Published online Mar 28, 2007. doi: 10.3748/wjg.v13.i12.1847
Revised: December 30, 2006
Accepted: January 25, 2007
Published online: March 28, 2007
AIM: To screen for metronidazole (MTZ)-resistance associated gene fragments of H pylori by suppression subtractive hybridization (SSH).
METHODS: Five MTZ-resistant (tester, T) and 1 MTZ-susceptible (driver, D) clinical H pylori isolates were selected. Genomic DNAs were prepared and submitted to RsaIdigestion. Then two different adaptors were ligated respectively to the 5’-end of two aliquots of the tester DNA fragments and SSH was made between the tester and driver DNAs. The specific inserts of tester strains were screened and MTZ-resistance related gene fragments were identified by dot blotting.
RESULTS: Among the randomly selected 120 subtractive colonies, 37 DNA fragments had a different number of DNA copies (≥ 2 times) in resistant and susceptible strains and 17 of them had a significantly different number of DNA copies (≥ 3 times). Among the sequences obtained from the 17 DNA fragments, new sequences were found in 10 DNA fragments and duplicated sequences in 7 DNA fragments, representing respectively the sequences of depeptide ABC transporter periplasmic dipeptide-binding protein (dppA), permease protein (dppB), ribosomal protein S4 (rps4), ribonuclease III (rnc), protease (pqqE), diaminopimelate epimerase (dapF), acetatekinase (ackA), H pylori plasmid pHP51 and H pylori 1334.
CONCLUSION: Gene fragments specific to MTZ-resistant H pylori strains can be screened by SSH and may be associated with MTZ-resistant H pylori.
-
Citation: Li AQ, Dai N, Yan J, Zhu YL. Screening for metronidazole-resistance associated gene fragments of
H pylori by suppression subtractive hybridization. World J Gastroenterol 2007; 13(12): 1847-1850 - URL: https://www.wjgnet.com/1007-9327/full/v13/i12/1847.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i12.1847
H pylori is a bacterial organism causing chronic gastritis and peptic ulcers. It also plays an important role in the pathogenesis of gastric cancer and mucosa associated lymphoid tissue (MALT) lymphoma. Metronidazole (MTZ) is an important component of many currently used H pylori eradication regimens. Antibiotic resistance of H pylori is known as an important cause of treatment failure of H pylori. The mechanism of MTZ resistance in H pylori treatment remains unclear, although some genes such as rdxA, frxA and fdxB have been found to be associated with MTZ resistance[1-5].
Suppression subtractive hybridization (SSH), one of the new methods for phenotypic cloning, has been developed for identifying genomic differences between the genomes of close relatives[6]. It is a powerful approach for screening genes associated with drug resistance. Using SSH, we have identified some DNA fragments which are specific to the resistant strains. The identified DNA fragments were sequenced and compared with GenBank database to search for some genes associated with MTZ- resistance in H pylori.
Each of the clinical biopsy specimens was homogenized with a tissue grinder and then inoculated onto Columbia agar (bioMérieux) plates supplemented with 8.0% (V/V) sheep blood, 0.2% (W/V) cyclodextrin, 5 mg/L trimethoprim (Sigma), 10 mg/L vancomycin (Sigma), 2.5 mg/L amphotericin B (Sigma) and 2500 U/L polymyxin B (Sigma). The plates were incubated at 37°C under microaerobic conditions (5% O2, 10% CO2 and 85% N2) for 3-5 d. Isolates were identified as H pylori according to typical Gram stain morphology, biochemical tests positive for urease and oxidase.
The minimal inhibition concentration (MIC) was determined by agar dilution method of the National Committee for Clinical Laboratory Standards (NCCLS). Agar dilution plates were prepared with two-fold serial dilution of MTZ, ranging from 0.25 to 128 mg/L. The inoculation concentration of H pylori was 1 × 106-7 CFU/5 μL. Results were read after 72 h incubation and the MIC was determined as the lowest concentration of MTZ in which no visible growth occurred. Strains with MIC value ≥ 8 mg/L were classified as resistant[7]. NCTC11637 was used as a reference strain.
Genomic DNA was extracted from H pylori strains according to UNIQ-10 genomics DNA isolation kit user manual provided by Shanghai Sangon Biological Engineering & Technology and Service Co. Ltd.
The subtractive DNA library was established according to Clontech PCR-selectTM bacterial genome subtraction kit (PT3170-1) user manual. Genomic DNA from five MTZ-resistant clinical H pylori strains was used as the tester respectively, and DNA from one MTZ-susceptible clinical strain was used as the driver. The sequences of adaptors and primers are as followings: adaptor 1: 5’-CTAATACGACTCACTATAGGGCTCGAGCGGCCGCCCGGGCAGGT-3’; adaptor 2R: 5’-CTAATACGACTCACTATAGGGCAGCGTGGTCGCGGCCGAGGT -3’; P1: 5’-CTAATACGACTCACTATAGGGC-3’; NP1: 5’-TCGAGCGGCCGCCCGGGCAGGT-3’; NP2: 5’-AGCGTGGTCGCGGCCGAGGT-3’. The PCR products were analyzed by 2% agarose gel electrophoresis, purified by using the 3S PCR product purification kit (BBST).
The second PCR products were purified and cloned into the pUCm-T vector (BBST) following the protocols. The recombinant plasmids were transformed into E. coli DH5α, which was then cultured overnight on the selective agar plates. One hundred and twenty white colonies were randomly picked and cultured in Luria-Bertani medium containing ampicillin at 37°C for 8 h. The plasmids were extracted and used as templates and the inserts were amplified under condition as in the second PCR for 25 cycles. The sizes of the inserts were identified by 2% agarose gel electrophoresis.
Each insert PCR-purified products were dotted on the Hybond N+ membrane (BioRad) in duplicating forms and DNA fixation was carried out by baking the Hybond N+ membrane at 80°C for two hours. In addition, 23S rRNA fragment of H pylori and pBR328 were dotted on the membrane as positive and negative control respectively. The RsaI-digested genomic DNA fragments of four resistant and four susceptible H pylori strains (not tester and driver strains) were used as probes and dot blotting was preformed using the DIG DNA labeling and detection kit (Roche). Pre-hybridization and hybridization were carried out in the hybridization oven (HYBAID) at 50°C for 2 and 20 h respectively. After the Hybond N+ membrane was stringently washed and blocked, the Anti-Dig-AP mixtures were added and the signals were detected by colorsubstrate solution (NBT/BCIP). The inserts that gave positive results (that is, obvious difference in gene copies between the DNA fragments of resistant and susceptible strains) were sequenced by BBST Company. The sequences were then submitted to gene homologous analysis based on GenBank database.
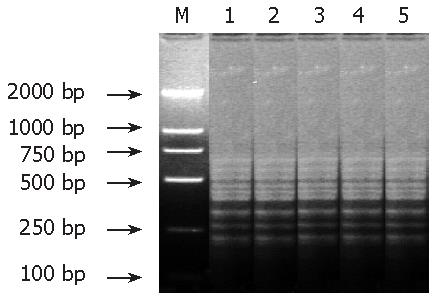
After subtractive hybridization of genomic DNA in resistant and susceptible isolates, PCR amplified products presented several similar and tight striples, indicating that most tester sequences formed with the driver were excluded, while tester-specific sequences were self- hybridized to form amplifiable fragments that were then enriched by PCR (Figure 1).
After SSH between the tester and driver DNA fragments, about 450 colonies grew on the ampicillin plates and one third of them were white in color. One hundred and twenty white colonies were randomly chosen and 101 positive colonies containing the target inserts were confirmed by nested primer PCR. The size of inserts ranged from 150 to 750 bp (the average was about 280 bp).
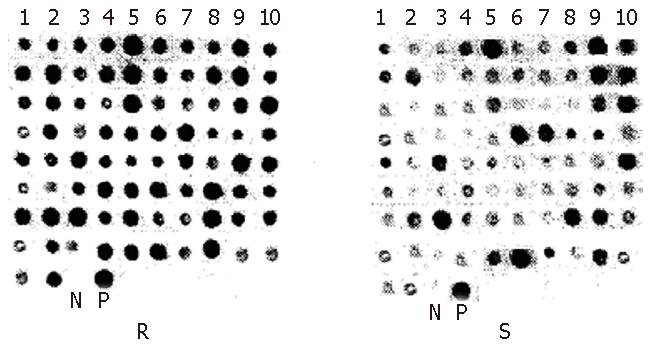
From 101 positive subtractive colonies, 82 with their size longer than 180 bp were picked out for dot blotting with genomic DNA fragments of RsaIdigested resistant and susceptible strains. Thirty-seven DNA fragments with a different number of DNA-copies (≥ 2 times) in resistant and susceptible strains were obtained and 17 of them differed significantly in the number of DNA-copies (≥ 3 times) (Figure 2).
Among the sequences obtained from the 17 specific DNA fragments, new sequences were found in 10 DNA fragments and duplicated sequences in DNA fragments, representing respectively the sequences of depeptide ABC transporter permease protein (dppB), periplasmic dipeptide-binding protein (dppA), ribosomal protein S4 (rps4), ribonuclease III (rnc), protease (pqqE), diaminopimelate epimerase (dapF), acetatekinase (ackA), H pylori plasmid pHP51, H pylori gene 1334 and replication protein B (Table 1).
| No. | GenBank accession No. | Homolog genes | DNA matches |
| R1 | AE000548 | Dipeptide ABC transporter periplasmic dipeptide-binding protein (dppA), S = 433, E = 0.0, I = 554/591 | 6142-6349 |
| R2 | AE000548 | Depeptide ABC transporter, permease protein (dppB), S = 433, E = 0.0, I = 554/591 | 6349-6728 |
| R3 | AE000609 | Protease (pqqE), S = 273, E = e-151, I = 309/321 | 9322-9642 |
| R4 | AE000579 | Ribonuclease III (rnc), S = 216, E = e-117, I = 265/280 | 10907-11185 |
| R5 | AY267368 | PHP51, S = 115, E = 4e-57, I = 146/154 | 3018-3169 |
| R6 | AB078638 | Replication protein B (REPB), S = 30, E = 2e-06, I = 55/62 | 1320-1381 |
| R7 | AE000633 | Ribosomal protein S4 (rps4), S = 354, E = 0.0, I = 406/422 | 7294-7714 |
| R8 | AE000635 | predicted coding region HP1334, S = 258, E = e-142, I = 457/523 | 1529-2050 |
| R9 | AE000570 | Diaminopimelate epimerase (dapF), S = 215, E = e-116, I = 247/260 | 7955-8214 |
| R10 | AE000599 | This region contains an authentic frame shift and is not the result of a sequencing artifact, acetate kinase (ackA), S = 99, E = 4e-47, I = 183/210 | 8555-8764 |
Genes that are present in certain isolates of a given bacterial species and absent or substantially different in others can be of great biological interest. Some may determine strain-specific traits such as drug resistance, pathogenicity, bacterial surface structure, or restriction-modification. SSH is a powerful technique that has been applied to many different fields, such as identification of PIs/genomic islands, mobile genetic elements, drug resistance associated genes and variations in gene expression, etc[8-12]. Here we used this method in H pylori to search MTZ-resistance associated gene fragments.
In this study, some gene fragments specific for MTZ-resistant H pylori strains were identified by SSH. Of them, 10 were identified by dot blotting, including depeptide ABC transporter permease protein (dppB), periplasmic dipeptide-binding protein (dppA), ribosomal protein S4(rps4), ribonuclease III (rnc), protease(pqqE), diaminopimelate epimerase (dapF), acetatekinase(ackA), H pylori plasmid pHP51, H pylori gene 1334 and replication protein B. These gene fragments may be associated with MTZ-resistance of H pylori.
Dipeptide ATP-binding cassette (ABC) transporters dppA and dppB which are relative to transportation of bi-peptide and polypeptide, like the multidrug resistance protein (MDR), belong to bacterial periplasmic transport system and ABC transporters or traffic ATPase superfamily, take part in active efflux of intracelluar compounds, and function as a drug efflux pump[13]. It was reported that antibiotic efflux pumps exist in almost all bacteria and are one of the reasons for MDR. Our previous study showed that verapamil could reduce the MIC of MTZ-resistant H pylori, suggesting that drug efflux pump in the membrane of H pylori might be inhibited by verapamil[14]. In this study, the number of gene copies encoding DppA and DppB was obviously higher in the resistant H pylori isolates than in susceptible ones, indicating that DppA and DppB may play an important role in the active MTZ efflux and result in the resistance to MTZ.
Plasmid is a close-circular extra-chromosomal DNA form of bacteria in vitro. Antibiotic resistant plasmids widely exist in Gram-negative or positive bacteria. The resistant rate of H pylori to MTZ is very high and has an increasing trend. Researchers suspect that resistant plasmids exist in H pylori, but no relative evidence has been found. Our study showed that the number of gene copies in cryptic plasmid PHP051 was much higher in MTZ-resistant isolates, which may be due to its random insertion into the H pylori DNA, thus leading to some gene shift-code mutations and MTZ resistance.
In summary, the 10 new gene fragments identified in this study may be associated with H pylori resistance to MTZ. However, the relationship between these genes and their resistance to MTZ needs to be further investigated.
S-Editor Zhu LH L-Editor Wang XL E- Editor Chin GJ
| 1. | Goodwin A, Kersulyte D, Sisson G, Veldhuyzen van Zanten SJ, Berg DE, Hoffman PS. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol Microbiol. 1998;28:383-393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 271] [Cited by in RCA: 260] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 2. | Kwon DH, El-Zaatari FA, Kato M, Osato MS, Reddy R, Yamaoka Y, Graham DY. Analysis of rdxA and involvement of additional genes encoding NAD(P)H flavin oxidoreductase (FrxA) and ferredoxin-like protein (FdxB) in metronidazole resistance of Helicobacter pylori. Antimicrob Agents Chemother. 2000;44:2133-2142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 97] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 3. | Marais A, Bilardi C, Cantet F, Mendz GL, Mégraud F. Characterization of the genes rdxA and frxA involved in metronidazole resistance in Helicobacter pylori. Res Microbiol. 2003;154:137-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 60] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 4. | Chisholm SA, Owen RJ. Mutations in Helicobacter pylori rdxA gene sequences may not contribute to metronidazole resistance. J Antimicrob Chemother. 2003;51:995-999. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 21] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 5. | Dai N, Zhou G, Yan J. Correlation of rdxA gene mutation and metronidazole resistance of Helicobacter pylori. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2003;32:37-40. [PubMed] |
| 6. | Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci U S A. 1996;93:6025-6030. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2262] [Cited by in RCA: 2003] [Article Influence: 69.1] [Reference Citation Analysis (0)] |
| 7. | Guidelines for clinical trials in Helicobacter pylori infection. Working Party of the European Helicobacter pylori Study Group. Gut. 1997;41 Suppl 2:S1-S9. [PubMed] |
| 8. | Agron PG, Macht M, Radnedge L, Skowronski EW, Miller W, Andersen GL. Use of subtractive hybridization for comprehensive surveys of prokaryotic genome differences. FEMS Microbiol Lett. 2002;211:175-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 36] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 9. | Zhang YL, Ong CT, Leung KY. Molecular analysis of genetic differences between virulent and avirulent strains of Aeromonas hydrophila isolated from diseased fish. Microbiology. 2000;146:999-1009. [PubMed] |
| 10. | Bogush ML, Velikodvorskaya TV, Lebedev YB, Nikolaev LG, Lukyanov SA, Fradkov AF, Pliyev BK, Boichenko MN, Usatova GN, Vorobiev AA. Identification and localization of differences between Escherichia coli and Salmonella typhimurium genomes by suppressive subtractive hybridization. Mol Gen Genet. 1999;262:721-729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 11. | Tian HS, Zhu CL, Gao XH, Ma L, Shen B, Li XL, Wu GL. Cloning and identification of deltamethrin-resistance or susceptibility associated genes of Culex pipiens pallens. Zhongguo Jishengchongxue Yu Jishengchongbing Zazhi. 2001;19:193-197. |
| 12. | Han FC, Gong M, Ng HC, Ho B. Identification of H. pylori strain specific DNA sequences between two clinical isolates from NUD and gastric ulcer by SSH. World J Gastroenterol. 2003;9:1747-1751. [PubMed] |
| 13. | Ames GF, Mimura CS, Shyamala V. Bacterial periplasmic permeases belong to a family of transport proteins operating from Escherichia coli to human: Traffic ATPases. FEMS Microbiol Rev. 1990;6:429-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 126] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 14. | Dai N, Zhu YL, Qian KD. The effect of Varapamil on metronidazole resistance in helicobacter pylori. Zhonghua Xiaohua Zazhi. 1998;18:352-354. |