Published online Jul 14, 2005. doi: 10.3748/wjg.v11.i26.4102
Revised: July 20, 2004
Accepted: July 22, 2004
Published online: July 14, 2005
AIM: To study the genetic alterations and their association with clinicopathological characteristics of hepatocellular carcinoma (HCC), and to find the tumor related DNA fragments.
METHODS: DNA isolated from tumors and corresponding noncancerous liver tissues of 56 HCC patients was amplified by random amplified polymorphic DNA (RAPD) with 10 random 10-mer arbitrary primers. The RAPD bands showing obvious differences in tumor tissue DNA corresponding to that of normal tissue were separated, purified, cloned and sequenced. DNA sequences were analyzed and compared with GenBank data.
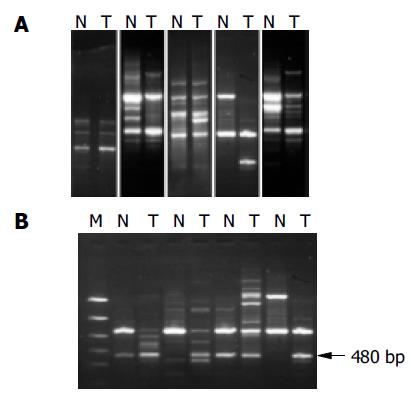
RESULTS: A total of 56 cases of HCC were demonstrated to have genetic alterations, which were detected by at least one primer. The detestability of genetic alterations ranged from 20% to 70% in each case, and 17.9% to 50% in each primer. Serum HBV infection, tumor size, histological grade, tumor capsule, as well as tumor intrahepatic metastasis, might be correlated with genetic alterations on certain primers. A band with a higher intensity of 480 bp or so amplified fragments in tumor DNA relative to normal DNA could be seen in 27 of 56 tumor samples using primer 4. Sequence analysis of these fragments showed 91% homology with Homo sapiens double homeobox protein DUX10 gene.
CONCLUSION: Genetic alterations are a frequent event in HCC, and tumor related DNA fragments have been found in this study, which may be associated with hepatocarcin-ogenesis. RAPD is an effective method for the identification and analysis of genetic alterations in HCC, and may provide new information for further evaluating the molecular mechanism of hepatocarcinogenesis.
- Citation: Xian ZH, Cong WM, Zhang SH, Wu MC. Genetic alterations of hepatocellular carcinoma by random amplified polymorphic DNA analysis and cloning sequencing of tumor differential DNA fragment. World J Gastroenterol 2005; 11(26): 4102-4107
- URL: https://www.wjgnet.com/1007-9327/full/v11/i26/4102.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i26.4102
Hepatocellular carcinoma (HCC) is one of the most common cancers worldwide and a leading cause of death in many countries, mainly in Asia and Africa[1]. It is commonly associated with chronic hepatitis B virus (HBV) and hepatitis C virus (HCV) infections, with chronic exposure to mycotoxin, aflatoxin B1 (AFB1), and is a complication of alcoholic cirrhosis[2]. The development of HCC is a multistep process associated with multiple genetic alterations, including the genetic alteration of cancer cells[3,4]. In the past several years, many molecular genetic studies have been extensively performed to clarify the molecular events related with initiation, development and progression of human cancers[4-7]. Like other cancers, a variety of genetic changes have frequently been found in HCC[8-13], including p53 mutations[1,14], loss of heterozygosity (LOH) on chromosome arms 1p, 4q, 5q, 6q, 8p, 10q, 11p, 16p, 16q, 22q[7-12]. Microsatellite instability has also been recognized in some HCCs[12,15-17]. However, no information on genetic alterations in the entire genome of HCC is available.
The random amplified polymorphic DNA (RAPD) method is a DNA fingerprinting technique based on polymerase chain reaction (PCR) amplification of random fragments of genomic DNA with single short primers of arbitrary nucleotide sequences[18]. Unlike the microsatellite instability analysis which can only detect specific microsatellite loci, the RAPD method can simply and rapidly detect genetic alterations in the entire genome without knowledge of specific DNA sequence information[19]. The RAPD method is a powerful tool for population and pedigree analysis, phylogenetic studies, and bacterial strain identification[20]. Recently, the RAPD method was used as a means for identifying the genetic alterations in human tumors and revealed that genetic alterations occurred frequently in lung cancer[21], squamous cell carcinoma of the head and neck[22], brain tumor[23], ovarian cancer[24], breast cancer[25], or human aberrant crypt foci and colon cancer[26]. In the present study, genetic alterations in HCC were investigated using RAPD in order to determine the relationship between the frequency of genetic alterations and the clinicopathological characteristics of HCC, and to find the tumor related DNA fragments.
Fifty-six liver cancer tissues were obtained from surgically resected samples in Eastern Hepatobiliary Surgery Hospital, Second Military Medical University. All patients did not receive any prior treatment. All specimens were confirmed with histopathological examination. This study included 43 men and 13 women. The age ranged from 29 years to 78 years. Thirty-five of fifty-six patients had serum AFP ≥ 20 µg/L. There were 39 cases with positive HBsAg. Twelve cases had small (≤ 3 cm) and 44 were advanced (> 3 cm) HCCs. Tumor differentiation was graded according to WHO criteria (2000). Twenty HCCs were well differentiated, 28 were moderately differentiated and 8 were poorly differentiated. Of the 56 patients, 45 had evidence of intrahepatic metastasis (portal vein invision and/or intrahepatic dissemination). Forty-seven HCCs were detected accompanied with liver cirrhosis.
Fresh samples were obtained, immediately frozen in liquid nitrogen and stored at -70°C until analysis. A microdissection technique with a cryostat was used to separate tumor cells from corresponding noncancerous liver tissues. Genomic DNA was extracted from carcinoma tissues and corresponding noncancerous liver tissues using the standard phenol/ chloroform extraction and ethanol precipitation method[24]. The concentration of DNA was determined with both spectrophotometric and fluorometric methods.
For RAPD analysis, 10 arbitrary 10-mer primers were used. These primers were designed, referred to sequences reported previously[19-25] and synthesized commercially. The sequences of these primers were:
P1, 5’-CCGGCTACGG-3’;
P2, 5’-CAGGCCCTTC-3’;
P3, 5’-TACGGACACG-3’;
P4, 5’-AGCTTCAGGG-3’;
P5, 5’-AGGCATTCCC-3’;
P6, 5’-GGTCTGAACC-3’;
P7, 5’-TAGGCTCACG-3’;
P8, 5’-ACGGTACACT-3’;
P9, 5’-GTCCTCAACG-3’;
P10, 5’-CTTCACCCGA-3’
In a total volume of 25 µL, 50 ng of genomic DNA extracted from carcinoma tissues or corresponding noncan-cerous liver tissues was amplified in 10 mmol/L Tris-HCl (pH 8.3), 1.5 mmol/L MgCl2, 200 µmol/L each of dNTP, 0.4 µmol/L of each arbitrary primer, and 1.0 units of AmpliTaq Gold DNA polymerase (Perkin-Elmer, Norwalk, CT). Forty cycles of denaturation (at 94°C for 30 s), annealing (at 36-40°C for 1 min), and extension (at 72°C for 2 min) were carried out in a DNA thermocycler (Biometr). Five microliters of the PCR products mixed with loading buffer was loaded in 1.5% agarose gels and electrophoresed with 100 V for 1 h. The gels were stained with ethidium bromide (0.5 mg/mL) and visualized under UV light.
Loss or gain of band(s) and clearly detectable changes in intensity were scored. Scoring was done by two independent observers. A change of band intensities was defined as an increase or decrease of the signal intensity by 50% in the tumor DNA compared to normal DNA by gray scanning with an EDAS290 digital camera system (Kodak) and image analysis with the PDQuest 6.2 software (Bio-Rad).
The amplified 480 bp or so fragments from HCC DNA using the primer P4 were excised from the agarose gel and purified. The purified DNAs were reamplified with the primer P4 using the same concentration of reaction mixture constituents and the PCR cycle conditions as described above. The PCR products were analyzed on agarose gels to confirm their size and purity. The reamplified 480 bp DNA fragments were cloned using the TA cloning kit (Invitrogen, Carlsbad, CA, USA) following the protocol provided by the manufacturer. Restriction analysis of the recombinant plasmid DNA prepared by the alkaline lysis purification method was carried out to determine the appropriate insert Size[27-29]. The pure clone amplified DNA insert was sequenced with an automated DNA sequencer through the MegaBACE-500 capillary array electrophoresis instrument in our department. Sequences obtained from our clones were compared with the known sequences in the GenBank database using the BLASTn and BLASTx programs.
The relationship between the frequency of genetic alterations and clinicopathological features was assessed by using the χ2 test, P < 0.05 was considered statistically significant.
We examined 56 HCC tissues and corresponding noncancerous liver tissues using the RAPD method to detect the genetic alterations in HCC. Our results indicated that RAPD-PCR was an effective tool for identifying genetic alterations in cancer tissues. All the cases of HCC were demonstrated to have genetic alterations in at least one of ten primers. The incidence of genetic alterations ranged from 20% to 70% in each case, and 17.9% (15/56) to 50% (28/56) in each primer. The primer with the highest frequency of genetic alteration was P4. Genetic alterations were identified as band loss, gain, intensity change and shift in the tumor DNA lane as compared to the paired normal DNA lane (Figure 1A).
The correlation between the frequency of genetic alterations and clinicopathological features in HCC is shown in Table 1. No significant correlation was observed between the genetic alterations and serum AFP concentration, HBV infection, tumor size, cirrhosis, histological grade, tumor capsule, as well as tumor intrahepatic metastasis. As to the primers, the frequencies of genetic alterations on primer P1, P4, P8 and P10 were significantly higher in patients with positive HBsAg than in those with negative HBsAg (P < 0.01). Genetic alterations on P3 were more frequent in tumors less than 3 cm in size (P < 0.05 or P < 0.01). Genetic alterations on P1 and P4 were more frequent in poorly or moderately differentiated tumors than in well-differentiated tumors (P < 0.01 and P < 0.05), but genetic alterations on P3 were more frequent in well-differentiated tumors than in poorly or moderately differentiated tumors (P < 0.05). Genetic alterations on P3 were significantly higher in patients without intact tumor capsules than in those with intact capsules (P < 0.01). Genetic alterations on P4 were more frequently detected in tumors with intrahepatic metastasis than in those without. Similarly, genetic alterations on P3 were significantly lower in patients with intrahepatic metastasis than in those without (P < 0.01).
| Clinicopathological features | No. | P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | P9 | P10 | |
| Sex | M | 43 | 14 | 14 | 7 | 20 | 19 | 9 | 18 | 10 | 10 | 22 |
| F | 13 | 3 | 5 | 3 | 7 | 3 | 6 | 5 | 4 | 3 | 7 | |
| Age (yr) | < 50 | 24 | 6 | 12 | 6 | 11 | 8 | 7 | 9 | 8 | 5 | 14 |
| 50 | 32 | 11 | 7 | 4 | 16 | 14 | 8 | 14 | 6 | 8 | 15 | |
| Serum AFP | < 20 | 21 | 3 | 6 | 5 | 9 | 5 | 7 | 9 | 9 | 6 | 8 |
| level (µg/L) | 20 | 35 | 14 | 13 | 5 | 18 | 17 | 8 | 14 | 5 | 7 | 21 |
| HBsAg | Positive | 39 | 17 | 12 | 8 | 24 | 16 | 10 | 13 | 13 | 8 | 27 |
| Negative | 17 | 0b | 7 | 2 | 3a | 6 | 5 | 10 | 1a | 5 | 2b | |
| Tumor | 3 | 12 | 3 | 3 | 7 | 4 | 5 | 2 | 6 | 3 | 3 | 7 |
| size (cm) | > 3 | 44 | 14 | 16 | 3a | 23 | 17 | 13 | 17 | 11 | 10 | 22 |
| Histological | Well | 20 | 0 | 4 | 7 | 5 | 8 | 5 | 10 | 7 | 3 | 13 |
| grade | Moderately | 28 | 12b | 12 | 2 | 17a | 12 | 8 | 10 | 5 | 8 | 13 |
| Poorly | 8 | 5b | 3 | 1 | 5a | 2 | 2 | 3 | 2 | 2 | 3 | |
| Liver | Absent | 9 | 2 | 3 | 3 | 3 | 4 | 3 | 5 | 1 | 3 | 4 |
| cirrhosis | present | 47 | 15 | 16 | 7 | 24 | 18 | 12 | 18 | 13 | 10 | 25 |
| Tumor | Absent | 11 | 5 | 3 | 0 | 6 | 5 | 2 | 5 | 1 | 2 | 4 |
| capsule | Not intact | 31 | 9 | 13 | 4a | 16 | 10 | 11 | 13 | 10 | 7 | 17 |
| Intact | 14 | 3 | 3 | 6a | 5 | 7 | 2 | 5 | 3 | 4 | 8 | |
| Intrahepatic | Not | 11 | 3 | 5 | 7 | 2 | 5 | 2 | 5 | 3 | 5 | 7 |
| metastasis | observed | |||||||||||
| Observed | 45 | 14 | 14 | 3b | 25a | 17 | 13 | 18 | 11 | 8 | 22 | |
| Total [n (%)] | 56 | 17 (30.4) | 19 (33.9) | 10 (17.9) | 27 (48.2) | 22 (39.3) | 15 (26.8) | 23 (41.1) | 14 (25) | 13 (23.2) | 28 (50) | |
The amplified 480 bp fragments from HCC DNA using the primer P4 were separated on the agarose gels, excised, eluted, and reamplified with the same primer (Figure 1B). After analysis of the PCR products on an agarose gel to confirm their size and purity, these amplified DNA products were cloned using the TA cloning kit. After confirmation of positively cloned DNA by hybridization with the purified RAPD-amplified fragments, sequences were obtained from several clones and compared with known sequences in GenBank. Figure 2 shows the electropherograms from sequencing with amplified 476 bp fragments using P4 primer. The followings were the nucleotide sequences of the fragments: TCCCCGCGATGGCCCTCCTGACAGCT- TCGGACAGCACCCACCCTGCAGAAGCACATTAAC- AGGAATGACGAAGGAGACTCCTTTGGAACC- CGAGCCAAAGCGAGGCCCTGAGAGCCTGCT- TTGAGCAAACCCATAGCCGGGTATCGCCACAAGAG- AATGGCTGGCCCAGGCCATCGGCATTCCAG- AACCCAGGGTCCAGATTTGGTTTCAGAATGAGA- GGTCACGCCAGCTGAGGCAGCACCGGCGGGA- ATCTCGCCCTGGCCTGGGAGACGCGGCCAGCAA- GAAGGCAGGTGAAAGTGGACCGCCGTCACCAGATCC- CAGACCGCCCTGCTCCTCCGAGCCTTTGAGAA-GGATCGCTTTCCAGGCATCGCAGCCAGGA-AGGCTGGCCAGAGAGACGGGCCTCCCG - G A G T C C A G G A T T C A T A T C T G T T T T C A - G A A T C A A AG G G C C T G G C AC C C G G G AC - AGGGTGGCAGGGCGAACACGCA.
BLAST analysis revealed that there were significant matches of the 480 bp or so RAPD fragments with any known gene in this database either at the DNA level or its corresponding amino acid sequence. The tumor-specific P4 amplified 476-bp fragment sequences showed 92% homology with homo sapiens 3 BAC RP11-413E6 (Roswell Park Cancer Institute Human BAC Library, accession number AC108724.4) from nt 84 643 to nt 84 168, and with human DNA sequence clones RP11-298A19 on chromosome 22 (accession number AL592170.21) from nt 84717 to nt 842981, and 91% homology with homo sapiens double homeobox protein DUX10 gene from nt 316 to nt 788 (accession number Ay044051.1), and with homo sapien putative proteins and FSHD region gene 2 protein (HSA10-FRG2) genes (accession number AY028079.1) from nt 118 939 to nt 119 411.
The RAPD method, a PCR-based method for nucleic acid fingerprinting, was originally reported by Williams et al[18] and it is thought to be suitable for investigation of genetic alterations. RAPD-PCR multiple bands amplified with arbitrary primers could generate a complex DNA fingerprint that can detect qualitative (structural) and quantitative (aneuploid) genetic differences between normal and tumor cell genomes. Mutations can affect the fingerprint by altering the distance between the two primers, the ability of the primer to anneal, and the relative amounts of target amplification[19]. In the present study, we identified genetic alterations in HCC at the level of ethidium bromide stained agarose gels. Genetic alterations indicating distance band shift or band loss/gain were observed. Mutations occurred at the primer-template interaction sites resulted in the loss or gain of a band, and mutations occurred between the primer-template interaction sites, such as deletions or insertions, resulted in a mobility shift of bands. However, single base situations between two primers could not be detected by this method. In addition to these qualitative alterations, we also observed decrease or increase in the relative intensities of the bands in tumor DNA compared to those in normal tissue DNA. Allelic losses due to their linkage to suppressor genes, might result in decrease of band intensities, and gene amplification or chromosomal aneuploidy might result in increase of band intensities. Furthermore, we found a significant correlation between the instability of certain primers and HBV infection, tumor size, cirrhosis, histological grade, tumor capsule, as well as tumor intrahepatic metastasis at some primers. These results suggest that the high frequency of genetic alterations identified by the RAPD method may be involved in the malignant potential of HCC.
Since this method permits the direct cloning and characterization of amplified DNA bands which represent genetic alterations specific in HCC genome during tumor progression, further research in this direction will find out new genes contributing to progression in liver cancer. The most important finding in the present study is that a band with a higher intensity of 480 bp or so amplified fragments in tumor DNA relative to normal DNA could be seen in 27 and 56 tumor samples using primer 4. An enhancement of the signal intensity of amplified DNA fragments may be related to localized overamplification of that gene locus in the genome, or could result from changes at the chromosome level, such as trisomy or tetrasomy. These events could play an important role in the development of HCC or they could occur during the clonal expansion of genetically unstable tumor cells. Sequence analysis of the fragments showed 92% homology with homo sapiens 3 BAC RP11-413E6 (Roswell Park Cancer Institute Human BAC Library) complete sequence, and with human DNA sequences from clone RP11-298A19 on chromosome 22, complete sequence. Also the fragment sequences showed 91% homology with homo sapiens double homeobox protein DUX10 gene, and with homo sapiens putative protein and FSHD region gene 2 protein (HSA10-FRG2) genes. Homeobox protein genes comprise a large and essential family of developmental regulators that are vital for all aspects of growth and differentiation. Although many studies have reported their deregulated expression in cancer, few studies have established direct functional roles for homeobox genes in carcinogenesis[30]. Our data suggest that homeobox protein gene family members are activated as an important pathway in hepatocarcinogenesis.
In summary, genetic alterations are a frequent event in HCC. Furthermore, serum AFP conce-ntration, HBV infection, tumor size, histological grade, tumor capsule, as well as tumor intrahepatic metastasis, may be correlated with allelic losses with certain primers. A band with a higher intensity of 480 bp or so amplified fragments in tumor DNA relative to normal DNA can be seen tumor samples and sequence analysis of the fragments can show 91% homology with homo sapiens double homeobox protein DUX10 gene. The RAPD method is an effective tool for the identification and analysis of genetic alterations in HCC tissues and it may provide some new information about the molecular mechanism of hepatocarcinogenesis.
Science Editor Wang XL and Li WZ Language Editor Elsevier HK
| 1. | Buendia MA. Genetics of hepatocellular carcinoma. Semin Cancer Biol. 2000;10:185-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 245] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 2. | Feitelson MA, Sun B, Satiroglu Tufan NL, Liu J, Pan J, Lian Z. Genetic mechanisms of hepatocarcinogenesis. Oncogene. 2002;21:2593-2604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 246] [Cited by in RCA: 243] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 3. | Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1097] [Cited by in RCA: 1104] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 4. | McGlynn KA, Edmonson MN, Michielli RA, London WT, Lin WY, Chen GC, Shen FM, Buetow KH. A phylogenetic analysis identifies heterogeneity among hepatocellular carcinomas. Hepatology. 2002;36:1341-1348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 5. | Tsopanomichalou M, Kouroumalis E, Ergazaki M, Spandidos DA. Loss of heterozygosity and microsatellite instability in human non-neoplastic hepatic lesions. Liver. 1999;19:305-311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 6. | Loeb KR, Loeb LA. Significance of multiple mutations in cancer. Carcinogenesis. 2000;21:379-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 273] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 7. | Suriawinata A, Xu R. An update on the molecular genetics of hepatocellular carcinoma. Semin Liver Dis. 2004;24:77-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 60] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 8. | Maggioni M, Coggi G, Cassani B, Bianchi P, Romagnoli S, Mandelli A, Borzio M, Colombo P, Roncalli M. Molecular changes in hepatocellular dysplastic nodules on microdissected liver biopsies. Hepatology. 2000;32:942-946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 54] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 9. | Okabe H, Ikai I, Matsuo K, Satoh S, Momoi H, Kamikawa T, Katsura N, Nishitai R, Takeyama O, Fukumoto M. Comprehensive allelotype study of hepatocellular carcinoma: potential differences in pathways to hepatocellular carcinoma between hepatitis B virus-positive and -negative tumors. Hepatology. 2000;31:1073-1079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 74] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 10. | Liao C, Zhao M, Song H, Uchida K, Yokoyama KK, Li T. Identification of the gene for a novel liver-related putative tumor suppressor at a high-frequency loss of heterozygosity region of chromosome 8p23 in human hepatocellular carcinoma. Hepatology. 2000;32:721-727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 71] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 11. | Tamura S, Nakamori S, Kuroki T, Sasaki Y, Furukawa H, Ishikawa O, Imaoka S, Nakamura Y. Association of cumulative allelic losses with tumor aggressiveness in hepatocellular carcinoma. J Hepatol. 1997;27:669-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 12. | Kondo Y, Kanai Y, Sakamoto M, Mizokami M, Ueda R, Hirohashi S. Microsatellite instability associated with hepatocarcinogenesis. J Hepatol. 1999;31:529-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 24] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 13. | Li SP, Wang HY, Li JQ, Zhang CQ, Feng QS, Huang P, Yu XJ, Huang LX, Liang QW, Zeng YX. Genome-wide analyses on loss of heterozygosity in hepatocellular carcinoma in Southern China. J Hepatol. 2001;34:840-849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 51] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 14. | Grisham JW. Interspecies comparison of liver carcinogenesis: implications for cancer risk assessment. Carcinogenesis. 1997;18:59-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 79] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 15. | Salvucci M, Lemoine A, Saffroy R, Azoulay D, Lepère B, Gaillard S, Bismuth H, Reynès M, Debuire B. Microsatellite instability in European hepatocellular carcinoma. Oncogene. 1999;18:181-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 55] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 16. | Kondo Y, Kanai Y, Sakamoto M, Mizokami M, Ueda R, Hirohashi S. Genetic instability and aberrant DNA methylation in chronic hepatitis and cirrhosis--A comprehensive study of loss of heterozygosity and microsatellite instability at 39 loci and DNA hypermethylation on 8 CpG islands in microdissected specimens from patients with hepatocellular carcinoma. Hepatology. 2000;32:970-979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 196] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 17. | Macdonald GA, Greenson JK, Saito K, Cherian SP, Appelman HD, Boland CR. Microsatellite instability and loss of heterozygosity at DNA mismatch repair gene loci occurs during hepatic carcinogenesis. Hepatology. 1998;28:90-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 60] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 18. | Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531-6535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7219] [Cited by in RCA: 4281] [Article Influence: 122.3] [Reference Citation Analysis (0)] |
| 19. | Papadopoulos S, Benter T, Anastassiou G, Pape M, Gerhard S, Bornfeld N, Ludwig WD, Dörken B. Assessment of genomic instability in breast cancer and uveal melanoma by random amplified polymorphic DNA analysis. Int J Cancer. 2002;99:193-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 20. | Hering O, Nirenberg HI. Differentiation of Fusarium sambucinum Fuckel sensu lato and related species by RAPD PCR. Mycopathologia. 1995;129:159-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 21. | Ong TM, Song B, Qian HW, Wu ZL, Whong WZ. Detection of genomic instability in lung cancer tissues by random amplified polymorphic DNA analysis. Carcinogenesis. 1998;19:233-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 42] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 22. | Maeda T, Jikko A, Hiranuma H, Fuchihata H. Analysis of genomic instability in squamous cell carcinoma of the head and neck using the random amplified polymorphic DNA method. Cancer Lett. 1999;138:183-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 23. | Dil-Afroze A, Sulaiman IM, Sinha S, Sarkar C, Mahapatra AK, Hasnain SE. Genetic alterations in brain tumors identified by RAPD analysis. Gene. 1998;206:45-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 24. | Sood AK, Buller RE. Genomic instability in ovarian cancer: a reassessment using an arbitrarily primed polymerase chain reaction. Oncogene. 1996;13:2499-2504. [PubMed] |
| 25. | Singh KP, Roy D. Identification of novel breast tumor-specific mutation(s) in the q11.2 region of chromosome 17 by RAPD/AP-PCR fingerprinting. Gene. 2001;269:33-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 26. | Luo L, Li B, Pretlow TP. DNA alterations in human aberrant crypt foci and colon cancers by random primed polymerase chain reaction. Cancer Res. 2003;63:6166-6169. [PubMed] |
| 27. | Navarro JM, Jorcano JL. The use of arbitrarily primed polymerase chain reaction in cancer research. Electrophoresis. 1999;20:283-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 28. | Misra A, Chosdol K, Sarkar C, Mahapatra AK, Sinha S. Alteration of a sequence with homology to human endogenous retrovirus (HERV-K) in primary human glioma: implications for viral repeat mediated rearrangement. Mutat Res. 2001;484:53-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 29. | Comes AM, Humbert JF, Laurent F. Rapid cloning of PCR-derived RAPD probes. Biotechniques. 1997;23:210-212. [PubMed] |
| 30. | Monica K, Galili N, Nourse J, Saltman D, Cleary ML. PBX2 and PBX3, new homeobox genes with extensive homology to the human proto-oncogene PBX1. Mol Cell Biol. 1991;11:6149-6157. [PubMed] |