC- and MAP kinase-dependent mechanism. Am J Physiol Gastrointest Liver Physiol. 2008;294:G1060-G1069. |
| 39. | Nerstedt A, Nilsson EC, Ohlson K, Håkansson J, Thomas Svensson L, Löwenadler B, Svensson UK, Mahlapuu M. Administration of Lactobacillus evokes coordinated changes in the intestinal expression profile of genes regulating energy homeostasis and immune phenotype in mice. Br J Nutr. 2007;97:1117-1127. |
| 40. | Martin FP, Wang Y, Sprenger N, Yap IK, Lundstedt T, Lek P, Rezzi S, Ramadan Z, van Bladeren P, Fay LB. Probiotic modulation of symbiotic gut microbial-host metabolic interactions in a humanized microbiome mouse model. Mol Syst Biol. 2008;4:157. |