Published online Jul 14, 2005. doi: 10.3748/wjg.v11.i26.4094
Revised: October 15, 2004
Accepted: October 20, 2004
Published online: July 14, 2005
AIM: To prepare and identify specific anti-mouse caspase-12 hammerhead ribozymes in vitro, in order to select a more effective ribozyme against mouse caspase-12 as a potential tool to rescue cells from endoplasmic reticulum stress induced apoptosis.
METHODS: Two hammerhead ribozymes directed separately against 138 and 218 site of nucleotide of mouse caspase-12 mRNA were designed by computer software, and their DNA sequences were synthesized. The synthesized ribozymes were cloned into an eukaryotic expression vector-neorpBSKU6 and embedded in U6 SnRNA context for further study. Mouse caspase-12 gene segment was cloned into PGEM-T vector under the control of T7 RNA polymerase promoter (containing gene sequence from positions nt 41 to nt 894) as target. In vitro transcription both the ribozymes and target utilize T7 promoter. The target was labeled with [α-32P]UTP, while ribozymes were not labeled. After gel purification the RNAs were dissolved in RNase free water. Ribozyme and target were incubated for 90 min at 37°C in reaction buffer (40 mmol/L Tris-HCL, pH 7.5, 10 mmol/L Mg2+). Molar ratio of ribozyme vs target was 30:1. Samples were analyzed on 6% PAGE (containing 8 mol/L urea).
RESULTS: Both caspase-12 and ribozyme gene sequences were successfully cloned into expression vector confirmed by sequencing. Ribozymes and caspase-12 mRNA were obtained by in vitro transcription. Cleavage experiment showed that in a physiological similar condition (37°C, pH 7.5), Rz138 and Rz218 both cleaved targets at predicted sites, for Rz138 the cleavage efficiency was about 100%, for Rz218 the value was 36.66%.
CONCLUSION: Rz138 prepared in vitro can site specific cleave mouse caspase-12 mRNA with an excellent efficiency. It shows a potential to suppress the expression of caspase-12 in vivo, thus provided a new way to protect cells from ER stress induced apoptosis.
- Citation: Jiang S, Xie Q, Zhang W, Zhou XQ, Jin YX. Preparation of anti-mouse caspase-12 mRNA hammerhead ribozyme and identification of its activity in vitro. World J Gastroenterol 2005; 11(26): 4094-4097
- URL: https://www.wjgnet.com/1007-9327/full/v11/i26/4094.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i26.4094
Apoptosis plays an important role in the pathogenesis of liver diseases[1]. Activation of death receptors and mitochondrial damage are well-described common apoptotic pathways. Recently, a novel pathway via endoplasmic reticulum (ER) stress was reported[2]. Following studies have demonstrated that many liver diseases are associated with ER stress, including nonalcoholic steatohepatitis, cholestasis, alcohol induced liver diseases, and viral hepatitis[3-5].
Among the death-specific enzymes in apoptosis is a family of cystein-dependent aspartate-specific proteases known as caspases[6]. Activation of caspases is a central mechanism in the apoptotic cell death process[7].
Caspase-12 resides in the endoplasmic reticulum and is specifically involved in the apoptosis that results from ER stress, Apoptosis triggered through pathways that do not involve the ER, such as serum deprivation or Fas-activation, do not result in activation of caspase-12. Cells isolated from transgenic mice that lack intact caspase-12 protein were more resistant to ER stress-induced apoptosis than wild type cells[2].
Caspases are central to both normal programmed cell death and injury-dependent apoptosis, so any therapy that manipulates caspase activity must take into account the possible effects on tissue homeostasis. In this regard, caspase-12 seems to have a strong advantage as a target over other caspases. In contrast to other caspase knockouts, caspase-12 deficient mice have no noticeable developmental or behavioral defects, and have a normal incidence of tumors[2]. So caspase-12 may be a promising target for treating ER stress associated diseases with few side effect.
Until now, no specific caspase-12 inhibitors are available. Ribozymes (Rz) are RNA molecules with enzymatic activity that can associate with a larger target RNA by base-pairing to cleave a specific phosphodiester bond[2,8]. In the past years, ribozyme-mediated gene inhibition has been demonstrated for many examples in cell lines. Selective downregulation of a particular caspase by ribozyme provides an alternative approach to rescue cells from apoptosis[9-11].
Catalytic cleavage of caspase-12 mRNA by ribozyme may block ER stress induced apoptotic pathway, thus protect liver cells from apoptosis. In the present study, ribozymes were designed and synthesized against the sequences of caspase-12 mRNA which cleave at nucleotide positions 138 and 218 respectively. Also mutant ribozymes were synthesized for future study. Transcription and cleavage reaction were performed in vitro to determine the activity of ribozymes designed and assess the potential value of them against caspase-12 mediated apoptosis.
PGEM-T vector kit, transcription kit, TRIzol kit, and T4 DNA ligase were purchased from Promega Company. RT-PCR kit, Restriction endonucleases were purchased from TaKaRa Company. [α -32p]UTP was the product of Amersham Biosciences UK. E coli DH5α and eukaryotic expression vector-neorpBSKU6 for ribozyme were kind gifts from Dr. YouXin Jin. The PCR primers and ribozyme gene sequences were synthesized in the Beckman Oligo-1000 DNA synthesizer.
Cloning of Caspase-12 cDNA Mouse caspase-12 mRNA gene sequence was obtained from online Genebank of NCBI (GI: 2094805). Total RNA of fresh mouse liver tissue was extracted using Trizol kit. The up stream primer for RT-PCR amplification of caspase-12 gene segment was F: 5’-TCT AGA CCA GGA GGA CAC ATG AAA GA-3’ (41-60 bp), the 5’ extension underlined is XbaI site, the downstream primer was R: 5’-GGA TCC TCT CAG ACT CCG ACA GTT AG-3’(894-875 bp), the 5’ extension underlined is BamHI site. PCR product contained gene sequence from position nt 41 to nt 894 with XbaI site ahead and BamHI site behind. Purified PCR product were inserted into pGEM-T vector through A-T pairing under the control of T7 RNA polymerase promoter. The recombinants were transfected into competent JM109 E Coli cells for blue/ white screening on LB plate. The selected clone containing Caspase-12 cDNA was confirmed by sequencing and named pCaspase-12.
Ribozyme Construction The hammerhead ribozymes were designed according to the computer software compiled by Professor Chen Nong-An (Shanghai Institute of Biochemistry of the Chinese Academy of Science). The specificity of ribozymes designed for mouse caspase-12 mRNA were determined by sequence analysis with other RNA sequences of mouse cells in NCBI GenBank using blastn. The sequences of the two ribozymes are shown in Table 1. G in the catalytic core for active ribozyme, A for inactive ribozyme. The inactive ribozymes allow binding to the target RNA, but lack cleavage ability[12]. Oligonucleotides encoding Rz138 and Rz218 were designed to generate XbaI/BamHI ends upon annealing and were ligated with the BamHI and XbaI digested neorpBSKU6 plasmid embedded in U6 SnRNA context containing U6 SnRNA promoter/enhancer and terminator. The reconstructed plasmids were named pRz138, pRz138m(mutant), pRz218, pRz218m respectively according to the confirmation of DNA sequencing, and the reconstructed Rz plasmids were prepared for further in vivo study.
| 5’ binding arm | Catalytic core | 3’ binding arm | |
| Rz138 | 5’ TCCATTTAA | CT(G/A)ATGAGTCCGT GAGGACGAA | ACATTCTT 3’ |
| Rz218 | 5’ TCTCTAAG | CT(G/A)ATGAGTCCGT GAGGACGAA | AGTTCTC 3’ |
In vitro transcription of ribozymes and target RNAIn vitro transcription were prepared according to the supplier (Promega). Both the ribozymes and substrate were transcribed with T7 RNA polymerase. Transcription of target RNA started with an additional 46 nt derived from the vector and terminated just at the end of BamHI linearized pCaspase-12. The total length was 910 nt. Substrate transcription was performed in 20 uL volume in the presence of 1μL [α-32p] UTP (10 uci/uL), while ribozymes were not labeled with isotope. Ribozyme templates were produced by PCR, primers were complementary to the flanking U6 sequence adjacent to the embedded ribozyme, T7 promoter was pulsed to the F primer 5’-TCT AGA GTA ATA CGA CTC ACT ATA GGG C CTT CGG CAG CAC ATA TAC-3’, the underlined sequence represents T7 promoter. Primer R was 5’-TAT GGA ACG CTT CAG GAT-3’. PCR products were purified by 12 g/L agarose gel electrophoresis as templates for transcription. The products of transcription were purified with 6% denaturing polyacrylamide gels (PAGE), the bands were cut off from the gel and soaked in NES (0.5 mol/L NH4Ac, 1 mmol/L EDTA, 0.1% SDS ) at 45°C overnight. After centrifuge, the supernatant were precipitated by ethanol and 3 mol/L sodium acetate, washed twice by 75% ethanol, then dissolved in RNase free water. The [α-32p] UTP labeled substrate was counted in a Beckman-LS 6500 counter[13]. As many studies show that inactive mutant ribozymes have no cleavage activity[12,14,15], we preferred to use the normal Rz in vitro study.
Identification of ribozyme activity by cleavage reaction The molar quantity of target RNA was estimated according to the cpm value combined with the UTP number in the RNA. Rz molar concentration was estimated according to the spectrophotometric A260 value. Ribozyme and substrate were incubated for 90 min at 37°C in reaction buffer (40 mmol/L Tris-HCl, pH 7.5, 10 mmol/L Mg2+). Molar ratio of ribozyme versus substrate was 30:1. Samples were analyzed on 6% PAGE (containing 8 mol/L urea). The cleavage efficiency [CE] was calculated from cpm values of the bands of undigested substrate (S) and product (P) separated from denaturing PAGE. CE = [P/(P+S)]•100%.
RT-PCR amplified caspase-12 cDNA segment was separated by 1.2% agarose gel and stained with ethidium bromide. The length of product containing restriction endonuclease site was 866bp. (Figure 1).
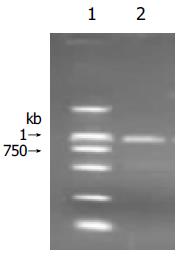
Purified caspase-12 PCR product was inserted into pGEM-T vector by A-T base pairing. After blue/white screening on LB plate and DNA sequencing, pCaspase-12 was confirmed. Then, BamHI linearized pCaspase-12 was used as transcription template. The length of transcribed target RNA as shown in Figure 2 was 910 nt.
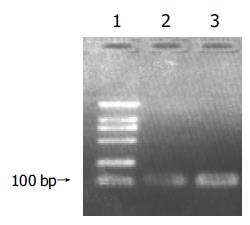
The length of Rz templates for transcription produced by PCR were about 100 bp (Figure 3). Transcription products were also purified through 6% PAGE, using upper ultraviolet to determine the position of the bands. The bands were excised and dissolved in RNase free water and reserved under -20°C.
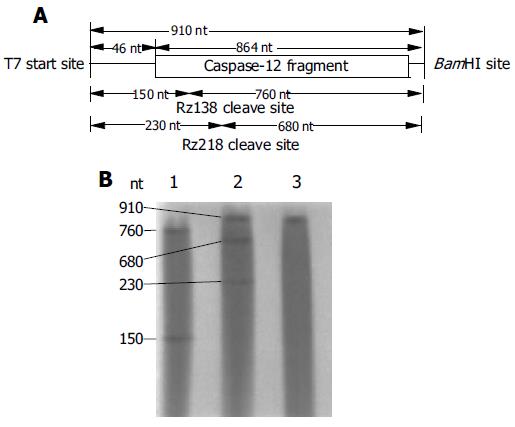
As shown in Figure 4A, The length of target RNA transcribed from BamHI linearized pCaspase-12, which contains the mouse caspase-12 cDNA segment, should be 910 nt, and the expected fragments cleaved by Rz138 in vitro should be 150 nt and 760 nt. Rz218 cleavage would generate 230 nt and 680 nt. The cleavage result showed that at a 30:1 ribozyme-to-target ratio, both Rz138 and Rz218 cleaved target at the predicted sites which were consistent with our design and proved to be correct (Figure 4B). The cleavage efficiency of Rz138 was about 100%, while 36.66% for Rz218.
Apoptosis is essential to development of multicellular organisms as well as to physiologic cell turn over. Excessive apoptosis may contribute to organ injury[16]. A novel apoptotic pathway via ER stress was recently testified in mouse. Prolonged ER stress leads to cell death and is linked to the pathogenesis of some neurodegenerative disorders. ER stress also plays an important role in several liver diseases[3-5,17]. Activation of caspase-12 from procaspase-12 is specifically induced by insult to the ER, ER stress triggers a specific cascade involving caspase-12,-9,-3. Although it was reported that human caspase-12 may have no significant effect on apoptotic sensitivity[18,19], many evidences show that functional caspase-12 exists in many human cell types and is associated with ER stress induced apoptosis[20-22]. It was knowing that cells lack of intact caspase-12 protein were more resistant to ER stress-induced apoptosis than wild type cells. Inhibition of caspase-12 expression by ribozyme may block ER stress induced apoptotic pathway, thus protect liver cells from apoptosis.
The ribozymes are divided into two groups: small ribozymes and large ribozymes. Small ribozymes, approximately those less than 100 nucleotides, include the hammerhead ribozyme, hairpin ribozyme, and hepatitis delta virus ribozyme. Large ribozymes contain ribonuclease P RNA and group I and group II introns. Hammerhead ribozyme is simple in structure with high turnover in cleavage reaction. The activity of the hammerhead ribozyme is significant higher than that of all other ribozymes in vitro. It’s being widely used to inhibit endogenous gene expression of basic biochemical pathways such as angiogenesis and apoptosis. It’s the first to be approved to use in clinical trial[23]. It contains a tripartite structure consisting of a central catalytic core that is flanked on both sides by two antisense side arms that can form base pairs with the RNA substrate, thus providing the sequence specificity of the endonuclease action. Sequences 5’ to the catalytic domain form helix I and sequences 3’ to it form helix III when complexed with the target RNA[24].
Ribozyme used as molecular tool for specific inhibition of gene translation is affected by many factors including the mRNA secondary structures and target accessibility. Our studies show that computer aided energy minimization algorithms predicted regions in mouse caspase-12 mRNA are accessible to ribozyme cleavage. The cleavage reaction revealed that Rz138 and Rz218 prepared in vitro possessed perfect specific catalytic cleavage activity. Rz138 has an excellent cleavage efficiency. The results were consistent with our designed. This finding made it worthy to do further study on the cleavage activity of these ribozymes in vivo and to develop them as a therapeutic nucleic acid drug in the future. Considering that the in vitro activity does not represent exactly the efficiency of a ribozyme in degrading the target mRNA in intact cells, we’ve cloned the two ribozymes in eukaryotic expression vector for further study in vivo.
Co-first-authors: Dr. You-Xin Jin
Science Editor Guo SY Language Editor Elsevier HK
| 1. | Ockner RK. Apoptosis and liver diseases: recent concepts of mechanism and significance. J Gastroenterol Hepatol. 2001;16:248-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 2. | Nakagawa T, Zhu H, Morishima N, Li E, Xu J, Yankner BA, Yuan J. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature. 2000;403:98-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2547] [Cited by in RCA: 2610] [Article Influence: 104.4] [Reference Citation Analysis (0)] |
| 3. | Ji C, Kaplowitz N. Betaine decreases hyperhomocysteinemia, endoplasmic reticulum stress, and liver injury in alcohol-fed mice. Gastroenterology. 2003;124:1488-1499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 424] [Cited by in RCA: 448] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 4. | Xu Z, Jensen G, Yen TS. Activation of hepatitis B virus S promoter by the viral large surface protein via induction of stress in the endoplasmic reticulum. J Virol. 1997;71:7387-7392. [PubMed] |
| 5. | Foo NC, Ahn BY, Ma X, Hyun W, Yen TS. Cellular vacuolization and apoptosis induced by hepatitis B virus large surface protein. Hepatology. 2002;36:1400-1407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 54] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 6. | Wolf BB, Green DR. Suicidal tendencies: apoptotic cell death by caspase family proteinases. J Biol Chem. 1999;274:20049-20052. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 674] [Cited by in RCA: 682] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 7. | Cryns V, Yuan J. Proteases to die for. Genes Dev. 1998;12:1551-1570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 943] [Cited by in RCA: 945] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 8. | Kato Y, Kuwabara T, Toda H, Warashina M, Taira K. Suppression of BCR-ABL mRNA by various ribozymes in HeLa cells. Nucleic Acids Symp Ser. 2000;44:283-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 9. | Eldadah BA, Ren RF, Faden AI. Ribozyme-mediated inhibition of caspase-3 protects cerebellar granule cells from apoptosis induced by serum-potassium deprivation. J Neurosci. 2000;20:179-186. [PubMed] |
| 10. | Xu R, Liu J, Chen X, Xu F, Xie Q, Yu H, Guo Q, Zhou X, Jin Y. Ribozyme-mediated inhibition of caspase-3 activity reduces apoptosis induced by 6-hydroxydopamine in PC12 cells. Brain Res. 2001;899:10-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 11. | Yaghootfam A, Gieselmann V. Specific hammerhead ribozymes reduce synthesis of cation-independent mannose 6-phosphate receptor mRNA and protein. Gene Ther. 2003;10:1567-1574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 12. | Mendoza-Maldonado R, Zentilin L, Fanin R, Giacca M. Purging of chronic myelogenous leukemia cells by retrovirally expressed anti-bcr-abl ribozymes with specific cellular compartmentalization. Cancer Gene Ther. 2002;9:71-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 13. | Grassi G, Forlino A, Marini JC. Cleavage of collagen RNA transcripts by hammerhead ribozymes in vitro is mutation-specific and shows competitive binding effects. Nucleic Acids Res. 1997;25:3451-3458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 14. | Pennati M, Colella G, Folini M, Citti L, Daidone MG, Zaffaroni N. Ribozyme-mediated attenuation of survivin expression sensitizes human melanoma cells to cisplatin-induced apoptosis. J Clin Invest. 2002;109:285-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 71] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 15. | Sriram B, Thakral D, Panda SK. Targeted cleavage of hepatitis E virus 3' end RNA mediated by hammerhead ribozymes inhibits viral RNA replication. Virology. 2003;312:350-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 16. | Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456-1462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4724] [Cited by in RCA: 4686] [Article Influence: 156.2] [Reference Citation Analysis (0)] |
| 17. | Schuchmann M, Galle PR. Apoptosis in liver disease. Eur J Gastroenterol Hepatol. 2001;13:785-790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 48] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 18. | Fischer H, Koenig U, Eckhart L, Tschachler E. Human caspase 12 has acquired deleterious mutations. Biochem Biophys Res Commun. 2002;293:722-726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 247] [Cited by in RCA: 257] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 19. | Saleh M, Vaillancourt JP, Graham RK, Huyck M, Srinivasula SM, Alnemri ES, Steinberg MH, Nolan V, Baldwin CT, Hotchkiss RS. Differential modulation of endotoxin responsiveness by human caspase-12 polymorphisms. Nature. 2004;429:75-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 307] [Cited by in RCA: 301] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 20. | Morishima N, Nakanishi K, Takenouchi H, Shibata T, Yasuhiko Y. An endoplasmic reticulum stress-specific caspase cascade in apoptosis. Cytochrome c-independent activation of caspase-9 by caspase-12. J Biol Chem. 2002;277:34287-34294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 689] [Cited by in RCA: 702] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 21. | Rao RV, Hermel E, Castro-Obregon S, del Rio G, Ellerby LM, Ellerby HM, Bredesen DE. Coupling endoplasmic reticulum stress to the cell death program. Mechanism of caspase activation. J Biol Chem. 2001;276:33869-33874. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 444] [Cited by in RCA: 448] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 22. | Xie Q, Khaoustov VI, Chung CC, Sohn J, Krishnan B, Lewis DE, Yoffe B. Effect of tauroursodeoxycholic acid on endoplasmic reticulum stress-induced caspase-12 activation. Hepatology. 2002;36:592-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 255] [Cited by in RCA: 262] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 23. | Wong-Staal F, Poeschla EM, Looney DJ. A controlled, Phase 1 clinical trial to evaluate the safety and effects in HIV-1 infected humans of autologous lymphocytes transduced with a ribozyme that cleaves HIV-1 RNA. Hum Gene Ther. 1998;9:2407-2425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 100] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 24. | Chen S, Song CS, Lavrovsky Y, Bi B, Vellanoweth R, Chatterjee B, Roy AK. Catalytic cleavage of the androgen receptor messenger RNA and functional inhibition of androgen receptor activity by a hammerhead ribozyme. Mol Endocrinol. 1998;12:1558-1566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 1.1] [Reference Citation Analysis (0)] |