Published online Jul 14, 2005. doi: 10.3748/wjg.v11.i26.3985
Revised: November 16, 2004
Accepted: November 19, 2004
Published online: July 14, 2005
AIM: To identify the scFv antibody fragments specific for hepatocellular carcinoma by biopanning from a large human naive scFv phage display library.
METHODS: A large human naive scFv phage library was used to search for the specific targets by biopanning with the hepatocellular carcinoma cell line HepG2 for the positive-selecting and the normal liver cell line L02 for the counter-selecting. After three rounds of biopanning, individual scFv phages binding selectively to HepG2 cells were picked out. PCR was carried out for identification of the clones containing scFv gene sequence. The specific scFv phages were selected by ELISA and flow cytometry. DNA sequences of positive clones were analyzed by using Applied Biosystem Automated DNA sequencers 3730. The expression proteins of the specific scFv antibody fragments in E.coli HB2151 were purified by the affinity chromatography and detected by SDS-PAGE, Western blot and ELISA. The biological effect of the soluble antibody fragments on the HepG2 cells was investigated by observing the cell proliferation.
RESULTS: Two different positive clones were obtained and the functional variable sequences were identified. Their DNA sequences of the scFv antibody fragments were submitted to GenBank (accession nos: AY686498 and AY686499). The soluble scFv antibody fragments were successfully expressed in E.coli HB2151. The relative molecular mass of the expression products was about 36 ku, according to its predicted Mr value. The two soluble scFv antibody fragments also had specific binding activity and obvious growth inhibition properties to HepG2 cells.
CONCLUSION: The phage library biopanning permits identification of specific antibody fragments for hepatocellular carcinoma and affords experiment evidence for its immunotherapy study.
- Citation: Yu B, Ni M, Li WH, Lei P, Xing W, Xiao DW, Huang Y, Tang ZJ, Zhu HF, Shen GX. Human scFv antibody fragments specific for hepatocellular carcinoma selected from a phage display library. World J Gastroenterol 2005; 11(26): 3985-3989
- URL: https://www.wjgnet.com/1007-9327/full/v11/i26/3985.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i26.3985
Hepatocellular carcinoma is the third leading cause of cancer deaths in the world and the prognosis of patients remains poor. Immunotherapeutic strategies against cancer have been developed during the last 20 years. In this respect, mouse monoclonal antibodies against tumor marks have been used for tumor targeting and imaging[1]. The use of murine monoclonal antibodies for therapy in humans has limitations because of the human anti-mouse antibody response. Moreover, whole antibodies are large molecules and have poor tumor penetration. Several advances made during the past years will probably facilitate the development of therapeutic antibodies[2]. Indeed, a chimeric anti-CD20[3] antibody and a humanized anti-HER2[4] antibody have been approved by FDA for the treatment of non-Hodgkins lymphoma and metastatic breast cancer respectively. These successes indicate that immunotherapeutic modalities are effective[5]. On the other hand, the antibody-mediated tumor immunotherapy has become critical parts of biotherapy and is used for the diagnosis of cancer and metastasis, fine staging, and decisions regarding therapeutic approaches[6]. So, it is urgent to obtain new humanized antibody specific for tumor targeting, and it is the same state in the therapy of hepatocellular carcinoma[7]. Phage libraries displaying antibody fragments provide the fastest route to obtain human antibody fragments[8]. The advent of antibody fragment display on phage and the development of large (> 6 × 109 clones) phage display libraries[9,10] offer a potential way of detecting new targets by biopanning using tumor cells and counter-selecting using non-tumor cells[11-13]. Significant progress has been made using this method to screen the antibodies against cell surface antigens[14-17]. Targeting with scFv antibody fragments may overcome some of the limitations of murine monoclonal antibodies, and provide greater targeting specificity[18-20]. Here, we report the selection of scFv antibodies binding specifically to HepG2 cells by using L02 as counter-selecting cells biopanning from a large human naive scFv phage library (Griffin. 1 library)[21].
The human naive scFv library (Griffin. 1 library) used in this study was a generous gift from Professor Winter, University of Cambridge, UK. E.coli TG1 (suppressor strain for propagation of phage particles), E.coli HB2151 (non-suppressor strain for expression of antibody fragments) and helper phage M13K07 were purchased from Pharmacia-Biotech. Mouse anti-M13 antibody was from Amersham. Goat anti-mouse IgG conjugated HRP was provided by Kirkegaard Perry Laboratories. Goat anti-mouse IgG conjugated with FITC was obtained from Zhongsheng, Beijing. 9E10 antibody, Ni-NTA, FACS were purchased from Santa Cruz Biotechnology, Qiagen, and BD, respectively.
Cell culture The hepatocellular carcinoma cell line HepG2 and liver cell line L02 were incubated in RPMI 1640 supplemented with 100 mL/L FBS at 37°C with 50 mL/L CO2.
Screening of scFv phages from phage library using whole cells An aliquot containing 1 × 1012 cfu from a large human scFv phage library was added to 1 × 106 L02 cells and mixed gently for 30 min at room temperature (RT). The phage-containing supernatant was used to resuspend a fresh pellet of 1 × 106 L02 cells and incubated for 30 min at RT, followed by pelleting the cells. Then, the resultant subtracted phage supernatant was incubated with 5 × 106 HepG2 cells for 1 h at RT with gentle mixing. The cell-bound phages were eluted with 0.5 mL of PBS containing 100 mmol/L citric acid (pH 2.2) for 10 min and neutralized with 0.5 mL of 1.0 mol/L Tris-HCl (pH 7.5). E.coli TG1 was infected with the eluted phages and plated on 2 × TY agar containing 1% glucose and 100 μg/mL ampicillin. The resultant colonies were propagated and used to prepare phages. Biopanning was performed in triplicate using 1 × 1012 cfu.
FCM for polyclonal scFv phages The polyclonal scFv phages were blocked with 6% BSA in PBS. The blocked scFv-phage supernatants were added to parallel plates containing 1 × 105 HepG2 cells (1 h, 4°C, gentle agitation). The cells were washed twice and centrifuged. Pellets were then resuspended in 100 μL of mouse anti-M13 antibody and incubated for 20 min at 4°C. After being washed twice, the cells were resuspended in 100 μL of goat anti-mouse IgG conjugated with FITC and incubated for 20 min at 4°C. After being washed thrice, the cells were analyzed by flow cytometry.
ELISA and FCM for monoclonal scFv phagesE.coli TG1 was infected with the third round scFv phages and plated on 2 × TY agar to obtain the monoclonal bacteria. PCR was carried out for identifying clones containing the scFv gene sequence. The clones containing scFv gene sequence were infected with helper phage to prepare monoclonal scFv phages. The ELISA for monoclonal scFv phages was performed as the FCM for polyclonal scFv phages described above. After being washed the cells were resuspended in 100 μL of goat anti-mouse IgG conjugated with HRP instead of goat anti-mouse IgG conjugated with FITC, then incubated for 20 min at 4°C. Cell pellets were resuspended in 100 μL of TMB reagents. The ELISA plates were read (A405-A630) and data were analyzed using a spreadsheet program (Microsoft Excel). Monoclonal scFv phages binding to HepG2 cells specifically were prepared for FCM.
Sequencing and analyzing of scFv DNA The positive monoclonal bacteria were isolated and sent to BoYa Shanghai Company for sequencing of DNA. The results of sequencing were Blast in GenBank and analyzed using IMGT/V-Quest software.
IMAC purification of soluble scFv antibody fragments Bacterial clones were cultured in 1 L of 2 × TY, 100 μg/mL ampicillin, 0.1% glucose and induced with 1 mmol/L final concentration of IPTG for 20 h at 30°C. The scFv antibody fragments were harvested from the periplasm and purified by IMAC of pentahistidine tag fused to the scFv fragments. The samples were dialyzed in PBS, filtered, concentrated and stored at 4°C in about 1 mg/mL PBS.
Western blot Five micrograms of purified scFv antibody fragments was run on a 10% SDS-PAGE gel and transferred onto nitro-cellulose. Filters were blocked for 1 h at RT in 10% Marvel/PBS. The scFv antibody fragments were detected with the murine monoclonal antibody 9E10 followed by anti-mouse IgG horseradish peroxidase conjugate. Horseradish peroxidase was visualized with diaminobenzidine tablets in the presence of cobalt ions.
Cell ELISA for purified soluble scFv antibody fragments L02, HepG2, Hela, MCF-7 and K562 cell lines were selected for cell ELISA to identify the specificity of the purified soluble scFv antibody fragments binding to HepG2. The method of cell ELISA was the same as the monoclonal scFv-phage ELISA described above. The purified SLH04 or SLH10 was used as the first antibody, and antibody 9E10 was used as the second antibody.
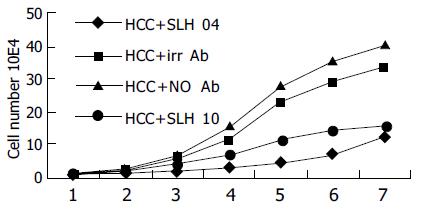
Test of HepG2 cell proliferation A total of 104 HepG2 cells were cultured in a 24-well cell culture plate. Two wells were incubated with 500 μL of purified soluble scFv antibody fragments, SLH04 or SLH10. One well was incubated with 500 μL unrelated antibody, the other well was not treated with any antibody. Cells were cultured for a week and counted everyday.
An aliquot containing 1 × 1012 cfu scFv phage was selected on the HepG2 and L02 cell lines. The recovery rate of the third round biopanning was close to that of the second round indicating that positive clones were fully enriched (Table 1).
| Round | Input phage (cfu) | Eluted phage (cfu) | Recall rate (%) |
| 1 | 1 × 1012 | 1.9 × 106 | 1.9 × 10-6 |
| 2 | 1 × 1012 | 1.2 × 107 | 1.2 × 10-5 |
| 3 | 1 × 1012 | 1.5 × 107 | 1.5 × 10-5 |
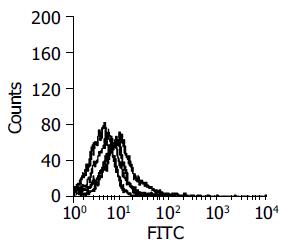
The selected and amplified polyclonal scFv phages were tested by FCM (Figure 1). The result also showed that positive clones were enriched well.
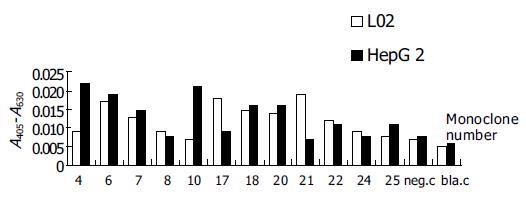
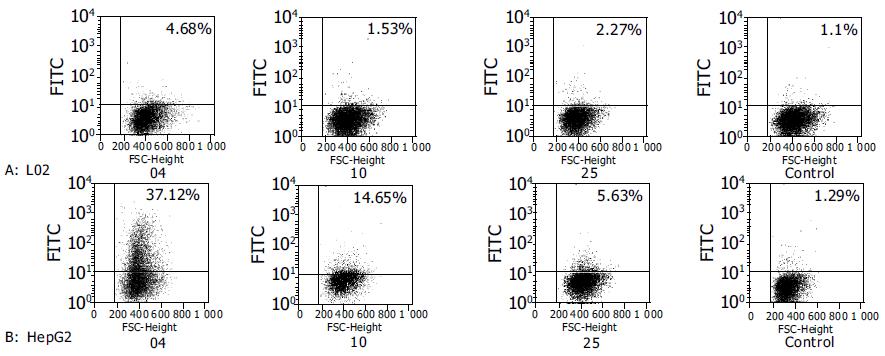
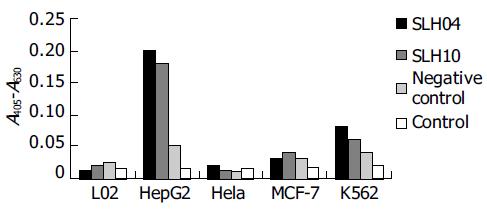
After E.coli TG1 was infected with scFv phages of the third round and plated on 2×TY agar, 31 clones were isolated. PCR was carried out for identification of the clones combined with scFv gene sequence. Twelve positive clones were isolated to prepare monoclonal scFv phages. By ELISA, three scFv phages showed obviously higher binding activity in HepG2 cells according to the data of A405-A630 (Figure 2). By FCM, clones 4, 10 and 25 showed more extensive activity in HepG2 cells (Figure 3).
The results of sequencing and Blast showed that clones 4 and 10 had homologous sequence of human immunoglobulin in GenBank. The scFv DNA sequences were analyzed by IMGT/V-Quest software and submitted to GenBank (accession nos: AY686498 and AY686499). The region of CDR3 of VH was absent in clone 25 and the terminator codon was found in its open reading frame.
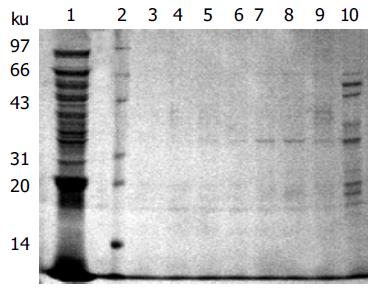
The scFv antibody fragments were harvested from the periplasm of 1 L E.coli HB2151 infected with positive phage SLH04 or SLH10 and the scFv antibody eluted effectively in PBS with 40 or 60 mmol/L imidazole by IMAC purification (Figure 4). SLH10 had the same result as SLH04 (data not shown).
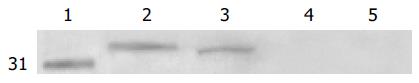
Five micrograms of purified scFv antibody fragments was run on a 10% SDS-PAGE gel and transferred to nitro-cellulose and detected with the antibody 9E10 (Figure 5).
The two positive soluble scFv antibody fragments showed higher ability to bind to HepG2 cells than to other cell lines (Figure 6).
The soluble scFv antibody fragments SLH04 and SLH10 showed growth inhibitory effects (Figure 7).
Antibodies displayed on phages are powerful tools for the identification of ligand specificity to targets of interest[22]. Binding ligands from this library are usually selected by repetitive biopanning rounds usually performed on pure antigens on a solid phase[23]. The isolation of antibodies for cellular receptors using whole cells as affinity matrix has a number of advantages. The receptors are likely to be in their native conformation compared to their purified one. Furthermore, a large number of known and novel surface antigens can be screened at the same time[24,25]. In this respect, some investigators have selected a number of phage antibodies that bind to known and novel surface antigens using whole cells[26,27].
In this study, two scFv antibody fragments were identified which could bind more extensively to HepG2 than to L02 cell line by biopanning of liver cells from a large naive antibody phage library. After three rounds of biopanning, the result indicated that positive clones were fully enriched. Then 12 monoclonal TG1 were picked out from the clones combined with scFv gene sequence. By ELISA, seven of them showed no diversity binding to the two kinds of cells, two of them bound more extensively to L02 cells, three of them showed more extensive binding to HepG2 cells. The three clones were identified as the final positive clones at the end of biopanning. By sequencing and analyzing these three scFv DNA sequences, two clones were discovered having the right full open reading frame; the terminator codon was found in the open reading frame of clone 25 and its region of CDR3 of VH was absent and mutated. Probably this is why the affinity of binding to HepG2 showed no more specificity than the two other positive scFv antibody fragments. Blast in GenBank indicates that heavy chain of the scFv-04 belongs to IGHV2 family and its light chain belongs to Vκ2 family, while the heavy chain of the scFv-10 belongs to IGHV1 family and its light chain belongs to Vλ3 family. Our data indicate that most soluble scFv antibody fragments are expressed in periplasm of the positive E.coli HB2151, few of them can be detected in bacterial supernatant. Therefore, we cannot obtain more soluble scFv antibodies purified from bacterial supernatant by IMAC as described previously[28]. By cell ELISA the two soluble scFv antibody fragments showed higher ability to bind to HepG2 cells than to other cell lines, suggesting that scFv antibodies can bind to living HepG2 cells specifically. Furthermore, our data demonstrate that the two soluble scFv antibody fragments have growth inhibitory effects when cultured with HepG2 cells. Some antibodies have such biological effects on target cells because they are able to mimic the receptor-ligand interaction by triggering specific intracellular cascades of reactions, which eventually lead to biological signals such as inhibition of cell growth. In this regard, anti-transferrin receptor monoclonal antibody has the above characteristics[29,30]. However, the fact that the transferrin receptor is expressed in most cell types makes these antibodies less interesting for cancer therapy. According to our results, the two scFv antibody fragments binding specifically to HepG2 cells can be used as target molecules in the treatment of hepatocellular carcinoma. Antibody phage biopanning method offers a potential application in the identification of human antibodies suitable for antitumor therapy[31]. In summary, large human naive scFv antibody phage library is a useful tool for the selection of antibodies against antigens. The ability of the human scFv antibody fragments to bind to HepG2 cell surfaces will allow them to become a diagnostic and therapeutic reagent of HCC.
Science Editor Wang XL and Guo SY Language Editor Elsevier HK
| 1. | Robinson MK, Weiner LM, Adams GP. Improving mono-clonal antibodies for cancer therapy. Drug Development Res. 2004;61:172-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 2. | Romanov VI. Phage display selection and evaluation of cancer drug targets. Curr Cancer Drug Targets. 2003;3:119-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 3. | Reff ME, Carner K, Chambers KS, Chinn PC, Leonard JE, Raab R, Newman RA, Hanna N, Anderson DR. Depletion of B cells in vivo by a chimeric mouse human monoclonal antibody to CD20. Blood. 1994;83:435-445. [PubMed] |
| 4. | Carter P, Presta L, Gorman CM, Ridgway JB, Henner D, Wong WL, Rowland AM, Kotts C, Carver ME, Shepard HM. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc Natl Acad Sci USA. 1992;89:4285-4289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1299] [Cited by in RCA: 1315] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 5. | Kim ES, Khuri FR, Herbst RS. Epidermal growth factor receptor biology (IMC-C225). Curr Opin Oncol. 2001;13:506-513. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 176] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 6. | Schneebaum S, Troitsa A, Avital S, Haddad R, Kashtan H, Gitstein G, Baratz M, Brazovsky E, Papo J, Skornick Y. Identification of lymph node metastases in recurrent colorectal cancer. Recent Results Cancer Res. 2000;157:281-292. [PubMed] |
| 7. | Butterfield LH. Immunotherapeutic strategies for hepatocellular carcinoma. Gastroenterology. 2004;127:S232-S241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 72] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 8. | Clackson T, Hoogenboom HR, Griffiths AD, Winter G. Making antibody fragments using phage display libraries. Nature. 1991;352:624-628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 869] [Cited by in RCA: 820] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 9. | Waterhouse P, Griffiths AD, Johnson KS, Winter G. Combinatorial infection and in vivo recombination: a strategy for making large phage antibody repertoires. Nucleic Acids Res. 1993;21:2265-2266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 104] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 10. | Sheets MD, Amersdorfer P, Finnern R, Sargent P, Lindquist E, Schier R, Hemingsen G, Wong C, Gerhart JC, Marks JD. Efficient construction of a large nonimmune phage antibody library: the production of high-affinity human single-chain antibodies to protein antigens. Proc Natl Acad Sci USA. 1998;95:6157-6162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 337] [Cited by in RCA: 324] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 11. | Marks JD, Ouwehand WH, Bye JM, Finnern R, Gorick BD, Voak D, Thorpe SJ, Hughes-Jones NC, Winter G. Human antibody fragments specific for human blood group antigens from a phage display library. Biotechnology (N Y). 1993;11:1145-1149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 131] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 12. | Portolano S, McLachlan SM, Rapoport B. High affinity, thyroid-specific human autoantibodies displayed on the surface of filamentous phage use V genes similar to other autoantibodies. J Immunol. 1993;151:2839-2851. [PubMed] |
| 13. | de Kruif J, Terstappen L, Boel E, Logtenberg T. Rapid selection of cell subpopulation-specific human monoclonal antibodies from a synthetic phage antibody library. Proc Natl Acad Sci USA. 1995;92:3938-3942. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 162] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 14. | Van Ewijk W, de Kruif J, Germeraad WT, Berendes P, Röpke C, Platenburg PP, Logtenberg T. Subtractive isolation of phage-displayed single-chain antibodies to thymic stromal cells by using intact thymic fragments. Proc Natl Acad Sci USA. 1997;94:3903-3908. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 60] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 15. | Cai X, Garen A. Anti-melanoma antibodies from melanoma patients immunized with genetically modified autologous tumor cells: selection of specific antibodies from single-chain Fv fusion phage libraries. Proc Natl Acad Sci USA. 1995;92:6537-6541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 137] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 16. | Cai X, Garen A. A melanoma-specific VH antibody cloned from a fusion phage library of a vaccinated melanoma patient. Proc Natl Acad Sci USA. 1996;93:6280-6285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 31] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 17. | Cai X, Garen A. Comparison of fusion phage libraries displaying VH or single-chain Fv antibody fragments derived from the antibody repertoire of a vaccinated melanoma patient as a source of melanoma-specific targeting molecules. Proc Natl Acad Sci USA. 1997;94:9261-9266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 22] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 18. | Bird RE, Hardman KD, Jacobson JW, Johnson S, Kaufman BM, Lee SM, Lee T, Pope SH, Riordan GS, Whitlow M. Single-chain antigen-binding proteins. Science. 1988;242:423-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1070] [Cited by in RCA: 1033] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 19. | Yokota T, Milenic DE, Whitlow M, Schlom J. Rapid tumor penetration of a single-chain Fv and comparison with other immunoglobulin forms. Cancer Res. 1992;52:3402-3408. [PubMed] |
| 20. | Mayer A, Chester KA, Flynn AA, Begent RH. Taking engineered anti-CEA antibodies to the clinic. J Immunol Methods. 1999;231:261-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 21. | Winter G, Griffiths AD, Hawkins RE, Hoogenboom HR. Making antibodies by phage display technology. Annu Rev Immunol. 1994;12:433-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1182] [Cited by in RCA: 1095] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 22. | Hoogenboom HR. Overview of antibody phage-display technology and its applications. Methods Mol Biol. 2002;178:1-37. [PubMed] |
| 23. | Griffiths AD, Williams SC, Hartley O, Tomlinson IM, Waterhouse P, Crosby WL, Kontermann RE, Jones PT, Low NM, Allison TJ. Isolation of high affinity human antibodies directly from large synthetic repertoires. EMBO J. 1994;13:3245-3260. [PubMed] |
| 24. | Kristensen P, Ravn P, Jensen KB, Jensen K. Applying phage display technology in aging research. Biogerontology. 2000;1:67-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 25. | Sawyer C, Embleton J, Dean C. Methodology for selection of human antibodies to membrane proteins from a phage-display library. J Immunol Methods. 1997;204:193-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 26. | Pereira S, Maruyama H, Siegel D, Van Belle P, Elder D, Curtis P, Herlyn D. A model system for detection and isolation of a tumor cell surface antigen using antibody phage display. J Immunol Methods. 1997;203:11-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 32] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 27. | Figini M, Obici L, Mezzanzanica D, Griffiths A, Colnaghi MI, Winter G, Canevari S. Panning phage antibody libraries on cells: isolation of human Fab fragments against ovarian carcinoma using guided selection. Cancer Res. 1998;58:991-996. [PubMed] |
| 28. | Casey JL, Keep PA, Chester KA, Robson L, Hawkins RE, Begent RH. Purification of bacterially expressed single chain Fv antibodies for clinical applications using metal chelate chromatography. J Immunol Methods. 1995;179:105-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 80] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 29. | Kovar J, Naumann PW, Stewart BC, Kemp JD. Differing sensitivity of non-hematopoietic human tumors to synergistic anti-transferrin receptor monoclonal antibodies and deferoxamine in vitro. Pathobiology. 1995;63:65-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 30. | Valentini M, Gregorini A, Bartolucci M, Porcellini A, Papa S. The blockage of the human transferrin receptor by a monoclonal antibody, EA.3, induces growth inhibition in leukemia cell lines. Eur J Histochem. 1994;38 Suppl 1:61-68. [PubMed] |
| 31. | Cooke SP, Boxer GM, Lawrence L, Pedley RB, Spencer DI, Begent RH, Chester KA. A strategy for antitumor vascular therapy by targeting the vascular endothelial growth factor: receptor complex. Cancer Res. 2001;61:3653-3659. [PubMed] |