Published online Nov 21, 2007. doi: 10.3748/wjg.v13.i43.5731
Revised: August 23, 2007
Accepted: October 8, 2007
Published online: November 21, 2007
AIM: To establish the role of enteric glial cells during infection with Mycobacterium avium subspecies paratuberculosis (MAP) in Crohn’s disease.
METHODS: In order to establish the role of enteric glial cells during infection with M. avium subspecies paratuberculosis (MAP) in Crohn’s disease, Map adhesion experiments on enteric glial cells were performed as well as expression analysis of Map sigma factors during infection.
RESULTS: In this study, for the first time, we found a high affinity of MAP to enteric glial cells and we analyzed the expression of MAP sigma factors under different conditions of growth.
CONCLUSION: The fact that Map showed a high affinity to the glial cells raises concerns about the complicated etiology of the Crohn’s disease. Elucidation of the mechanisms whereby inflammation alters enteric neural control of gut functions may lead to novel treatments for Crohn’s disease.
-
Citation: Sechi LA, Ruehl A, Ahmed N, Usai D, Paccagnini D, Felis GE, Zanetti S.
Mycobacterium avium subspeciesparatuberculosis infects and multiplies in enteric glial cells. World J Gastroenterol 2007; 13(43): 5731-5735 - URL: https://www.wjgnet.com/1007-9327/full/v13/i43/5731.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i43.5731
Several cell lines of the gut have been extensively studied such as intestinal epithelial cells, immune cells, smooth muscle cells and enteric neurons[1-4]. However, very little is known about enteric glial cells which belong to the enteric nervous system (ENS) along with the neurons[5]. The ENS, comprised of several plexuses, is located alongside the intestinal wall. The enteric glia are localised within the mucosal plexuses and the glial processes make close contact with the epithelial cell layers. The epithelial crypt bases, in particular, are surrounded by a dense network of glial cells[6]. These glial cells are small, star shaped with different processes of various length and shape[6]; they are morphologically and immunohistochemically different from microglia of the central nervous system and from all other peripheral glia and appear more closely related to astrocytes although functionally different[6]. Evidence for functional heterogeneicity of enteric glia has recently been described[7]. The enteric glial cells (EGC) could participate in neurotransmitter synthesis/inactivaction or in synaptic transmission. Moreover, they may also interact with intestinal capillaries to modulate endothelial permeability[8].
It has been shown that the enteric nervous system regulates intestinal barrier functions by regulating the zonula occludens-tight junctions[9]. Regarding EGC, it has been shown in a mouse model that EGC ablation results in fulminant intestinal inflammation due to disregulation and rupture of the epithelial intestinal barrier[8,10]. It has been suggested that there is a direct interaction between EGC (releasing soluble factors) and epithelial cells to enhance the intestinal epithelial cells[5]. Moreover they could have a role in the prevention of epithelial barrier disorganization and hyperproliferation during bacterial infection, inflammatory processes and neoplasia[11-13]. All these findings indicate that glial cells may be an important component of the intestinal mucosal defense system.
On the other hand, Mycobacterium avium subspecies paratuberculosis is the causative agent of Johne’s disease, a chronic and incurable disease affecting ruminants and other animals[14]. Moreover, there is increasing evidence of its involvement in the enteric granulomatous syndrome of humans called Crohn’s disease[15-17]. For all these reasons we analyzed the interactions between EGC and M. paratuberculosis in vitro.
Sigma factors are part of the transcriptional regulators family and are responsible for binding to the RNA polymerase complex (composed of four distinct subunits); to recognize promoters and separate DNA strands[18]. In fact, every sigma factor has its own specificity, allowing the initiation of transcription of different subset of genes[18,19]. Some sigma factors (sigD, sigE, sigC, sigH and sigL) play a role in the virulence of M. tuberculosis[20,21]. The number and abundance of sigma factors reflect the ability of the bacterium to cope with various environmental conditions, stresses and insults[18,22].
In this study the expression of all 19 sigma factors of M. paratuberculosis were also tested during EGC infection.
M. paratuberculosis ATCC 43015, of human source was obtained from The RIVM, Bilthoven, The Netherlands. M. paratuberculosis for DNA extraction was grown in Mycobacteria Growth Indicator Tube (MGITl) medium supplemented with Mycobactin J and egg yolk. Murine enteric glial cells were previously isolated and characterized by Dr. Anne Ruhel[23].
Identification of individual sigma factors in M. avium subspecies. paratuberculosis was obtained by text annotation searches and by BlastP similarity searches using M. smegmatis and M. tuberculosis H37Rv Sigma ORF as queries. Primers used in real time PCR are described in Table 1.
| Target | Primer name | 5'-3' sequence | Position1 |
| 16S | MAP16Sfor | ATCATGCCCCTTATGTCCAG | 1179-1198 |
| MAP16Srev | TGAGACCGGCTTTAAAAGGA | 1259-1278 | |
| sigA | sigAfor | GTACGCCACCCAGCTGATGTCG | 714-735 |
| sigArev | CGTCGCGGCAGATCCACAT | 787-805 | |
| sigB | sigBfor | GACCTGCTCGAGCACAGC | 646-663 |
| sigBrev | CAGCACGCTGCGGATGTCGGTG | 783-804 | |
| sigC | sigCfor | ACATCCGTCACCTGCAGTC | 1088-1106 |
| sigCrev | GTCACCTCGACCAGATCCTC | 1177-1195 | |
| sigD | sigDfor | CTTCCTGGCTTTCCTGTACG | 249-268 |
| sigDrev | GATGGACTCGGTCGGGTAG | 324-242 | |
| sigE | sigEfor | CACCCAGGAGACCTTCATCC | 327-346 |
| sigErev | GACCATGTCCAGGAACAG | 418-435 | |
| sigF | sigFfor | GGCAGCTCCTACAACACCTT | 493-512 |
| sigFrev | ACTCCTGGTCCTCGATCCGG | 597-616 | |
| sigF-like | sigF-likefor | ATGACCAACGCAATCGCTCC | 1-20 |
| sigF-likerev | GGCATCCGGCGCAGTTCCA | 88-107 | |
| sigG | sigGfor | GCGTTCGAAAGCTACGACAT | 622-641 |
| sigGrev | CTGATACCACCCGGTGTACG | 692-711 | |
| sigH | sigHfor | AATCTCAAGGCGTGGCTCTA | 385-404 |
| sigHrev | TGATTTCCTCGGTCGGATAC | 465-484 | |
| sigI | sigIfor | GGGCGACATCGACGACGTGC | 150-169 |
| sigIrev | GACATCGCCCGGACGTTCGTG | 235-255 | |
| sigJ | sigJfor | GCATCTACACGGCGGGCCTG | 740-759 |
| sigJrev | GGCGAACCGGTGAACTTGT | 857-875 | |
| sigL | sigLfor | CGTGATCGAACGGTCCTACT | 399-418 |
| sigLrev | CCGCACCGCATAGTGTAGT | 491-504 | |
| sigM | sigMfor | TGGCTGCACCGCATCGTG | 238-255 |
| sigMrev | TCGGCGACCGGATAGTAGTCT | 315-335 | |
| Other ECF-1 | sigECF1for | GTTCTCCGCCGAGTCGATTT | 310-328 |
| sigECF1rev | GTCGAATCCGAACACCTCAC | 492-411 | |
| Other ECF-2 | sigECF2for | GCATCCACACGATCGACAT | 839-857 |
| sigECF2rev | GGTTGTGATGTTCCTGAACC | 914-933 | |
| Other ECF-3 | sigECF3for | GTCGGTCATGGGTTTCGT | 780-797 |
| sigECF3rev | GCACCCAGCTCCAGTTTC | 855-873 | |
| Other ECF-4 | sigECF4for | GATCTCGTCGGCATCTCG | 502-519 |
| sigECF4rev | TCCAATTCGTTTCGGAGATT | 592-611 | |
| Other ECF-5 | sigECF5for | GCAATTGACCCGTTCACC | 945-962 |
| sigECF5rev | CTCCTCCAAAGCGGCTAAG | 1020-1038 | |
| Other ECF-6 | sigECF6for | TGCAAGGTAATTCGATCAAGG | 419-439 |
| sigECF6rev | TCCCTCGTTGACCTGTGC | 511-528 |
DNA extraction was performed using the Ribolyser system (HYBAID, USA) and purified as previously described[17].
Oligonucleotide sequences and position are reported in Table 1. Specificity of primer pairs was ascertained with PCR with the following conditions: MgCl2 1.5 mmol/L, deossi Nucleotide Triphosphate (dNTPS) 150 μmol/L, primers 0.2 μmol/L each, Taq 0.025 U/μL reaction volume. After an initial denaturation of 3 min at 94°C, 30 cycles of 1 min at 94°C, 1 min at 58°C, 1 min at 72°C and a final extension at 72°C for 5 min were performed. Only the amplification products of the expected length were obtained.
RNA was extracted using the Ribolyser instrument[17]. RNA was extracted from MAP cells after different conditions: (1) growth in 7H9 medium plus Mycobactin J, (2) after addition of lysozyme at 24 h and 7 d; (3) after infection of EGC line, after 6 h, 24 h, 48 h and 7 d. RNA was quantified for further experiments and samples were stored at -80°C until further use. cDNA was prepared by an initial incubation at 65°C for 10 min of the reaction mixture containing 100 μmol/L oligo-dT and 10 μL of extracted RNA; this first step was followed by a second incubation of 1 h at 42°C of the mixture containing 50 U/reaction of reverse transcriptase (Mooloney-Murine Leukemia Virus M-MLV 200 U/μL), 0.1 mol/L buffer 5 × Tris HCl pH 8.3 (Life Technologies) with 150 mmol/L KCl, 7 mmol/L MgCl2 and 20 mmol/L dithiothreitol (DTT); 0.1 mol/L DTT, 10 mmol/L dNTPs, 40 U/μL RNase inhibitor.
Real Time PCR was performed using the iCycler Detection System (Biorad) with the SyberGreen assay (Applied Biosystems). Gene expression of all the Sigma factors (except Extracytoplasmic function 3 ECF3) was determined using quantitative Real-Time PCR by comparing the fluorescence produced by test samples to that of the 16S rRNA gene expression (house-keeping gene). Five micro liters of cDNA was subjected to PCR (1 cycle at 94°C for 1 min; 50 cycles at 94°C for 30 s and at 60°C for 40 s) with primers that amplified 16S rRNA, all 19 sigma factor genes. PCR was carried out with the following reaction-mixture in a total volume of 30 μL: 1 × of 10 × buffer, MgCl2 1.5 mmol/L, dNTPs 0.2 mmol/L, primers 0.5 μmol/L each, DNA polymerase 0.5 U/sample. PCR was performed in a 96 well plate, for each sample 2 wells were utilized to guarantee uniformity in results. The quantitative analysis of the data obtained was performed by Biorad's method of relative quantitation of gene expression (Bio-Rad Laboratories, USA).
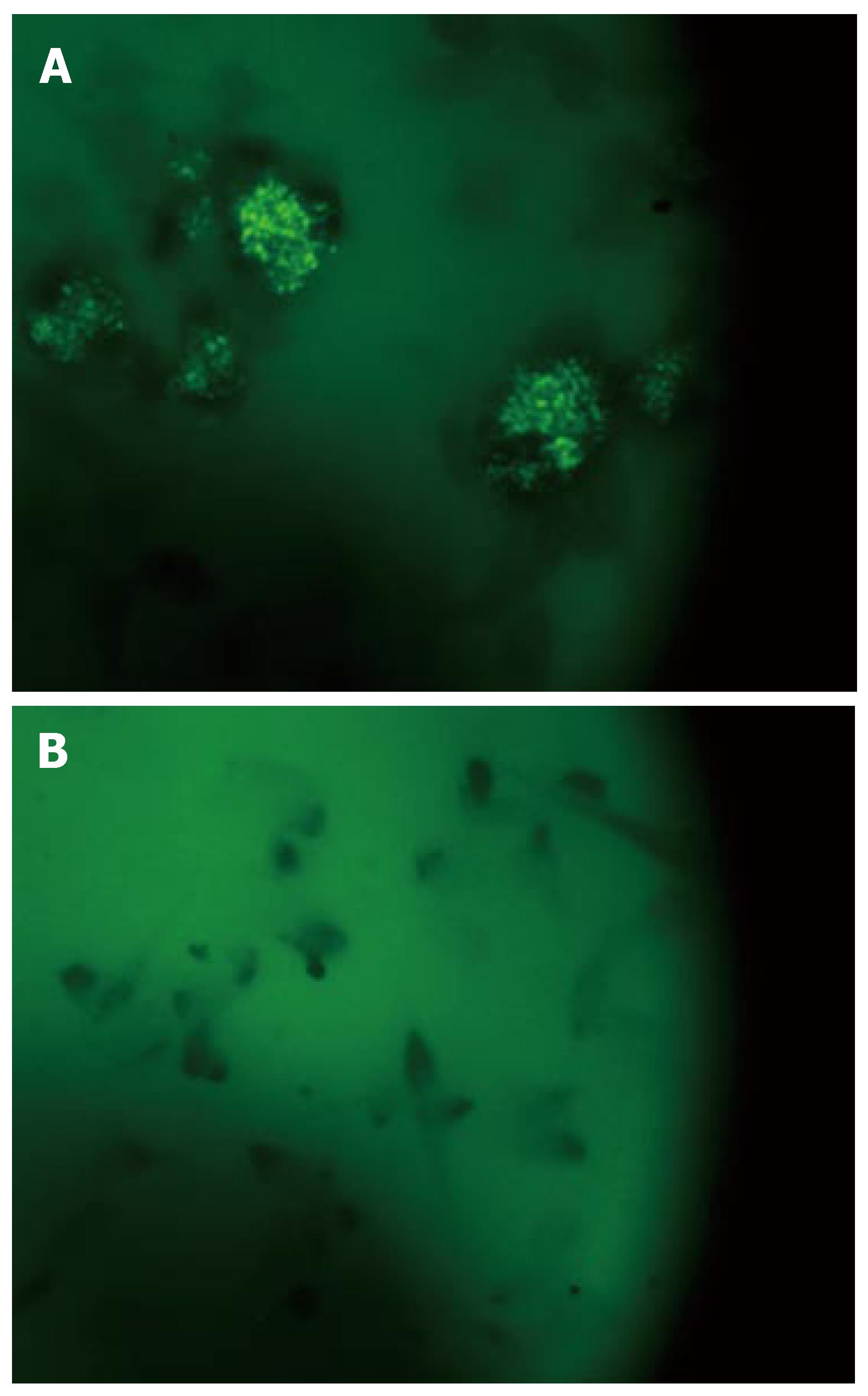
Glial Enteric cell lines were cultivated in 24 well plastic dishes. The EGCs were cultivated in Dulbecco's modified Eagle medium supplemented with 15% fetal bovine serum and 4 mmol of L-glutamine per liter. All tissue culture reagents were obtained from Sigma (Sigma Chemical Co.). Cells were seeded at 2 × 104 cells per well and incubated at 37°C in 5% CO2. EGC cells were used when they were semiconfluent. MAP grown over night in 7H9 broth (with mycobactin J) was diluted in cell medium. One milliliter containing 106 bacteria was added to each well containing EGCs (cells were infected at a ratio of bacteria/cells of 10:1). After two hours infected EGCs were washed with cell medium containing Kanamycin 100 μg/mL and incubated with the same medium. Cells were stained with auramine rodhamine at the different incubation times (6 h, 48 h and 7 d at 37°C) to visualize MAP, and counted as previously described[17].
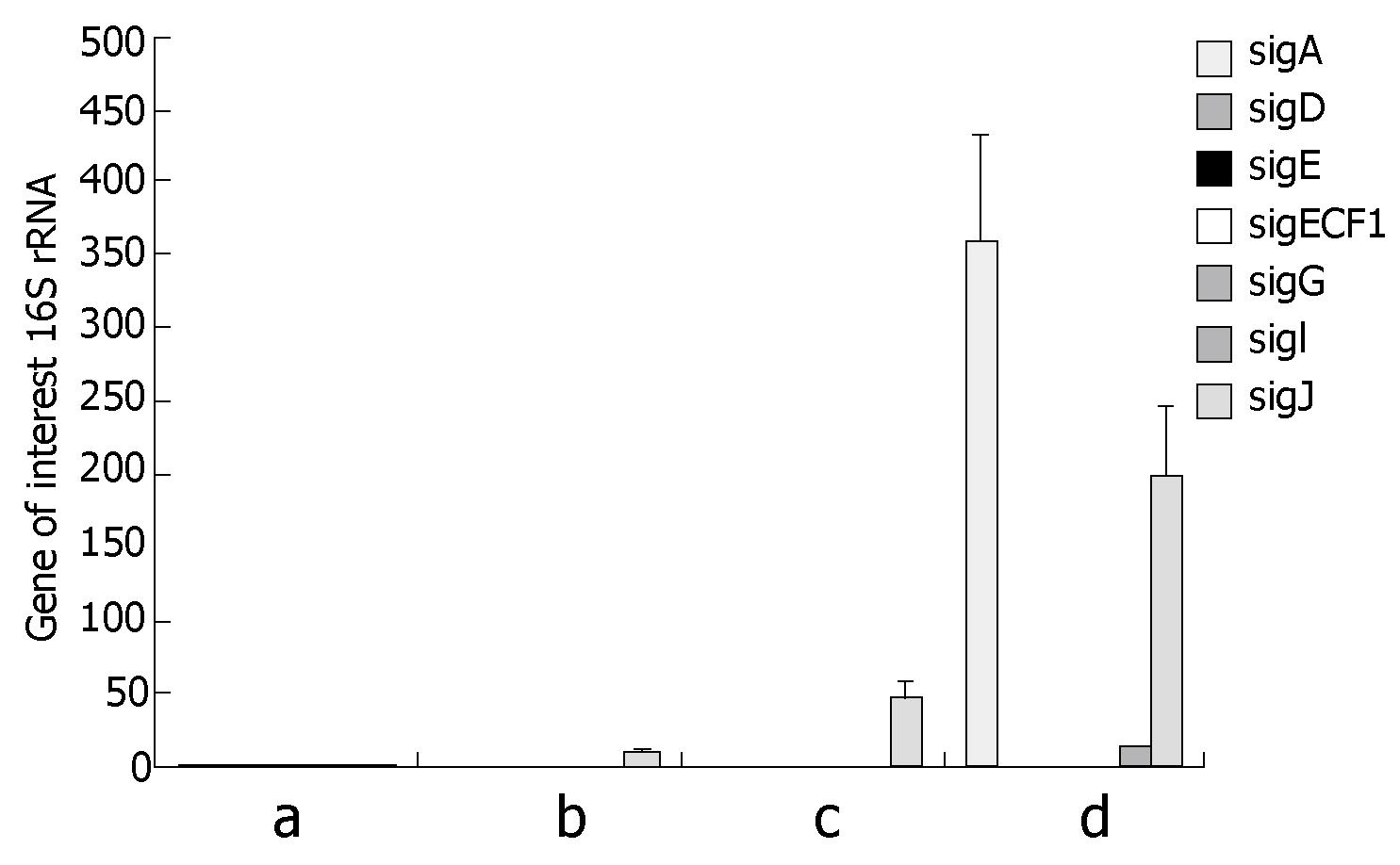
7H9 broth plus mycobactin J: MAP during growth in broth 7H9 supplemented with Mycobactine J, expressed several sigma factors as expected (Figure 1A). SigA, Sig D, SigE, Sig G and SigIwere among the most expressed ones as compared to the 16S rRNA expression taken as house keeping gene whereas ECF1 and sig J were expressed in lower quantity (Figure 1A).
Infection of EGC: The expression of the 19 sigma factors of MAP was observed and quantified after growth infection of EGC at 6 h, 48 h and 7 d (Figure 1B-D). Soon after infection, expression of all sigma factors shut down, except for sig J which was expressed in high quantity (Figure 1B). The expression of sig J increased after 48 h (Figure 1C) and 7 d. SigA was overexpressed after seven days (Figure 1D), at the same time an expression of sigIwas also detected. Figure 2 reveals an example of EGC infection by MAP where bacteria were visualized within the intestinal glial cells after staining with auramine rodamine as compared to control cells not infected.
Enteric glias are distinct from all other glial cell types[5,7]. No gastrointestinal disorder has been reported linked to glial defect yet, most probably because subtle changes in glial function might be involved in the etiopathogenesis of enteric disorders. Crohn’s disease with neuroinflammation and neurodegeneration components may be associated with EGC alteration[23-25]. Indeed EGC interact with enteric neurons, endothelial cells, immune cells and the intestinal epithelium; all these factors can contribute to the pathogenesis of Crohn’s disease. In our experiments, we found that MAP has a high affinity to EGCs. Experiments “in vitro” show a high adhesion and intracellular multiplication as confirmed by the active expression of sigA (the housekeeping sigma factor) after seven days of infection along with the expression of sigJ and sigI (expressed in different conditions of cellular stress).
The fact that this dangerous intestinal pathogen has a demonstrated affinity to the glial cells and that MAP has recently been reported in substantial percentages of Crohn’s patients[15-17] raises concerns about the complicated etiology of the Crohn's disease.
MAP expression of sigma factors in EGC is very similar to sigma factor expression after infection of the Caco2 intestinal epithelial cell line (manuscript in preparation) and shows how there is a rapid change in gene expression after cell infection.
EGCs are an active part of an intestinal network system essential for a healthy and functional gut[13,26,27]. The role of EGC in Crohn's and other enteric diseases is not well studied and this is the first report that attempts to unravel interaction between an intestinal pathogen and EGC. Future work is certainly needed to elucidate this complex interaction.
Enteric glia might play a role in regulating barrier functions in mucosal epithelia. Mycobacterium avium subspecies paratuberculosis (MAP) is the causative agent of Johne’s disease, a chronic and incurable disease affecting ruminants and other animals. Moreover, evidence of its involvement in Crohn’s disease is accumulating. MAP enter the host through Peyer’s patches and intestinal epithelium and are sampled by intestinal dendritic cells; they survives inside macrophages. Nothing is known about the role of enteric glial cells during MAP infection thus we performed this study to establish the role of enteric glial cells during infection with Mycobacterium avium subspecies paratuberculosis (MAP) in Crohn’s disease.
It is firmly established that intestinal inflammation is accompanied by functional and structural alterations of the Enteric Nervous System to which belong Enteric Glial Cells. In Crohn’s disease patients, a compromised glial network that responds poorly to inflammatory stimuli has been suggested. Our report will open a new frontier on the interaction of MAP and enteric glia cells supporting the idea that Crohn’s disease is associated with the invasion and persistence of a multi host pathogen as it is MAP.
This is the first research that highlights the interaction of a true enteric pathogen such as Map with Enteric glial cells. This study will open the research field in this matter.
Enteric glial cells may be the target of a series of cytokine modulators or drugs. Map will be recognized as a pathogen associated with intestinal disorders.
EGCs: enteric glial cells; Map: Mycobacterium avium subs. Paratuberculosis; ENS: Enteric nervous system; Johne’s disease: Enteric granulomatous infection of ruminants caused by Map; Crohn’s disease: Enteric granulomatous chronic inflammation in humans with ulcerative colitis (UC) forms inflammatory bowel disease (IBD).
The manuscript by Sechi et al describes the expression of MAP sigma factors during infection of enteric glial cells. The first findings that sigma factors are differentially expressed during infection are very promising.
S- Editor Zhu LH L- Editor Alpini GD E- Editor Yin DH
| 1. | Liévin-Le Moal V, Servin AL. The front line of enteric host defense against unwelcome intrusion of harmful microorganisms: mucins, antimicrobial peptides, and microbiota. Clin Microbiol Rev. 2006;19:315-337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 359] [Cited by in RCA: 370] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 2. | Niess JH, Reinecker HC. Dendritic cells in the recognition of intestinal microbiota. Cell Microbiol. 2006;8:558-564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 61] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 3. | Ozaki H, Hori M, Kinoshita K, Ohama T. Intestinal dysmotility in inflammatory bowel disease: mechanisms of the reduced activity of smooth muscle contraction. Inflammopharmacology. 2005;13:103-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 4. | Grundy D, Schemann M. Enteric nervous system. Curr Opin Gastroenterol. 2005;21:176-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 50] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 5. | Rühl A. Glial cells in the gut. Neurogastroenterol Motil. 2005;17:777-790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 179] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 6. | Rühl A, Nasser Y, Sharkey KA. Enteric glia. Neurogastroenterol Motil. 2004;16 Suppl 1:44-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 102] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 7. | Rühl A. Glial regulation of neuronal plasticity in the gut: implications for clinicians. Gut. 2006;55:600-602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 8. | Neunlist M, Aubert P, Bonnaud S, Van Landeghem L, Coron E, Wedel T, Naveilhan P, Ruhl A, Lardeux B, Savidge T. Enteric glia inhibit intestinal epithelial cell proliferation partly through a TGF-beta1-dependent pathway. Am J Physiol Gastrointest Liver Physiol. 2007;292:G231-G241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 130] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 9. | Aubé AC, Cabarrocas J, Bauer J, Philippe D, Aubert P, Doulay F, Liblau R, Galmiche JP, Neunlist M. Changes in enteric neurone phenotype and intestinal functions in a transgenic mouse model of enteric glia disruption. Gut. 2006;55:630-637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 181] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 10. | Cabarrocas J, Savidge TC, Liblau RS. Role of enteric glial cells in inflammatory bowel disease. Glia. 2003;41:81-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 141] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 11. | Bassotti G, Villanacci V, Maurer CA, Fisogni S, Di Fabio F, Cadei M, Morelli A, Panagiotis T, Cathomas G, Salerni B. The role of glial cells and apoptosis of enteric neurones in the neuropathology of intractable slow transit constipation. Gut. 2006;55:41-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 147] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 12. | Lomax AE, Fernández E, Sharkey KA. Plasticity of the enteric nervous system during intestinal inflammation. Neurogastroenterol Motil. 2005;17:4-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 142] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 13. | Tjwa ET, Bradley JM, Keenan CM, Kroese AB, Sharkey KA. Interleukin-1beta activates specific populations of enteric neurons and enteric glia in the guinea pig ileum and colon. Am J Physiol Gastrointest Liver Physiol. 2003;285:G1268-G1276. [PubMed] |
| 14. | Chacon O, Bermudez LE, Barletta RG. Johne's disease, inflammatory bowel disease, and Mycobacterium paratuberculosis. Annu Rev Microbiol. 2004;58:329-363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 170] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 15. | Bull TJ, McMinn EJ, Sidi-Boumedine K, Skull A, Durkin D, Neild P, Rhodes G, Pickup R, Hermon-Taylor J. Detection and verification of Mycobacterium avium subsp. paratuberculosis in fresh ileocolonic mucosal biopsy specimens from individuals with and without Crohn's disease. J Clin Microbiol. 2003;41:2915-2923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 286] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 16. | Naser SA, Ghobrial G, Romero C, Valentine JF. Culture of Mycobacterium avium subspecies paratuberculosis from the blood of patients with Crohn's disease. Lancet. 2004;364:1039-1044. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 440] [Cited by in RCA: 429] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 17. | Sechi LA, Scanu AM, Molicotti P, Cannas S, Mura M, Dettori G, Fadda G, Zanetti S. Detection and Isolation of Mycobacterium avium subspecies paratuberculosis from intestinal mucosal biopsies of patients with and without Crohn's disease in Sardinia. Am J Gastroenterol. 2005;100:1529-1536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 149] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 18. | Wösten MM. Eubacterial sigma-factors. FEMS Microbiol Rev. 1998;22:127-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 162] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 19. | Dove SL, Darst SA, Hochschild A. Region 4 of sigma as a target for transcription regulation. Mol Microbiol. 2003;48:863-874. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 83] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 20. | Manganelli R, Voskuil MI, Schoolnik GK, Smith I. The Mycobacterium tuberculosis ECF sigma factor sigmaE: role in global gene expression and survival in macrophages. Mol Microbiol. 2001;41:423-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 333] [Cited by in RCA: 338] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 21. | Manganelli R, Provvedi R, Rodrigue S, Beaucher J, Gaudreau L, Smith I. Sigma factors and global gene regulation in Mycobacterium tuberculosis. J Bacteriol. 2004;186:895-902. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 173] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 22. | Li L, Bannantine JP, Zhang Q, Amonsin A, May BJ, Alt D, Banerji N, Kanjilal S, Kapur V. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc Natl Acad Sci USA. 2005;102:12344-12349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 335] [Cited by in RCA: 339] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 23. | Rühl A, Franzke S, Collins SM, Stremmel W. Interleukin-6 expression and regulation in rat enteric glial cells. Am J Physiol Gastrointest Liver Physiol. 2001;280:G1163-G1171. [PubMed] |
| 24. | Bush TG. Enteric glial cells. An upstream target for induction of necrotizing enterocolitis and Crohn's disease? Bioessays. 2002;24:130-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 38] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 25. | Cornet A, Savidge TC, Cabarrocas J, Deng WL, Colombel JF, Lassmann H, Desreumaux P, Liblau RS. Enterocolitis induced by autoimmune targeting of enteric glial cells: a possible mechanism in Crohn's disease? Proc Natl Acad Sci USA. 2001;98:13306-13311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 252] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 26. | Nasser Y, Ho W, Sharkey KA. Distribution of adrenergic receptors in the enteric nervous system of the guinea pig, mouse, and rat. J Comp Neurol. 2006;495:529-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 68] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 27. | von Boyen GB, Steinkamp M, Reinshagen M, Schäfer KH, Adler G, Kirsch J. Proinflammatory cytokines increase glial fibrillary acidic protein expression in enteric glia. Gut. 2004;53:222-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 177] [Article Influence: 8.4] [Reference Citation Analysis (0)] |