Published online Apr 7, 2006. doi: 10.3748/wjg.v12.i13.1991
Revised: November 11, 2005
Accepted: November 18, 2005
Published online: April 7, 2006
The inflammatory bowel disease, Crohn’s disease and ulcerative colitis, are polygenic disorders with important environmental interactions. To date, the most widely adopted approach to identifying susceptibility genes in complex diseases has involved genome wide linkage studies followed by studies of positional candidate genes in loci of interest. This review encompasses data from studies into novel candidate genes implicated in the pathogenesis of inflammatory bowel disease. Novel techniques to identify candidate genes-genome wide association studies, yeast-two hybrid screening, microarray gene expression studies and proteomic profiling, are also reviewed and their potential role in unravelling the pathogenesis of inflammatory bowel disease are discussed.
- Citation: Noble C, Nimmo E, Gaya D, Russell RK, Satsangi J. Novel susceptibility genes in inflammatory bowel disease. World J Gastroenterol 2006; 12(13): 1991-1999
- URL: https://www.wjgnet.com/1007-9327/full/v12/i13/1991.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i13.1991
Following the identification of the NOD2/CARD15 gene in 2001[1,2], the impetus has grown to identify novel genetic determinants of susceptibility and phenotype in the inflammatory bowel diseases, Crohn’s disease and ulcerative colitis. The inflammatory bowel disease are now considered to be non-Mendelian polygenic disorders with important environmental interactions[3], and as such the accepted approach to identifying candidate genes in complex disease genetics has been adopted, namely genome wide scanning followed by studies of positional candidate genes in sub-chromosomal regions of association (loci).
A number of candidate genes and susceptibility loci have been discussed elsewhere in this issue, so in this review we first summarise recent data with regards to genome wide scanning and candidate gene studies, which may lead to gene identification. Novel techniques to identify candidate genes - genome wide association studies, yeast- two hybrid screening, microarray gene expression studies and proteomic profiling, are subsequently discussed.
Genome-wide linkage analysis using highly polymorphic microsatellite markers identified during the course of the Human Genome Project has led to success in identifying genetic determinants both in single gene disorders and in complex genetic diseases. To date, successful genome scans have typically involved several hundred microsatellite markers and a large number of multiply affected inflammatory bowel disease families (typically sibling pairs), with the aim of examining whether the degree of sharing variant alleles between affected individuals exceeds that as expected by chance alone[4].
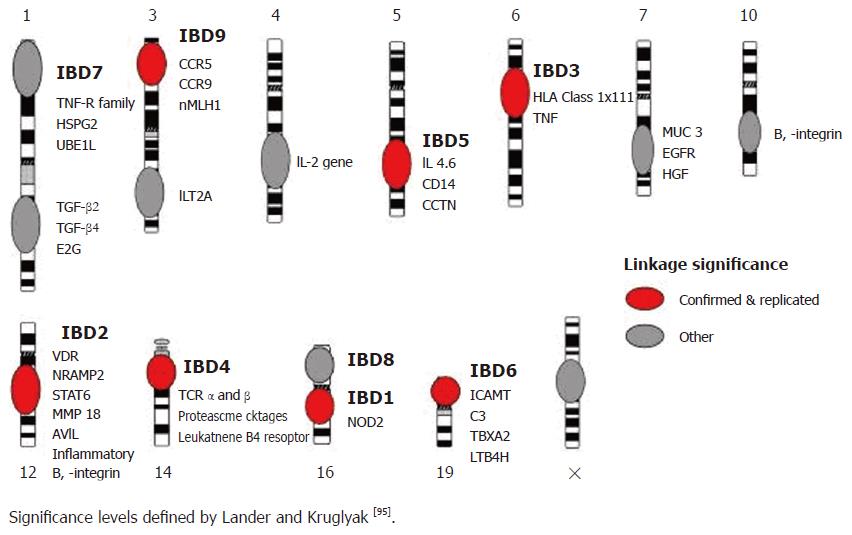
To date, fourteen genome wide scans have been carried out in patients with inflammatory bowel disease. The most widely accepted guidelines for assessing the results are those defined in 1995 by Lander and Kruglyak who proposed criteria for reporting areas of linkage[5], with areas of ‘suggestive linkage’ having LOD scores of 2.2 and above and P values of less than 7 × 10-4, areas of ‘significant linkage’ having LOD scores above 3.6 and P values of less than 2 × 10-5, and areas of ‘highly significant linkage’ having LOD scores of 5.4 and above and P values of less than 3 × 10-7. Areas of’ confirmed linkage defined as areas of significant linkage have been replicated in an independent cohort, with a nominal P value of less than 0.01. Using these criteria, loci with confirmed linkage have been identified on chromosomes 1[6], 3 (IBD9)[7,8], 5 (IBD5)[9-11], 6 (IBD3; HLA)[10,12], 12 (IBD2)[13], 14 (IBD4)[9,14], 16 (IBD1)[13,15] and 19 (IBD6)[10] (Figure 1).
These confirmed regions of linkage need to be further narrowed by fine mapping of these areas, as each spans a large genomic region. The classical positional cloning approach is used by Hugot and colleagues to identify NOD2/CARD15 as the gene confers the critical mutations in IBD1 on chromosome 16[1]. Alternative strategies for fine mapping these regions or providing a short-cut to the critical genes are discussed in the following.
The positional candidate gene approach to identifying association involves the use of data concerning the chromosomal location of the candidate gene as well as functional or expression data. In inflammatory bowel disease genes involved in the regulation of the innate immune system, mucosal integrity and cell-cell interactions are all clearly plausible candidate genes. By comparing the allelic frequencies of variants in these genes between patients with IBD and matched controls (case control analysis), or by investigating intra-familial association/ linkage, genes containing critical disease causing mutations may be identified.
Using this approach, Ogura and colleagues[2] identified the frameshift mutation in the NOD2/CARD15 gene (Leu1007fsinsC). Stoll and colleagues[16] also identified the DLG5 (Drosophila Discs Large Homolog 5) gene located on chromosome 10q23 by candidate gene approach allied to its position as being associated with inflammatory bowel disease in the German population[16].
However, whilst these approaches have had notable success, especially with the identification of the NOD2/ CARD15 gene within the IBD1 locus, there have been difficulties in identifying candidate genes within other loci, notably the IBD5 locus as discussed elsewhere. Due to the tight linkage disequilibrium across the IBD5 locus and the wealth of candidate genes in the region[17], it has been impossible to identify with certainty the causative mutations in this region. Although Peltekova and colleagues[18] have suggested that the OCTN1/2 genes contain the causative mutations, considerably larger cohorts of inflammatory bowel disease patients (around 3000 patients) are needed to confirm these findings using genetic studies alone and functional and expression data can help delineate this region[19].
The function of the immune system is to be able to recognise the vast number of antigens that are present in our environment, to mount an appropriate response to invasive pathogens and also to limit damage during inflammation and immune response to these pathogens. Dysregulation of the immune system can lead to immune-mediated diseases such as inflammatory bowel disease. Following the discovery of NOD2/CARD15, attention has focussed on the innate immune system[20]. From an evolutionary prospective, the innate immune response is more ancient than the adaptive immune response present in nearly all taxa[21]. As a further measure of the importance of the innate immune response in plants, genes involved in plant disease resistance occupy >1% of the genome of primitive species, such as Rockcress[22].
Toll like receptors (TLRs) are transmembrane proteins which play a pivotal role in mediating the innate immune response to viral, bacterial and fungal pathogens[23]. TLRs were initially identified in Drosophila and they are widely conserved across animal species[24]. In mammals TLRs are integral membrane glycoproteins which recognise conserved products unique to microbial metabolism and signal via a number of downstream signalling molecules (MyD88, Il-1R-associated kinases, TGF-β, and TNF- receptor associated factor 6)[25]. To date 11 members of the TLR family have been identified in mammals[24].
TLR4 functions as a sensor for lipopolysaccharide (LPS), part of the cell membrane of Gram negative bacteria[26], and as such has been considered a strong functional candidate gene in the pathogenesis of inflammatory bowel disease as well as in other immunoinflammatory diseases. Variant alleles of the TLR4 Asp299Gly polymorphism have been associated with decreased bronchial responsiveness to LPS in humans and reduced activation in transfection experiments[27]. The Asp299Gly polymorphism has been associated with Crohn’s disease and ulcerative colitis both in a cohort of Belgian patients[28] and in cohorts from Greece[29] (Table 1).
| Healthy controls (%) | Ulcerative colitis (%) | Crohn’s disease (%) | P value for controls versus Crohn’s disease(NS: not significant) | |
| Franchimont et al, Belgium (28) | 139 (5.0) | 163 (10.0) | 334 (11.0) | 0.007 |
| Arnott et al, Scotland (32) | 189 (8.8) | 246 (6.8) | 234 (10.3) | NS |
| Torok et al, Germany (30) | 145 (4.0) | 98 (9.0) | 102 (7.0) | NS |
| Gazouli et al, Greece (29) | 100 (3) | 85 (3.5) | 120 (7.9) | 0.026 |
| Brand et al, Germany ( 31) | 199 (7.5) | 204 (14.2) | 0.038 | |
| Oostenbrug et al, Netherlands (33) | 296 (4.6) | 179 (5.9) | 393 (6.7) | NS |
An association was observed between ulcerative colitis and the TLR4 Thr399Ile polymorphism but not the Asp299Gly variant in one German cohort[30]. However, in a further cohort of German patients Asp299Gly variants were associated with Crohn’s disease[31]. No association between inflammatory bowel disease and the TLR4 Asp299Gly polymorphism has been observed in cohorts of patients from Scotland[32]. In the light of these discordant data-sets, it is difficult to ascertain whether these two polymorphisms represent the critical disease causing mutations in the TLR4 gene[33]. Recent data from two independent mouse models have shown that treatment with TLR4 antagonists may prevent the development of colitis, further suggesting that this innate immune receptor may be central to the pathogenesis of inflammatory bowel disease[34].
TLR5 functions by recognising flagellin, a common bacterial antigen that is present in enteric bacteria.[35, 36]. Strong serological response to flagellin in multiple animal models of colitis has been observed by Lodes and colleagues[37], who demonstrated that colitis can be induced by transferring flagellin-specific T cells to immunodeficient animals. Further evidence implicating TLR5 polymorphisms in the pathogenesis of inflammatory bowel disease suggests that carriage of a dominant negative TLR5 polymorphism (TLR5-stop) appears to protect against Crohn’s disease and results in a significant reduction in flagellin-specific circulating concentrations of IgA and IgG[38]. These data linking a genetic defect in TLR5 to an alteration in the acquired immune response are intriguing and of great pertinence as recent studies report synergism between NOD2/CARD15 and TLR5 signalling[39].
NOD (nucleotide- binding oligomerization domain) - LLR (leucine rich region) proteins are part of the CATERPILLER (CARD, transcription enhancer, R [purine]- binding, pyrin, lots of purine repeats) family, and these proteins are key regulators of innate immunity and apoptosis in mammals and plants[40]. NOD- LLR proteins play a central role in recognising pathogens on the cell surface and in the cytosol, and a family of greater than twenty human proteins possessing a NOD domain has been identified[41]. NOD- LLR proteins are comprised of three distinct functional domains: the amino terminal effector domain involved in signalling, the centrally located NOD domain and the LLR ligand-sensing domain[42]. Three of the human NOD-LLR proteins have effector domains, namely caspase- recruitment domains (CARD): NOD1/CARD4, NOD2/CARD15 and Apaf-1.[40]
With the established role of NOD2/CARD15 in the pathogenesis of Crohn’s disease, attention has focused on NOD1/CARD4 identified by two groups in 1999[43,44]. NOD1/CARD4 has a similar structure to NOD2/CARD15 with it having only one CARD domain, a central NOD domain and a LLR[45]. NOD1 detects intracellular diaminopimelic acid, a tripeptide motif found in many Gram negative bacteria and unique to Gram negative proteoglycans[46,47]. NOD1/CARD4 plays a role in colonic epithelial defence against the intracellular pathogens E. coli and Shigella flexeneri[48,49]. Its effector domain is associated with RipK2( a CARD-containing interleukin-1 beta-converting enzyme-associated kinase), thus mediating NFκB activation[43,44,50].
Linkage to the area that encompasses the NOD1/CARD4 gene, chromosome 7p14, was first found on a genome wide scan of the UK population in 1996[13]. This initial finding is replicated in two further genome wide scans involving patients in North America (USA and Canada)[6,10]. In a pan-European study involving 381 inflammatory bowel disease families from France, Sweden, Belgium, Spain, Denmark, Italy and Ireland (235 CD and 58 UC), no association between susceptibility to IBD and the E266K variant which lies within the CARD domain of NOD1/CARD4 was observed[51].
However, a further recent UK study of 556 IBD trios (294 CD and 252 UC) a significant association was observed between a complex insertion deletion allele (CCCCCCAC/ CCCCCCCCC) of NOD1/CARD4 at nucleotides 30, 258, 950 (Ensembl build 35) which is partially identified by rs6958571 and is located within intron 9, and predisposition to inflammatory bowel disease (P = 0.002) and, also to ulcerative colitis (P = 0.01)[52]. By creating a sliding two marker haplotype using this insertion deletion allele and the rs2907748 (T → C) variant at nucleotides 30,246,263, the association was strengthened with P values of 3 × 10-6 for inflammatory bowel disease, 7 × 10-4 for Crohn’s disease and 3 × 10-5 for ulcerative colitis. The deletion allele was significantly associated with an early age of onset of IBD (<25 years) and there was no evidence of an epistatic effect when NOD1/CARD4, NOD2/CARD15 and IBD5 are investigated.
Interestingly allelic variation of the same insertion deletion polymorphism (CCCCCCAC/ CCCCCCCCC) at nucleotides 30, 258, 950, which appear to alter the protein binding of NOD1/CARD4, is also associated with susceptibility to childhood onset of asthma and elevated serum IgE levels[53]. Further genetic and functional data with regards to NOD1/CARD4 are required to discover whether these variants play a role in the pathogenesis of inflammatory bowel disease in racially diverse populations.
CC chemokine ligand (CCL) 20 is responsible for the chemoattraction of immature dendritic cells (DC) expressing the CCR6 receptor in the epithelium and Peyer’s patches in the bowel mucosa[54]. The ligand- receptor pair of CCL20 and CCR6 also play a role in the chemoattraction of effector/memory T-cells and B-cells under homeostatic and inflammatory conditions, in diseases including cancer and rheumatoid arthritis[55]. Microarray and real-time PCR analysis of endoscopic colonic biopsies of patients with IBD have revealed increased levels of CCL20 mRNA in histologically inflamed biopsies when compared to non-inflamed IBD biopsies and inflamed non-IBD colonic biopsies[56,57].
Using microarray analysis of peripheral blood mononuclear cells in 8 patients with ulcerative colitis and 8 healthy controls, Choi and colleagues showed that the CCL20 gene is upregulated in ulcerative colitis samples[58]. Five SNPs in the promoter region of CCL20 are identified and analysed in a case control study of 118 ulcerative colitis patients and 300 healthy controls in the Korean population. Three of the CCL20 variants are significantly associated with ulcerative colitis (P < 0.0038) and further replication data in ethnically diverse cohorts of patients with IBD are required.
The identification of other genetic variants in loci identified by genome wide scan continues to yield other potential determinants of inflammatory bowel disease. These genes may play a role that is limited to specific populations and disease phenotypes. However, their identification can unravel the pathogenesis of inflammatory bowel disease and may reveal new therapeutic targets.
Other loci of current interest include the IBD2 locus on chromosome 12 first described in the UK[13] and subsequent analysis suggests that this locus is most strongly associated with colonic disease[59]. In the Flemish population the IBD4 region on chromosome 14 appears to be an important determinant of Crohn’s disease[60] and this association has been replicated in the USA[14] and by the International IBD genetics consortium[61]. Fine mapping of the IBD2 and the IBD4 loci are required to establish which genes in these regions contain critical disease causing mutations.
With the advent of the HapMap project[62] and plans to include information on around 300 million genotypes, genome-wide association studies have been proposed as a powerful tool in identifying common variants that contribute to complex genetic traits[63]. Genome wide association studies compare the frequency of alleles and genotypes between cases and controls on a genome-wide scale, thus creating a comprehensive unbiased method to identify candidate genes. Advances in technology lowering the price of genotyping on a large scale and genome wide linkage disequilibrium mapping using data from the HapMap project have made this method available. However, significant concerns remain about the power of these studies and it has been proposed that levels of significance of P < 5 × 10-8 are required to constitute significance in genome wide association studies[64].
Using this approach Tamiya and colleagues[65] placed 27 039 microsatellite markers across the human genome at 100kb intervals in 470 patients with rheumatoid arthritis and 470 controls. Forty seven candidate regions were identified and the previously implicated major histocompatibility complex gene HLA- DRB1 in the chromosomal region 6p21.3 was shown to be associated with rheumatoid arthritis. In the UK,the Wellcome Trust has provided initial funding for a phase I study including 1 000 patients with Crohn’s disease and 3 000 controls, 675 000 SNPs will be genotyped to cover the entire genome and results are expected in 12 months.
The yeast two-hybrid assay is an elegant means of investigating protein-protein interactions, which have become increasingly important in our understanding of biological systems and pathways. The yeast two- hybrid model can also be used to characterize interactions already known to occur, thus helping delineate the protein domains responsible for interaction and the environmental conditions involved[66].
The yeast two-hybrid assay is performed in the budding yeast, S cerevisiae using two fusion proteins: the target protein of interest, known as ‘bait’ is fused to a DNA binding domain attached to its N-terminus. The second protein, the ‘prey’ is fused to an activation domain. If the bait protein interacts with the prey, these bring the binding domain and the activation domain of transcriptional activator together, which in turn switches on the expression of the reporter genes. The reporter genes are constructed to allow growth of the yeast in a selective medium when the interaction occurs. When investigating ‘protein- protein’ interactions, a single bait protein is used to search for interaction with a library of proteins fused to the activation domain. The choice of library is determined by the tissue of interest, e.g. intestinal cell library for inflammatory bowel disease[67].
Schizophrenia is a disease of polygenic genetic susceptibility where the yeast two hybrid systems have been successfully used to identify candidate genes. The disrupted-in- schizophrenia 1 (DISC1) gene was identified in 2 000[68] and confirmed in other cohorts,[69,70] was used as bait. DISC1 encodes a novel protein of unknown function and full-length human DISC1 protein was used to screen human adult and foetal brain libraries for interacting proteins, using the yeast two-hybrid system[71]. Twenty-one proteins from a variety of locations have been identified implicating DISC1 in several aspects of central nervous system signalling and confirming data from other yeast two- hybrid scans using DISC1 as bait[72,73]. From these data the authors are able to identify a number of potential DISC1 interactions and to speculate that DISC1 may be at the centre of an extensive protein interaction network.
Barnich and colleagues[74] used NOD2/CARD15 gene as ‘bait’ to screen a bone marrow library and identified GRIM19, a protein with homology to the NADPH dehydrogenase which interacts with endogenous NOD2[74]. GRIM19 is required for NFκB activation following NOD2-mediated recognition of bacterial muramyl dipeptide and the authors hypothesised that GRIM19 is a key component of the CARD15/NOD2 signalling pathway which currently remains under detailed investigation[75-77]. In our own protein yeast two- hybrid studies a series of 12 candidate genes which interact with CARD15/NOD2 (Nimmo, Satsangi, unpublished data) have been identified.
Gene expression technology using microarray allows a comprehensive picture of gene expression at the tissue and cellular level, thus helping understand the underlying physiological and pathological processes. Microarray technology has developed from spotted nylon array technology used to identify genomic inserts in bacterial colonies by hybridisation with preidentified cDNAs[78]. In the seminal microarray experiment in 1995, a two-colour fluorescent pattern of differential gene expression is generated when comparing 48 genes in the root and the shoot of Arabidopsis[79].
Since 1995 there has been a rapid increase in the number of papers published using microarray technology from 7 in 1995-1996 to 139 in 1999 and to 3 000 in 2003[80]. The initial optimism set out by Mark Schena, one of the authors of the Arabidopsis microarray experiment suggesting that all human diseases can be studied by microarray technology, With the ultimate goal of this work to develop effective treatments and cures for every human disease by 2 050 has not yet fully been born out by the published data. However, a number of exciting and novel observations have been generated.
RNA isolated from the tissue sample or cells can be used to generate cRNA or cDNA[78]. The probe set is then designed with transcripts targeting the genes of interest and care needs to be taken to prevent cross- hybridization. It is worth noting at this point that the probe refers to the reporter sequence placed at a particular position on the array which interrogates the sample and not the other way around. The probes are hybridised on the chip with the target unknown cRNA or cDNA. Following this process which can take several hours, the unbound target is washed off and the arrays are fluorescently labelled, so that they can be analyzed by confocal laser scanning. Expression of the entire human genome can be analyzed on one chip and complex computational and statistical techniques have been developed to analyze expression data because of the large amount of data microarrays[81].
Probably the most provocative and clinically relevant data generated by microarray have been in the field of cancer research. An example of the potential of microarray has demonstrated by Alizadeh and colleagues[82]. By comparing expression analysis on a ‘lymphochip’ panel of 3186 genes between patients with diffuse large B- cell lymphoma (DLBCL) prior to treatment, the investigators are able to group these patients into two discrete groups: germinal centre DLBCL and activated DLBCL. When the clinical progress of these two groups of patients were examined, the germinal cell DLBCL had a higher five year survival rate than the activated DLBCL (75% n = 25 versus 16% n = 37 respectively, P ≤ 0.01). Gene expression profiles have also been successfully used to predict prognosis in 295 patients with breast cancer[83]. However, a meta- analysis of 84 studies found that DNA microarrays have a variable performance in measuring prognosis in a number of different cancers[84].
Microarray has been used to compare synovial tissue obtained from patients with severe rheumatoid arthritis and macroscopically affected bowel of patients undergoing surgery for Crohn’s disease[85]. A number of inflammatory genes were commonly expressed in diseased tissues. In 2000 Dieckgraefe and colleagues[86] published a study using microarray technology to compare patients with ulcerative colitis undergoing colectomies for disease refractory to medical management and a control group. Six thousand five hundred genes were analysed and the results confirmed an increase in a number of genes previously implicated in the pathogenesis of ulcerative colitis (IL-1, IL-1 RA and IL8) and suggested that multiple members of the chemokine subfamily may play a role in disease pathogenesis.
In a further microarray experiment using surgically resected inflammatory bowel disease tissue, 7 070 genes were examined and 170 genes were differentially expressed in ulcerative colitis and Crohn’s disease with almost an equal number up regulated and down regulated[87]. Twenty percent of the differentially regulated genes were common to both forms of inflammatory bowel disease and when the locations of these genes were mapped several were found to lie within IBD2 locus on chromosome 12.
More recently Langman and colleagues[88] used microarray to analyze biopsies taken from patients with Crohn’s disease, ulcerative colitis and control patients, 22 283 genes were analysed. They found that genes involved in cellular detoxification and biotransformation (pregnane X receptor and MDR1) are significantly down regulated in the colon of patients with ulcerative colitis.
Endoscopic investigation of inflammatory bowel disease with the ability to take pinch mucosal biopsies has allowed investigators to take microarray tissue from a larger range of patients including those with less severe disease compared to patients requiring rescectional surgery. In a study of 24 patients with ulcerative colitis, Okahara and colleagues[89] investigated the difference in gene expression between endoscopic biopsies taken from inflamed and non inflamed areas using a 1300 gene microarray, and found that migration inhibitory factor- related protein 14 (MRP14), growth-related oncogene gamma (GROγ) and serum amyloid A1 (SAA1) were upregulated whereas TIMP1 and PDZ and LIM domain 1 (elfin) were down regulated in the inflamed biopsies when compared to the non- inflamed biopsies.
In a study involving endoscopic biopsies of patients with Crohn’s disease, ulcerative colitis and controls, Costello and colleagues[90] found that 500 and 272 transcripts are differentially regulated in CD and UC, respectively. Candidate genes are confirmed by real-time PCR and immunohistochemistry, and a number of genes involved in immune regulation were identified.
Microarray has been useful in studies thus far and continues to be a powerful tool in investigation of the pathogenesis of inflammatory bowel disease. Further studies need to be designed in order to allow the collection of accurate clinical data describing the phenotype and activity of inflammatory bowel disease at the time of sample collection as these data are critical in analysis and interpretation of the results. The confounding problems associated with the heterogeneous mixture of cells, which is inevitable in the analysis of entire biopsy samples, can be reduced by using laser capture micro-dissection, in order to analyze only the cells of interest. By obtaining painstakingly accurate clinical information at the time of biopsy collection and narrowing down cell heterogeneity, the amount of background ‘noise’ that has hampered previous microarray studies can be reduced[91]. Minimum standards of information reported on microarray data have been proposed with the aim of establishing a standard for recording micoarray-based gene expression data[92], and replication of results with real-time PCR helps to validate these studies.
Proteomic analysis is another relatively new investigative tool that has not been used to any large extent in the field of inflammatory bowel disease. Proteomics refers to the study of the total protein content in cells and the products of genes, so as to identify differences between normal and diseased tissue[93]. This is achieved by combining the techniques of protein electrophoresis and mass spectrometry. Expression profiles of proteomes may be generated from samples of serum or secreted fluid, and may be able to differentiate disease progression, response to therapy and to identify novel therapeutic targets.
Preliminary data have been generated in our unit by comparing serum of patients with severe ulcerative colitis who responded to corticosteroid therapy and matched patients who were resistant to and failed corticosteroid therapy[94]. Proteomic profiles of corticosteroid resistant and responsive groups are significantly different at 19 protein biomarkers: 12 proteins were up-regulated and 7 proteins were down-regulated in the corticosteroid resistant group. These results suggest that protein profiling may be useful in predicting patient response to corticosteroid therapy and identification of these proteins is currently underway.
The application of novel technologies has catalyzed the search for novel determinants in inflammatory bowel disease. Proof of the principle for genome wide studies is provided by the NOD2/CARD15 discovery and investigation of genes at other loci is underway. Rigorous attention to statistical design and phenotypic classification of disease is of paramount importance in gene discovery.
With the advent of new powerful investigative tools such as microarray and proteomic analysis, large amounts of data can be generated from small studies in inflammatory bowel disease. Among the clinical challenges in harnessing the power of these new investigative tools will be to accurately define clinical phenotype in patients studied, given the heterogeneity inevitable in these studies.
S- Editor Guo SY L- Editor Wang XL E- Editor Ma WH
| 1. | Hugot JP, Chamaillard M, Zouali H, Lesage S, Cézard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599-603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4223] [Cited by in RCA: 3901] [Article Influence: 162.5] [Reference Citation Analysis (0)] |
| 2. | Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603-606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3555] [Cited by in RCA: 3474] [Article Influence: 144.8] [Reference Citation Analysis (1)] |
| 3. | Schreiber S, Rosenstiel P, Albrecht M, Hampe J, Krawczak M. Genetics of Crohn disease, an archetypal inflammatory barrier disease. Nat Rev Genet. 2005;6:376-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 220] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 4. | Satsangi J, Sutherland LR, Duerr RH. Inflammatory bowel diseases. 1st edition. Oxford. Churchill Livingstone. 2003;29-43. |
| 5. | Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3803] [Cited by in RCA: 3642] [Article Influence: 121.4] [Reference Citation Analysis (0)] |
| 6. | Cho JH, Nicolae DL, Gold LH, Fields CT, LaBuda MC, Rohal PM, Pickles MR, Qin L, Fu Y, Mann JS. Identification of novel susceptibility loci for inflammatory bowel disease on chromosomes 1p, 3q, and 4q: evidence for epistasis between 1p and IBD1. Proc Natl Acad Sci U S A. 1998;95:7502-7507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 279] [Cited by in RCA: 272] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 7. | Duerr RH, Barmada MM, Zhang L, Achkar JP, Cho JH, Hanauer SB, Brant SR, Bayless TM, Baldassano RN, Weeks DE. Evidence for an inflammatory bowel disease locus on chromosome 3p26: linkage, transmission/disequilibrium and partitioning of linkage. Hum Mol Genet. 2002;11:2599-2606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 8. | Hampe J, Lynch NJ, Daniels S, Bridger S, Macpherson AJ, Stokkers P, Forbes A, Lennard-Jones JE, Mathew CG, Curran ME. Fine mapping of the chromosome 3p susceptibility locus in inflammatory bowel disease. Gut. 2001;48:191-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 55] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 9. | Ma Y, Ohmen JD, Li Z, Bentley LG, McElree C, Pressman S, Targan SR, Fischel-Ghodsian N, Rotter JI, Yang H. A genome-wide search identifies potential new susceptibility loci for Crohn's disease. Inflamm Bowel Dis. 1999;5:271-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 110] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 10. | Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, McLeod RS, Griffiths AM, Green T, Brettin TS, Stone V, Bull SB. Genomewide search in Canadian families with inflammatory bowel disease reveals two novel susceptibility loci. Am J Hum Genet. 2000;66:1863-1870. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 361] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 11. | Rioux JD, Daly MJ, Silverberg MS, Lindblad K, Steinhart H, Cohen Z, Delmonte T, Kocher K, Miller K, Guschwan S. Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Nat Genet. 2001;29:223-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 566] [Cited by in RCA: 544] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 12. | Hampe J, Schreiber S, Shaw SH, Lau KF, Bridger S, Macpherson AJ, Cardon LR, Sakul H, Harris TJ, Buckler A. A genomewide analysis provides evidence for novel linkages in inflammatory bowel disease in a large European cohort. Am J Hum Genet. 1999;64:808-816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 258] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 13. | Satsangi J, Parkes M, Louis E, Hashimoto L, Kato N, Welsh K, Terwilliger JD, Lathrop GM, Bell JI, Jewell DP. Two stage genome-wide search in inflammatory bowel disease provides evidence for susceptibility loci on chromosomes 3, 7 and 12. Nat Genet. 1996;14:199-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 476] [Cited by in RCA: 458] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 14. | Duerr RH, Barmada MM, Zhang L, Pfützer R, Weeks DE. High-density genome scan in Crohn disease shows confirmed linkage to chromosome 14q11-12. Am J Hum Genet. 2000;66:1857-1862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 134] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 15. | Hugot JP, Laurent-Puig P, Gower-Rousseau C, Olson JM, Lee JC, Beaugerie L, Naom I, Dupas JL, Van Gossum A, Orholm M. Mapping of a susceptibility locus for Crohn's disease on chromosome 16. Nature. 1996;379:821-823. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 649] [Cited by in RCA: 598] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 16. | Stoll M, Corneliussen B, Costello CM, Waetzig GH, Mellgard B, Koch WA, Rosenstiel P, Albrecht M, Croucher PJ, Seegert D. Genetic variation in DLG5 is associated with inflammatory bowel disease. Nat Genet. 2004;36:476-480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 339] [Cited by in RCA: 315] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 17. | Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES. High-resolution haplotype structure in the human genome. Nat Genet. 2001;29:229-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1212] [Cited by in RCA: 1059] [Article Influence: 44.1] [Reference Citation Analysis (0)] |
| 18. | Peltekova VD, Wintle RF, Rubin LA, Amos CI, Huang Q, Gu X, Newman B, Van Oene M, Cescon D, Greenberg G. Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat Genet. 2004;36:471-475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 572] [Cited by in RCA: 561] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 19. | Noble CL, Nimmo ER, Drummond H, Ho GT, Tenesa A, Smith L, Anderson N, Arnott ID, Satsangi J. The contribution of OCTN1/2 variants within the IBD5 locus to disease susceptibility and severity in Crohn's disease. Gastroenterology. 2005;129:1854-1864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 90] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 20. | Ting JP, Davis BK. CATERPILLER: a novel gene family important in immunity, cell death, and diseases. Annu Rev Immunol. 2005;23:387-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 264] [Cited by in RCA: 262] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 21. | Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1:135-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2861] [Cited by in RCA: 2899] [Article Influence: 120.8] [Reference Citation Analysis (0)] |
| 22. | Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND. Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 1999;20:317-332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 560] [Cited by in RCA: 443] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 23. | Hoffmann JA. The immune response of Drosophila. Nature. 2003;426:33-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1054] [Cited by in RCA: 1063] [Article Influence: 48.3] [Reference Citation Analysis (0)] |
| 24. | Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499-511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6034] [Cited by in RCA: 6310] [Article Influence: 300.5] [Reference Citation Analysis (0)] |
| 25. | Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4232] [Cited by in RCA: 4140] [Article Influence: 188.2] [Reference Citation Analysis (0)] |
| 26. | Beutler B. Tlr4: central component of the sole mammalian LPS sensor. Curr Opin Immunol. 2000;12:20-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 577] [Cited by in RCA: 581] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 27. | Arbour NC, Lorenz E, Schutte BC, Zabner J, Kline JN, Jones M, Frees K, Watt JL, Schwartz DA. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat Genet. 2000;25:187-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1504] [Cited by in RCA: 1472] [Article Influence: 58.9] [Reference Citation Analysis (0)] |
| 28. | Franchimont D, Vermeire S, El Housni H, Pierik M, Van Steen K, Gustot T, Quertinmont E, Abramowicz M, Van Gossum A, Devière J. Deficient host-bacteria interactions in inflammatory bowel disease The toll-like receptor (TLR)-4 Asp299gly polymorphism is associated with Crohn's disease and ulcerative colitis. Gut. 2004;53:987-992. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 438] [Cited by in RCA: 445] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 29. | Gazouli M, Mantzaris G, Kotsinas A, Zacharatos P, Papalambros E, Archimandritis A, Ikonomopoulos J, Gorgoulis VG. Association between polymorphisms in the Toll-like receptor 4, CD14, and CARD15/NOD2 and inflammatory bowel disease in the Greek population. World J Gastroenterol. 2005;11:681-685. [PubMed] |
| 30. | Török HP, Glas J, Tonenchi L, Mussack T, Folwaczny C. Polymorphisms of the lipopolysaccharide-signaling complex in inflammatory bowel disease: association of a mutation in the Toll-like receptor 4 gene with ulcerative colitis. Clin Immunol. 2004;112:85-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 164] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 31. | Brand S, Staudinger T, Schnitzler F, Pfennig S, Hofbauer K, Dambacher J, Seiderer J, Tillack C, Konrad A, Crispin A. The role of Toll-like receptor 4 Asp299Gly and Thr399Ile polymorphisms and CARD15/NOD2 mutations in the susceptibility and phenotype of Crohn's disease. Inflamm Bowel Dis. 2005;11:645-652. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 125] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 32. | Arnott ID, Nimmo ER, Drummond HE, Fennell J, Smith BR, MacKinlay E, Morecroft J, Anderson N, Kelleher D, O'Sullivan M. NOD2/CARD15, TLR4 and CD14 mutations in Scottish and Irish Crohn's disease patients: evidence for genetic heterogeneity within Europe. Genes Immun. 2004;5:417-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 152] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 33. | Oostenbrug LE, Drenth JP, de Jong DJ, Nolte IM, Oosterom E, van Dullemen HM, van der Linde K, te Meerman GJ, van der Steege G, Kleibeuker JH. Association between Toll-like receptor 4 and inflammatory bowel disease. Inflamm Bowel Dis. 2005;11:567-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 90] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 34. | Fort MM, Mozaffarian A, Stöver AG, Correia Jda S, Johnson DA, Crane RT, Ulevitch RJ, Persing DH, Bielefeldt-Ohmann H, Probst P. A synthetic TLR4 antagonist has anti-inflammatory effects in two murine models of inflammatory bowel disease. J Immunol. 2005;174:6416-6423. [PubMed] |
| 35. | Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, Eng JK, Akira S, Underhill DM, Aderem A. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099-1103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2603] [Cited by in RCA: 2574] [Article Influence: 107.3] [Reference Citation Analysis (0)] |
| 36. | Winstanley C, Morgan JA. The bacterial flagellin gene as a biomarker for detection, population genetics and epidemiological analysis. Microbiology. 1997;143:3071-3084. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 81] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 37. | Lodes MJ, Cong Y, Elson CO, Mohamath R, Landers CJ, Targan SR, Fort M, Hershberg RM. Bacterial flagellin is a dominant antigen in Crohn disease. J Clin Invest. 2004;113:1296-1306. [PubMed] |
| 38. | Gerwitz AT, Vijay-Kumar M, Swanson E, Duerr RH, Brant SR, Cho J. Common Dominant-Negative Tlr5 Polymorphism Reduces Adaptive Immune Response to Flagellin and Provides Protection from Crohn's Disease. Gastroenterology. 2005;128:A55. |
| 39. | Netea MG, Ferwerda G, de Jong DJ, Jansen T, Jacobs L, Kramer M, Naber TH, Drenth JP, Girardin SE, Kullberg BJ. Nucleotide-binding oligomerization domain-2 modulates specific TLR pathways for the induction of cytokine release. J Immunol. 2005;174:6518-6523. [PubMed] |
| 40. | Inohara C, Nuñez G. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu Rev Biochem. 2005;74:355-383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 728] [Cited by in RCA: 708] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 41. | Ting JP, Williams KL. The CATERPILLER family: an ancient family of immune/apoptotic proteins. Clin Immunol. 2005;115:33-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 42. | Russell RK, Nimmo ER, Satsangi J. Molecular genetics of Crohn's disease. Curr Opin Genet Dev. 2004;14:264-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 40] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 43. | Bertin J, Nir WJ, Fischer CM, Tayber OV, Errada PR, Grant JR, Keilty JJ, Gosselin ML, Robison KE, Wong GH. Human CARD4 protein is a novel CED-4/Apaf-1 cell death family member that activates NF-kappaB. J Biol Chem. 1999;274:12955-12958. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 267] [Cited by in RCA: 275] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 44. | Inohara N, Koseki T, del Peso L, Hu Y, Yee C, Chen S, Carrio R, Merino J, Liu D, Ni J. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-kappaB. J Biol Chem. 1999;274:14560-14567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 556] [Cited by in RCA: 560] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 45. | Inohara N, Ogura Y, Nuñez G. Nods: a family of cytosolic proteins that regulate the host response to pathogens. Curr Opin Microbiol. 2002;5:76-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 163] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 46. | Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L, Ogura Y, Kawasaki A, Fukase K, Kusumoto S. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4:702-707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 928] [Cited by in RCA: 953] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 47. | Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jéhanno M, Viala J, Tedin K, Taha MK, Labigne A, Zähringer U. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science. 2003;300:1584-1587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1178] [Cited by in RCA: 1159] [Article Influence: 52.7] [Reference Citation Analysis (0)] |
| 48. | Girardin SE, Tournebize R, Mavris M, Page AL, Li X, Stark GR, Bertin J, DiStefano PS, Yaniv M, Sansonetti PJ. CARD4/Nod1 mediates NF-kappaB and JNK activation by invasive Shigella flexneri. EMBO Rep. 2001;2:736-742. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 468] [Cited by in RCA: 475] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 49. | Kim JG, Lee SJ, Kagnoff MF. Nod1 is an essential signal transducer in intestinal epithelial cells infected with bacteria that avoid recognition by toll-like receptors. Infect Immun. 2004;72:1487-1495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 177] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 50. | Ogura Y, Inohara N, Benito A, Chen FF, Yamaoka S, Nunez G. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J Biol Chem. 2001;276:4812-4818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1034] [Cited by in RCA: 1040] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 51. | Zouali H, Lesage S, Merlin F, Cézard JP, Colombel JF, Belaiche J, Almer S, Tysk C, O'Morain C, Gassull M. CARD4/NOD1 is not involved in inflammatory bowel disease. Gut. 2003;52:71-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 53] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 52. | McGovern DP, Hysi P, Ahmad T, van Heel DA, Moffatt MF, Carey A, Cookson WO, Jewell DP. Association between a complex insertion/deletion polymorphism in NOD1 (CARD4) and susceptibility to inflammatory bowel disease. Hum Mol Genet. 2005;14:1245-1250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 238] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 53. | Hysi P, Kabesch M, Moffatt MF, Schedel M, Carr D, Zhang Y, Boardman B, von Mutius E, Weiland SK, Leupold W. NOD1 variation, immunoglobulin E and asthma. Hum Mol Genet. 2005;14:935-941. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 206] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 54. | Iwasaki A, Kelsall BL. Localization of distinct Peyer's patch dendritic cell subsets and their recruitment by chemokines macrophage inflammatory protein (MIP)-3alpha, MIP-3beta, and secondary lymphoid organ chemokine. J Exp Med. 2000;191:1381-1394. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 450] [Cited by in RCA: 445] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 55. | Schutyser E, Struyf S, Van Damme J. The CC chemokine CCL20 and its receptor CCR6. Cytokine Growth Factor Rev. 2003;14:409-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 544] [Cited by in RCA: 618] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 56. | Kaser A, Ludwiczek O, Holzmann S, Moschen AR, Weiss G, Enrich B, Graziadei I, Dunzendorfer S, Wiedermann CJ, Mürzl E. Increased expression of CCL20 in human inflammatory bowel disease. J Clin Immunol. 2004;24:74-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 57. | Puleston J, Cooper M, Murch S, Bid K, Makh S, Ashwood P, Bingham AH, Green H, Moss P, Dhillon A. A distinct subset of chemokines dominates the mucosal chemokine response in inflammatory bowel disease. Aliment Pharmacol Ther. 2005;21:109-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 100] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 58. | Choi S, Seo E, Lee C, Jun C, Kim T, Nah Y, Son Y, Kim C, Choi C, Kim S. Molecular Variations in the Promoter Region Of Mip-3a/CCL20 Gene and Relationship To Its mRNA Expression in Patients With Ulcerative Colitis. Gastroenterology. 2005;128:A137. |
| 59. | Crawford N, Uthoff S, Eichenberger M, Cobbs G, Petras R, Martin E, Galandiuk S. Characterization of genotype-phenotype correlations show that the IBD2 susceptibility locus is associated with colonic Crohn's disease and ulcerative colitis. Gastroenterology. 2003;124:A48. [DOI] [Full Text] |
| 60. | Vermeire S, Rutgeerts P, Van Steen K, Joossens S, Claessens G, Pierik M, Peeters M, Vlietinck R. Genome wide scan in a Flemish inflammatory bowel disease population: support for the IBD4 locus, population heterogeneity, and epistasis. Gut. 2004;53:980-986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 62] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 61. | Pierik M, Yang H, Barmada MM, Cavanaugh JA, Annese V, Brant SR, Cho JH, Duerr RH, Hugot JP, McGovern DP. The IBD international genetics consortium provides further evidence for linkage to IBD4 and shows gene-environment interaction. Inflamm Bowel Dis. 2005;11:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 43] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 62. | The International HapMap Project. Nature. 2003;426:789-796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4437] [Cited by in RCA: 4313] [Article Influence: 205.4] [Reference Citation Analysis (0)] |
| 63. | Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1893] [Cited by in RCA: 1770] [Article Influence: 88.5] [Reference Citation Analysis (0)] |
| 64. | Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516-1517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3635] [Cited by in RCA: 3344] [Article Influence: 115.3] [Reference Citation Analysis (0)] |
| 65. | Tamiya G, Shinya M, Imanishi T, Ikuta T, Makino S, Okamoto K, Furugaki K, Matsumoto T, Mano S, Ando S. Whole genome association study of rheumatoid arthritis using 27 039 microsatellites. Hum Mol Genet. 2005;14:2305-2321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 90] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 66. | Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. 4th Edition New York. Garland Publishing. 2002;. |
| 67. | Lodish H, Berk A, Zipursky S L, Matsudairsa P, Baltimore D, Darnell J. Molecular Cell Biology. 4th ed . New York WH. Freedman and Co. 2000;. |
| 68. | Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, Devon RS, St Clair DM, Muir WJ, Blackwood DH. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000;9:1415-1423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 966] [Cited by in RCA: 960] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 69. | Blackwood DH, Fordyce A, Walker MT, St Clair DM, Porteous DJ, Muir WJ. Schizophrenia and affective disorders--cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet. 2001;69:428-433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 562] [Cited by in RCA: 562] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 70. | Ekelund J, Hovatta I, Parker A, Paunio T, Varilo T, Martin R, Suhonen J, Ellonen P, Chan G, Sinsheimer JS. Chromosome 1 loci in Finnish schizophrenia families. Hum Mol Genet. 2001;10:1611-1617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 226] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 71. | Millar JK, Christie S, Porteous DJ. Yeast two-hybrid screens implicate DISC1 in brain development and function. Biochem Biophys Res Commun. 2003;311:1019-1025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 129] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 72. | Morris JA, Kandpal G, Ma L, Austin CP. DISC1 (Disrupted-In-Schizophrenia 1) is a centrosome-associated protein that interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: regulation and loss of interaction with mutation. Hum Mol Genet. 2003;12:1591-1608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 305] [Cited by in RCA: 306] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 73. | Ozeki Y, Tomoda T, Kleiderlein J, Kamiya A, Bord L, Fujii K, Okawa M, Yamada N, Hatten ME, Snyder SH. Disrupted-in-Schizophrenia-1 (DISC-1): mutant truncation prevents binding to NudE-like (NUDEL) and inhibits neurite outgrowth. Proc Natl Acad Sci U S A. 2003;100:289-294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 319] [Cited by in RCA: 318] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 74. | Barnich N, Aguirre JE, Reinecker HC, Xavier R, Podolsky DK. Membrane recruitment of NOD2 in intestinal epithelial cells is essential for nuclear factor-{kappa}B activation in muramyl dipeptide recognition. J Cell Biol. 2005;170:21-26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 215] [Cited by in RCA: 236] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 75. | Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nuñez G, Flavell RA. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731-734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1334] [Cited by in RCA: 1325] [Article Influence: 66.3] [Reference Citation Analysis (2)] |
| 76. | Maeda S, Hsu LC, Liu H, Bankston LA, Iimura M, Kagnoff MF, Eckmann L, Karin M. Nod2 mutation in Crohn's disease potentiates NF-kappaB activity and IL-1beta processing. Science. 2005;307:734-738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 583] [Cited by in RCA: 583] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 77. | Watanabe T, Kitani A, Murray PJ, Strober W. NOD2 is a negative regulator of Toll-like receptor 2-mediated T helper type 1 responses. Nat Immunol. 2004;5:800-808. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 615] [Cited by in RCA: 602] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 78. | Stoughton RB. Applications of DNA microarrays in biology. Annu Rev Biochem. 2005;74:53-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 279] [Cited by in RCA: 225] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 79. | Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6477] [Cited by in RCA: 5102] [Article Influence: 170.1] [Reference Citation Analysis (0)] |
| 80. | Marshall E. Getting the noise out of gene arrays. Science. 2004;306:630-631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 198] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 81. | Wu TD. Analysing gene expression data from DNA microarrays to identify candidate genes. J Pathol. 2001;195:53-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 82. | Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503-511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7015] [Cited by in RCA: 6301] [Article Influence: 252.0] [Reference Citation Analysis (0)] |
| 83. | van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999-2009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4800] [Cited by in RCA: 4422] [Article Influence: 192.3] [Reference Citation Analysis (0)] |
| 84. | Ntzani EE, Ioannidis JP. Predictive ability of DNA microarrays for cancer outcomes and correlates: an empirical assessment. Lancet. 2003;362:1439-1444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 217] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 85. | Heller RA, Schena M, Chai A, Shalon D, Bedilion T, Gilmore J, Woolley DE, Davis RW. Discovery and analysis of inflammatory disease-related genes using cDNA microarrays. Proc Natl Acad Sci U S A. 1997;94:2150-2155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 513] [Cited by in RCA: 481] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 86. | Dieckgraefe BK, Stenson WF, Korzenik JR, Swanson PE, Harrington CA. Analysis of mucosal gene expression in inflammatory bowel disease by parallel oligonucleotide arrays. Physiol Genomics. 2000;4:1-11. [PubMed] |
| 87. | Lawrance IC, Fiocchi C, Chakravarti S. Ulcerative colitis and Crohn's disease: distinctive gene expression profiles and novel susceptibility candidate genes. Hum Mol Genet. 2001;10:445-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 291] [Article Influence: 12.1] [Reference Citation Analysis (1)] |
| 88. | Langmann T, Moehle C, Mauerer R, Scharl M, Liebisch G, Zahn A, Stremmel W, Schmitz G. Loss of detoxification in inflammatory bowel disease: dysregulation of pregnane X receptor target genes. Gastroenterology. 2004;127:26-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 259] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 89. | Okahara S, Arimura Y, Yabana T, Kobayashi K, Gotoh A, Motoya S, Imamura A, Endo T, Imai K. Inflammatory gene signature in ulcerative colitis with cDNA macroarray analysis. Aliment Pharmacol Ther. 2005;21:1091-1097. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 34] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 90. | Costello CM, Mah N, Häsler R, Rosenstiel P, Waetzig GH, Hahn A, Lu T, Gurbuz Y, Nikolaus S, Albrecht M. Dissection of the inflammatory bowel disease transcriptome using genome-wide cDNA microarrays. PLoS Med. 2005;2:e199. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 147] [Cited by in RCA: 159] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 91. | Ioannidis JP. Microarrays and molecular research: noise discovery. Lancet. 2005;365:454-455. [PubMed] |
| 92. | Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365-371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2967] [Cited by in RCA: 2689] [Article Influence: 112.0] [Reference Citation Analysis (0)] |
| 93. | Sauer S, Lange BM, Gobom J, Nyarsik L, Seitz H, Lehrach H. Miniaturization in functional genomics and proteomics. Nat Rev Genet. 2005;6:465-476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 94] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 94. | Din S, Lennon AM, Hogarth C, Ho GT, Arnott ID, Hupp T, Satsangi J. Proteomic Profiling Identifies Corticosteroid Resistant Patients In Severe Ulcerative Colitis. Gastroenterology. 2005;128:A310. |
| 95. | Ahmad T, Tamboli CP, Jewell D, Colombel JF. Clinical relevance of advances in genetics and pharmacogenetics of IBD. Gastroenterology. 2004;126:1533-1549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 120] [Article Influence: 5.7] [Reference Citation Analysis (0)] |