INTRODUCTION
Eukaryotic cells are divided into different compartments and viruses have to get access to the compartment that provides the cellular machinery for replication. Unlike bacteriophages that “just” have to pass the bacterial wall and membrane all viruses that infect eukaryotic cells have to travel through the cell to reach the place of replication. Dependent upon the virus and the type of genome, the machinery of DNA replication, transcription and RNA processing may be required. Gaining access to the nucleus where these factors are found requires active transport since passive diffusion is ineffective.
Although summarized by the term “intracellular transport” one has to differentiate between the intracytoplasmic transport towards the nucleus and the passage through the nuclear envelope into the karyoplasm. Nucleic acids are not karyophilic per se. Thus proteins attached to the viral genome must interact with cellular factors that facilitate the different transport processes. As intracytosolic and nuclear transport are based on different mechanisms it is evident that the interacting domains on the viral proteins have to be exposed in a coordinated manner. Moreover an effective virus, meaning a virus with a good particle-infectious unit ratio, has to release its genome from surrounding proteins only after termination of the various transport steps preventing abortion of the infection process. It has to be considered that the analysis of the underlying principles is not only important for analysis of potential drug targets for treatment of individual viral infections but also for creation of efficient vectors in gene therapy.
OVERVIEW ON INTRACYTOSOLIC TRANSPORT PRINCIPLES
The need for active and directed intracytosolic transport results from the high viscosity of the cytoplasm. Protein concentrations of 170-350 mg/mL, RNA concentrations of 30 μg/mL[1] and micro compartmentalization evoke a viscosity 10 to 100 fold higher than the viscosity of water[2]. In consequence diffusion processes are enormously reduced so that only particles with diameters below 50 nm significantly diffuse[3]. Obviously such a passive movement is incompatible with efficient trafficking of organelles. Eukaryotic cells thus provide different active transport machineries that are not only used by large structures but even by small macromolecules as it was shown for the heat shock protein 90 (Hsp90)/glucocorticoid receptor β (GRβ) complex[4] and the human tumour suppressor protein p53[5]. It is thus likely that structures as the HBV capsid with a diameter of 36 nm (80 % of the capsids that show a T = 4 symmetry, 20 % exhibit a T = 3 symmetry and a diameter of 32 nm[6,7]) use the same active cellular transport pathways towards the nucleus.
Most investigations on intracytosolic transport of viruses are done by adding inhibitors to cells during infection. To prevent misinterpretations it is important to realize the variety of cellular processes that are affected. Eukaryotic cells provide two active cytoplasmic transport systems based on microfilaments (MF) and microtubules (MT). Microfilaments have diameters of 5-9 nm and are double-stranded helical polymers of the ATPase actin. They form linear bundles, 2D networks and 3D gels and are most highly concentrated underneath the plasma membrane. Microfilaments are dynamic polar structures with a fast-growing plus-end and a relatively inert, slow-growing minus-end. They stabilize the cell structure and are involved in cell movement e.g. by filopodia and lamellopodia. With a velocity of 2-6 mm/d transport via MF is slow[8,9] and generally thought to serve as the transport pathway for short distances.
Beside its role in transport actin is also involved in the internalization and/or formation of endocytic vesicles. In clathrin-mediated endocytosis a functional actin cytoskeleton enhances the internalization of the clathrin-coated vesicles but without being obligatory[10-12]. In phagocytosis and macropinocytosis local actin polymerization at the cytosolic site of the plasma membrane is required for vesicle formation as well as actin depolymerization[13-16]. Similarly, releasing of caveolin-coated vesicles into the cytoplasm depends on actin polymerization and depolymerization in caveolae-mediated endocytosis[17,18].
Microtubules are hollow cylinders of 11-13 protofila-ments made of the GTPase α- and β-tubulin. MT are 25 nm in diameter and like MF they have a highly dynamic, fast-growing plus-end and a less dynamic minus-end, which is typically attached to a microtubule-organizing centre (MTOC). MT surround the nucleus and extend from the perinuclear MTOC to the cell periphery. They are thought to be the major long-range transport system[9] allowing a velocity of 3-5 mm per hour (HSV 1 capsids, retrograde)[19].
For both MT and MF two different mechanisms of transport exist (Figure 1) using polymerization (filament growth)/depolymerization (filament shrinkage) or motor proteins[20]. In the first mechanism the cargo binds dynamically via adapter proteins to one end of a growing or shortening filament and can be pushed or pulled in the given direction. Mitotic chromosomes for example bind at its kinetochor via a dynein/dynactin-complex to the plus-end of the kinetochor-MT[21-23]. Depolymerization at the plus-end as well as depolymerization at the minus-end directs the chromosomes to the minus-end that is fixed at the spindle pole. Phagosomes[24,25], macropinosomes[26,27] endosomes and lysosomes[27,28] are able to cross the cytosol by the help of a polymerizing actin tail comparable to the movement of the bacteria listeria monocytogenes[29-31] and the nuclear polyhedrosis virus (NPV) capsid[32-34].
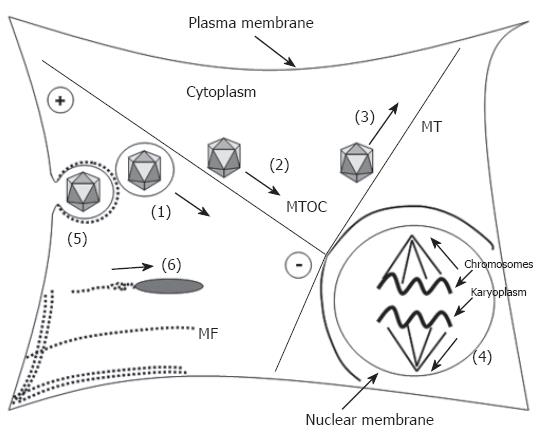
Figure 1 Participation of microtubules and microfilaments in transport processes.
Transport processes are indicated as bold arrows. Microtubules (MT, bold lines) have a highly dynamic plus-end and a less dynamic minus-end that is located at the microtubule-organizing (MTOC). They participate in the transport of (1) organelles, e.g. endosomes, (2) direct retrograde transport of capsids via the dynein motor protein complex (adenovirus, HSV 1, parvoviruses), (3) direct anterograde transport of progeny HSV 1 capsids by conventional kinesin, and (4) participate in chromosome segregation upon mitosis. Microfilaments (MF), depicted as dotted lines participate (5) in separation of endocytotic vesicles from the plasma membrane and (6) via polymerization in transport of e.g. Listeria monocytogenes and nuclear polyhedrosis virus (NPV).
However the most commonly used transport strategy for macromolecules, mRNA, RNPs (ribonucleoprotein complexes), cellular organelles and vesicles along polar MF or MT involves motor protein complexes, namely myosins, kinesins and dyneins[20,35-38]. These cargo specific filament binding proteins exhibit a motor domain (ATPase) and move ATP-dependent and unidirectional along the stable filaments. Dependent upon the involved filament and transport direction different motor protein complexes are used. Class VI myosins are unconventional myosins which move towards the minus-end of MF[39]. They are involved in the transport of clathrin-coated vesicles from the plasma membrane into the inner cell[40,41]. Class I and class V myosins in contrast migrate towards the plus-end of MF[20,35]. Myosin I mediates transport of membranes whereas myosin V is responsible for the transport of organelles, as e.g. recycling endosomes towards the plasma membrane[20,42,43]. In addition myosin V can transport cargos along MT[44] thus linking both transport systems.
The MT specific kinesins can be distinguished upon the localization of the conserved motor domain into conventional kinesins, which have an N-terminal motor domain and that direct cargos in the anterograde direction towards the plus-end of the MT (i.e. towards the cell periphery)[20,36]. They participate in the intracellular transport and localization of different cellular membrane organelles, as the extension of the ER (endoplasmatic reticulum) from the nucleus towards the cell periphery[45,46]. In addition, conventional kinesins are used in directing progeny capsids of herpes simplex virus 1 (HSV 1) in the anterograde direction[47].
Unconventional kinesins, which have a C-terminal motor domain move in contrast towards the minus-end of MT. They mediate axonal MT transport of vesicles and organelles[36].
The cytoplasmic dynein is commonly associated with the cofactor dynactin (dynactin-complex)[48] transporting cellular vesicles (endosomes and caveolin-negative vesicles[49-52] and macromolecules (e.g. tumor suppressor protein p53[5]) in the retrograde direction towards MT minus-end (the MTOC). Apparently the dynein-complex is frequently used in transport of viral capsids towards the nucleus as it was shown for adenoviruses[53-55], parvoviruses[56-59] and HSV 1 upon infection[47,60].
INTRACYTOSOLIC TRANSPORT OF HBV CAPSIDS
With the exception of NPV all viruses being analyzed so far make use of the MT transport system for their transport towards the nucleus[61]. This includes large viruses that have to travel long distances as the herpes simplex virus 1 (HSV 1;[47]) that has to be transported for centimeters between the axon end and the cell body and parvoviruses (18-26 nm)[56,58,62] that are below the diffusion limit inside the cytoplasm.
Evaluating the transport mode of hepatitis B virus is however not trivial as the entry mechanism is not fully understood. This is mainly caused by a lack of an appropriate and effective in vitro infection system that allows study of the early steps of the hepadnaviral life cycle. As described in “Viral and cellular determinants involved in hepadnaviral entry” hepadnaviruses enter the cells in vesicles[63] but do not need acidification for infection[64]. This step that normally occurs upon endocytosis has a major effect on viral structures as described for adeno- and parvoviruses[57,65,66]. The altered structure distinguishes capsids that have passed endocytosis from progeny capsids that are newly synthesized. The different exposed epitopes allow variant interactions so that incoming capsids are targeted to the nucleus while progeny capsids are not.
As such an acid-induced conformational change is missing in the hepadnaviral life cycle the viral capsids released from the endosomal pathway and the newly synthesized progeny capsids have the same structure. Consequently both types of capsid can participate in nuclear transport of the viral genome.
The restrictions of the experimental systems for analysis of hepadnaviral viral infections are most likely caused by an insufficient entry. It was thus a self suggested idea to replace the viral surface proteins by a lipid shell as it is done in protein transfection (lipofection)[67]. The lipids fuse with the plasma membrane and release the capsids into the cytoplasm. For studying the intracellular transport of the hepadnaviral capsid lipofection has the advantage that no cellular transport vesicles are involved.
In fact lipofection of hepatoma cells yielded in a highly productive HBV infection similar to the in vivo efficiency.
As lipofection is independent upon receptors high amounts of capsids could be loaded on the cells. This allows one to follow the fate of the capsids and of the viral genomes by microscopical techniques[67] showing that the capsids accumulated at the nuclear envelope after 15 min. As diffusion can be assumed to take 1 h (for calculation see[9]), these data imply a directed active capsid transport towards the nucleus. Released viral genomes occurred exclusively inside the karyoplasm suggesting site-specific disintegration at the nuclear envelope. The use of the MT depolymerizing drug nocodazole inhibits accumulation of capsids at the nuclear envelope and the release of genomes suggesting that genome liberation requires transport to the nucleus.
The microscopical finding of an active MT-mediated transport towards the nucleus was supported analysing the effect of nocodazole on the hepadnaviral life-cycle. It was demonstrated that MT are essential for formation of nuclear DNA (cccDNA that occurs only after repair of the partially double stranded DNA genome within the nucleus) and for amplification of viral DNA via synthesis of progeny mature capsids.
The use of the MT transport system was confirmed in another experimental system in which capsids were injected into the cytosol of Xenopus laevis oocytes. Due to the longer transport distances 30 min are required for the capsids to reach the nucleus upon injection at the pole of the oocyte opposed the nucleus. Assuming a distance of 0.5 mm that has to be bridged this period is consistent with the cytosolic transport of HSV 1 capsids that traverse the cytoplasm with 3-5 mm per hour (retrograde transport,[9]). As electron microscopy was used as read-out for capsid localization these data could show that the capsids did not bind to undefined sites of the nuclear envelope but to the nuclear pore complexes (NPC)[68]. However, when anti tubulin-antibodies were preinjected the arrival of the capsids at the NPCs was inhibited confirming that even in cells only distantly related to human hepatocytes the same transport system is used.
Although conclusive, it must be considered that all the observations described above were not done in the “authentic primary” cells and that a liver-specific factor may alter e.g. the place of genome release and lead to another transport model that does not involve the capsid. However, the generation of capsids with a translocation motif (TLM) fused to the N-terminus of the capsid protein recently gave further support[69]. These capsids were still capable of encapsidating the polymerase and the pregenome so that mature DNA capsids were generated being able to initiate an HBV infection in primary human hepatocytes. Although the mode of uptake by a TLM remains controversial-directly penetrating the plasma membrane or using transporters[70]-a clear uptake of the capsids could be observed, resulting in accumulation in the perinuclear region. This uptake was not observed when wild-type capsids were used being contradictory to most recent results of others[71]. Irrespectively of this divergence the location of the TLM capsids at the perinuclear region where the MTOC is situated supports that the MT were used for transport towards the nucleus.
An open question not being answered for any cargo that uses MT transport for reaching the nucleus is derived from the polarity and arrangement of the MT. As their minus-end is not directly located at the NPC but attached to the MTOC the cargo must cross the distance between the MTOC and the nucleus. There are observations that even for this short gap passive diffusion is not likely: HSV 1 capsids show an equal distribution around the nucleus after infection and do not accumulate at those NPCs adjacent to the MTOC[47]. However, the mechanism of this translocation remains open.
GENERAL MECHANISM OF NUCLEAR TRANSPORT
Viruses that replicate in the nucleus of non-dividing cells have to traverse the nuclear envelope. For this reason nuclear proteins pass the nuclear pore complexes (NPCs). NPCs are large proteinaceous structures consisting of 30 different proteins[72], collectively termed nucleoporins (Nups). Nucleoporins exist in multiples copies, forming a complex of estimated 125 MD[73]. Many nucleoporins contain distinct domains of phenylalanine-glycine (FG) repeats, which mediate the main interaction between nucleoporins and soluble transport receptors. The NPC consists of a central ring-like framework with 8-fold symmetry, representing the part of the complex that is embedded in the nuclear envelope (NE) Attached to a cytoplasmic ring moiety 8 cytoplasmic filaments form an initial docking side for transport complexes. On the karyoplasmic face, 8 fibres form the cage-like structure of the nuclear basket[74]. The central framework is a ring-like assembly built around a central pore through which the exchange of macromolecules occurs. The dimension of the nuclear pore restricts complexes to a diameter of 39 nm including their shell of transport receptors[68]; a size that is exceeded by most viruses or subviral particles.
NPCs regulate the traffic of proteins and nucleic acids into and out of the nucleus[75]. Substrates smaller than roughly 9 nm in size, including ions, metabolites and proteins, travel through the NPC in a diffusion-controlled and energy independent manner[76].
Most nuclear cargos exhibit signals that interact with nuclear transport receptors of the importin β superfamily, comprising importins and transportins. All members of this family exhibit an N-terminal RanGTP-binding domain which is important for dissociating receptor and cargo (reviewed by[77]). There is a variety of different signals that are recognized as exemplified by the M9 domain, bound by transportin, polypeptides of basic amino acids that represent an importin β binding domain (IBB) and “classical” nuclear localization signals (NLSs) that show the consensus sequence K(K/R)X(K/R)[78]. The classical NLS does not directly bind to the transport-mediating receptor importin β (Imp β) but requires an adapter molecule, importin α (Imp α), which connects NLS and - via its IBB-Imp β.
The driving force of nuclear import and export is determined by the different concentrations of RanGTP in the nucleus versus the cytoplasm. RanGTP that is enriched in the karyoplasm, interacts with the transport receptors of the import complex, leading to dissociation of cargo and receptor. While the cargo diffuses deeper into the karyoplasm, the RanGTP-receptor complex becomes exported to the cytoplasm.
Hepadnaviral genomes have to enter the nucleoplasm for replication. As hepatocytes are terminally differentiated cells that do not divide they cannot wait until the cell undergoes mitosis as most retroviruses-excluding HIV-do. As karyophilic proteins the capsids use the nuclear pore complex to get for access to the nucleoplasm.
NUCLEAR IMPORT OF HEPADNAVIRAL GENOMES
Nucleic acids are not karyophilic per se. Therefore one or more proteins attached to the genome must interact with cellular nuclear import receptors. In case of the hepadnaviruses three models, each involving a different mediator, may play a crucial role in the nuclear import of the HBV genome: (1) The viral polymerase of the Hepatitis B virus. The enzyme is covalently attached to the viral genome and probably contains a hidden NLS. Expression of the polymerase in eukaryotic cells revealed that the protein stays cytoplasmic[79] but extraction of this complex from mature virions showed that it enters the nucleus[80]. However, the procedure of extraction requires harsh treatment and thus structure altering methods. (2) Some of the heat shock proteins as Hsc70 or Hsp90 activating the polymerase[81-84] and (3) the capsid proteins surrounding the viral replication complex.
The lipofection experiments described above, show that released viral DNA is exclusively present within the nucleus. The release is combined with the accumulation of capsids at the nuclear envelope. It thus has to be concluded that if the polymerase mediates nuclear import the release of the import complex must occur at the nuclear envelope, probably after docking of the capsids to the nuclear pore. Examples for such a pathway are the HSV 1 capsid that becomes opened upon the interaction of a penton with the NPC and adenovirus 2 that releases the complex of DNA and associated proteins at the pore[47,85-88].
The polymerase-associated heat shock proteins may act in a similar manner. For example, the interaction with Hsc70 that exhibits a nuclear transport capacity is established[84]. The capsid-nuclear envelope interaction was more extensively investigated in cell biological assays and in microinjection experiments using oocytes of Xenopus laevis. Another experimental design is based on Digitonin permeabilized cells most commonly used in studying nuclear import reactions in detail. Digitonin permeabilizes only cholesterol containing membranes as e.g. the plasma membrane and membranes of mitochondria. Other membranes as the nuclear and ER membrane remain unaffected[89]. In general adhesive cells are analysed under conditions where they attach to the surface of a glass cover slip thus the Digitonin has to be removed by washing steps. The washing removes the soluble cytosolic proteins (and some small nuclear proteins that rapidly diffuse out of the nucleus) including the cellular nuclear transport factors. Consequently these factors have to be replaced either by addition of selected import factors or in form of a cytosolic extract. However, as the nuclear transport capacity is conserved this process can be transferred to the in vivo situation presupposed that the subjected cargo with its modification is physiological.
Microinjection in the cytoplasm is another established technique. For electron microscopy Xenopus laevis oocytes are frequently used as the huge dimensions of the nucleus allow the analysis of multiple sections. Since nuclear import is phylogenetically well conserved the results are transferable to other cell types as long as no embryogenesis related processes are affected.
These assays and biochemical analyses have been used to analyse the nuclear import of the hepadnaviral genome in more detail. Using permeabilized cells it was shown that the HBV capsid protein contains an NLS within its C-terminal domain. This domain is hidden in the lumen of RNA-containing capsids expressed in E. coli and in eukaryotically expressed capsids devoid of the polymerase[90,91]. These capsids failed to interact with nuclei of permeabilized cells. However, the exposure of the C-terminus on the capsid surface was shown to be linked to genome maturation as in vitro studies revealed[90]. Interestingly, the exposure could be initiated in RNA-containing capsids expressed in E. coli when the C-terminus was in vitro phosphorylated by protein kinase C[92] or protein kinase A[93]. These observations strengthen the idea that phosphorylation and genome maturation are linked. The impact of the exposed C-termini for nuclear pore complex association becomes evident by cleavage experiments in which the C-termini were removed from mature capsids followed by subjection of the capsids to Digitonin-permeabilized cells. According with the hypothesis of a capsid-mediated NPC docking the capsids and the necessity of exposed NLS to the digested capsids failed to interact with the nuclei. Consistently with the identification of an NLS on the capsid protein importin β was found to be the mediator of NPC interaction, requiring importin α as an adapter protein.
Most fascinating is the different import behaviour between capsids that have undergone genome maturation to a different extent. Capsids with an immature DNA genome interacted with the NPCs but remained associated with the pores while subjecting mature capsids to the permeabilized cells resulted in a nuclear capsid stain and released viral genomes within the nucleus.
Based on these data all three import models-polymerase, heat shock proteins or capsid mediation-could be true. The mature capsids could interact with the NPC releasing their genome followed by import of the genome mediated by the polymerase or heat shock proteins. The dissociated capsid subunits enter the nucleus as they apparently do in HBV infected individuals. The immature capsids could have been just more stable thus failing to disintegrate and to release the genome.
Alternatively, only the mature capsids that expose more NLS may have become surrounded by enough nuclear transport receptors to pass a hydrophobic mesh caused by the crosslinking of hydrophobe FxFG repeats of nucleoporins. Only these capsids interact with the factors required for genome release.
To differentiate between the models further investigations were initiated to follow the fate of the capsids at the NPC. Surprisingly, electron microscopy after microinjection into Xenopus laevis oocytes showed that not only the mature capsids passed the pore and entered the nuclear basket but also immature capsids that apparently failed to diffuse deeper into the karyoplasm. Consistent with the experiments in permeabilized cells, RNA containing capsids did not interact with the NPCs.
For the model of nuclear import these findings implied that apparently the capsids mediated the passage through the nuclear pore into the nuclear basket. Here only the mature capsids disintegrated while the immature capsid stay arrested. Hypothetically this arrest can increase efficiency of HBV infection, assuming that genome maturation proceeds in these capsids.
BEYOND NUCLEAR IMPORT
Hepadnaviral polymerases can only successfully synthesize the full length viral DNA when interacting with the capsid proteins. In fact, recent studies on the phosphorylation sites of the HBV capsid protein show that one serine residue (Ser 157) has to be phosphorylated for pregenome packaging while serine 164 is required for allowing DNA synthesis[94,95]. The highly efficient infections by hepadnaviruses thus imply that the genome release is well coordinated in that only capsids with a mature genome disintegrate. The factor however that apparently is only present within the nucleus remains unknown. Incubations of mature capsids with nuclear extracts failed to induce significant genome release while permeabilized hepatoma cells only require minutes to release thousands of HBV genomes per nucleus. Furthermore a liver-specific factor must be assumed as genome release is more efficient in permeabilized hepatoma- than in HeLa cells. In vivo, this conclusion is supported by observations of Untergasser et al[96], who observed that cccDNA generation does not occur upon infection of dendritic cells with chimera of HBV and adenovirus.
Another open question is raised by the high copy number of empty capsids found in the nuclei of HBV infected hepatocytes or in hepatocytes of mice that are transgenic for HBV. Apparently they are not derived from capsids that have transported the genome into the nucleus as the half-life of the genome-as different data in the literature are (ranging from 3 d[97], 55 d[98] to a non-relevant degradation[99])-do not explain their frequent abundance. However, the capsid protein is strongly over expressed with regard to the number of viruses and capsids that participate in nuclear entry of the viral genome. Apparently capsid proteins have a nuclear transport capacity by their NLS, which is not hidden as long as they are not assembled to particles. Based on the results described above one must thus conclude that not capsids but the supernumerous capsid proteins or their assembly intermediates are imported. In accordance with consistent biochemical data of all groups studying the assembly process, it must be proposed that the capsid proteins do not need any other protein for assembly. The only driving force is the affinity to each other being supported by their interaction with other components as e.g. RNA. It is thus likely that after import of high numbers of capsid proteins into the nucleus the threshold concentration is reached resulting in rapid assembly to particles.
Nonetheless, one has to ask why the capsid proteins do not become arrested in the nuclear basket as the immature capsids. One can assume that assembled capsids can interact with at least eight basket proteins, most likely the nucleoporin 153[100], that are arresting the capsid. Although the answer remains experimentally open a protein monomer in contrast is restricted to one interaction that is in competition with the thousands of proteins that pass the nuclear pore every second.
The meaning of the nuclear assembly is unsolved. It might be just a side effect caused by the intrinsic assembly ability of the capsid. However, as the capsid proteins interact preferentially with single stranded nucleic acids the assembly may prevent interference with the cellular transcription and RNA export machinery thus reducing toxicity of the virus and ensuring a long life of the infected cell for persistent virus production.
SUMMARIZING THE TRAFFICKING OF HEPADNAVIRAL CAPSID AND GENOME
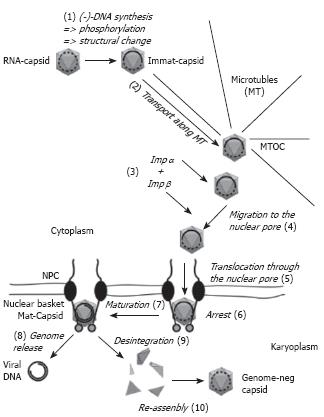
The summary of the current knowledge on the nuclear import of the hepadnaviral genome is depicted in Figure 2. It must be considered however that there are several elements as e.g. the co-ordination of the transport processes that are yet unknown. (1) Upon genome maturation and phosphorylation the hepadnaviral capsids undergo a structural change that leads to exposure of increasing numbers of the C-termini that are part of the capsid protein. (2) The capsids are transported towards the microtubule-organizing centre (MTOC), which is located at the perinuclear region. (3) As the exposed C-terminal domain exhibits a nuclear localization signal the probability of an interaction with the adaptor protein importin α increases. As in the physiological import of karyophilic proteins this complex is bound by importin β. Whether this acquisition already occurs during the MT-mediated transport or (4) during the unknown passage from the MTOC to the nucleus remains open. (5) Importin β next facilitates docking to the cytosolic fibres of the nuclear pore and translocation of the complex into the nuclear basket. (6) After dissociation of the nuclear import receptors from the capsid the capsid most likely interacts with a protein of the basket. (7) While immature capsids stay arrested and may continue with genome maturation, (8) mature capsids that are less stable release the genome with the associated proteins into the nucleus where genome repair takes place. (9) Supernumerous capsid proteins that result from disintegration can diffuse deeper into the karyoplasm where (10) they re-assemble after the capsid protein concentration reaches the threshold concentration for assembly.
Figure 2 Hepadnaviral trafficking within the cell.
Capsids are drawn as grey icosahedra. Immat-Capsid, immature capsid, Mat-Capsid, mature capsid. The nucleic acid found within the capsids is depicted as a dotted line (RNA) or a full line (DNA). The arrows present movements (2, 4, 5, 8) or changes of the capsid (1, 7, 9, 10). Further explanations are given in the text.
S- Editor Liu Y L- Editor Alpini GD E- Editor Ma WH