Published online Jan 14, 2005. doi: 10.3748/wjg.v11.i2.299
Revised: March 22, 2004
Accepted: April 21, 2004
Published online: January 14, 2005
AIM: To investigate the epidemiology of hepatitis B virus (HBV) strains with a mutation at nt551 in surface gene among hepatitis B patients in Nanjing and its neighbourhood.
METHODS: By using mutation-specific polymerase chain reaction (msPCR) established by our laboratory for amplifying HBV DNAs with a mutation at nt551, 117 serum samples taken from hepatitis B patients were detected.
RESULTS: The results showed that 112 samples were positive for nt551A, 4 samples were positive for nt551G. One sample was positive for nt551T. No nt551C of HBV DNA was found. The incidence of HBsAg mutants with G, C, T, A at nt551 among 117 samples was 3.42%, 0%, 0.85%, 95.73%, respectively.
CONCLUSION: In Nanjing and its neighbourhood, hepatitis B patients are mainly infected with wild genotype HBV. The incidence of mutants with a mutation at nt551 in HBV genome is significantly lower than that in wild genotype HBV DNA (P<0.01). The necessity of adding components of HBsAg mutants to HBV vaccine needs further investigation.
- Citation: Ma CL, Fang DX, Yao K, Li FQ, Jin HY, Li SQ, Tan WG. Incidence of HBV variants with a mutation at nt551 among hepatitis B patients in Nanjing and its neighbourhood. World J Gastroenterol 2005; 11(2): 299-302
- URL: https://www.wjgnet.com/1007-9327/full/v11/i2/299.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i2.299
Hepatitis B surface antigen (HBsAg) is considered to be one of the best markers for the diagnosis of acute and chronic HBV infection. But in some patients, this antigen cannot be detected by routine serological assays despite the presence of virus. One of the most important explanations for the lack of detectable HBsAg is that mutations which occur within the “a” determinant of HBV S gene can alter expression of HBsAg and lead to changes of antigenicity and immunogenicity of HBsAg accordingly. As a result, these mutants cannot be detected by diagnostic assays. Thus, it is essential to find specific and sensitive methods to detect the new mutants and further investigate their distribution[1,2].
HBV is a hepatotrophic DNA virus. In the reverse transcription process of DNA replication, HBV DNA template is transcribed by cellular RNA polymerase to pregenomic RNA, which is reversely transcribed to DNA by viral polymerase, and a consequence of the unique way of HBV replication is a significant tendency of mutation[1-3]. Substitution, deletion and frame-shift by insertion or deletion of short sequences have been found in four open reading frames[4-7]. The diversity is also shown in different serological subtypes such as adr, adw, ayr and ayw, which have a common “a” determinant. It is well known that “a” determinant is the common antigenic epitope of all subtypes of HBsAg. A large antigenic area of “a” determinant is now called the major hydrophilic region (MHR). Mutations within MHR of HBsAg have been considered to be associated with vaccine failure and chronic infection[1,2,6,8]. These mutations have been reported repeatedly since Carman et al[9] found the first case of immune escape mutants in 1990. The point mutation reported most commonly in immunized children causes a substitution from Arg to Gly at position 145 of HBsAg[1,3,8,9]. Other reports about substitutions in HBsAg such as 120, 121, 126, 129, 131, 133, 141, 144 are found repeatedly[8-12]. These findings of HBV immune escape mutants have caused attention of scientists all over the world in recent years. Immune escape of HBV mutants is best known to be associated with HBV genome itself, but the immune pressure is considered to be one of the most important factors that result in escape mutants[12-16].
The immune escape variants could influence the effect of HBV vaccine, it is argued that the components of mutant HBsAg should be added into the HBV vaccine in the future[3,12,16,17]. However, in order to achieve this aim, it is necessary to confirm firstly the mutants that are the big problems among hepatitis B patients. At present, it is very important to find new immune escape mutants and further investigate their distribution. Specific and sensitive assays are essential for investigating the distribution of mutants. To detect the mutant HBV DNA, msPCR is a potential method. Our laboratory has discovered a mutation of A-to-G at nt551 of HBV genome, which can result in the alteration of Met to Val at 133aa of HBsAg. To investigate the distribution of mutants with a mutation at nt551 among hepatitis B patients in Nanjing and its neighbourhood in China, we collected 326 serum samples from hepatitis B patients in different hospitals in Nanjing. One hundred and seventeen positive samples for HBV surface gene were detected by using msPCR which is sensitive and specific to the mutation at nt551 in HBV genome.
Three hundred and twenty-six serum samples from hepatitis B patients were provided by Nanjing Jinling Hospital, Bayi Hospital and Nanjing Children’s Hospital. Viral markers were tested by using the enzyme immune assay (EIA). ALT levels of all the samples were abnormal. Diagnosis of hepatitis B was made according to the revised standard of hepatitis B diagnosis established at the Tenth National Symposium on Viral Hepatitis in Xi’an in 2000.
By using the HBV DNA extraction method, all the 326 serum samples were prepared and stored at -20 °C.
After nested PCR amplification, we achieved 101 HBV S DNAs. Thirty-five samples were positive for HBsAg and anti-HBs, and the other 66 samples were negative for HBsAg and positive for anti-HBs.
Primers for nested PCR to amplify HBV S DNA were designed according to the known HBV genome sequences and the main popular subtypes, adr and adw in China.
Primers for the first-round: P1’: 5’ACATCATCTGTGGAAGGC3’, nt2756-nt2773; the upstream primer; P6’: 5’TATCCCATGAAGTTAAGG3’, nt884-nt867, the downstream primer.
Primers for the second-round: PEC: 5’CGGAATTCACCATATTCTTGGGAACAAG3’, nt2 823-nt2 844, the upstream primer; PPS: 5’GCTGCAGGTTTAAATGTATACCCAAAGAC3’, nt838-nt816, the downstream primer.
An EcoRI or a Pst I site was originally added at 5’-end respectively for cloning purpose.
To achieve HBV DNA fragments with an A at nt551, the wild genotype HBV S DNA as template was amplified by using the primer pair P551A-PPS under the condition of regular PCR. The HBsAg mutant with G at nt551 as template was amplified by using the primer pair P551G-PPS to achieve the HBV DNA fragments with a G at nt551. HBV DNA fragments with C or T at nt551 were achieved by introducing mutation in a PCR. The PCR primer sequences were as follows: P551A: 5’TCCTGCTCAAGGAACCTCTA3’, nt532-nt551, upstream primer; P551G: 5’TCCTGCTCAAGGAACCTCTG3’, nt532-nt551, upstream primer; P551C: 5’TCCTGCTCAAGGAACCTCTC3’, nt532-nt551, upstream primer; P551T: 5’TCCTGCTCAAGGAACC TCTT3’, nt532-nt551, upstream primer; PPS: See the above. P551C-PPS and P551T-PPS were used respectively to amplify HBV DNA fragments with C or T at nt551, which were 314 bp in length. Additionally, two upstream primers were designed respectively by introducing mutation to achieve the controls of HBV DNA with C or T at nt551. P551CM: TCCTGCTCAAGGAACCTCTCTGTTTC, nt532-nt557; P551TM: TCCTGCTCAAGGAACCTCTTTGTTTC, nt532-nt557.
In order to amplify HBV DNA specifically, 101 serum samples were amplified by primer pair P551A-PPS using msPCR method. The annealing temperature of PCR was at 71 °C and the concentration of primers was at 0.8 nmol/mL in 25 μL reaction volume. Thirty cycles of amplification were performed, each at 94 °C for 30 s, at 71 °C for 30 s, and at 72 °C for 1 min. Then the negative samples were amplified by primer pairs P551G-PPS, P551C-PPS and P551T-PPS respectively. For primer pairs P551C-PPS and P551T-PPS, the annealing temperature was at 72 °C and the concentration of primers was at 0.2 nmol/mL in 25 μL reaction volume, and other conditions were similar to the amplification by P551A-PPS and P551G-PPS.
χ2-test was used to assess the statistical significance of differences. P<0.01 was considered statistically significant.
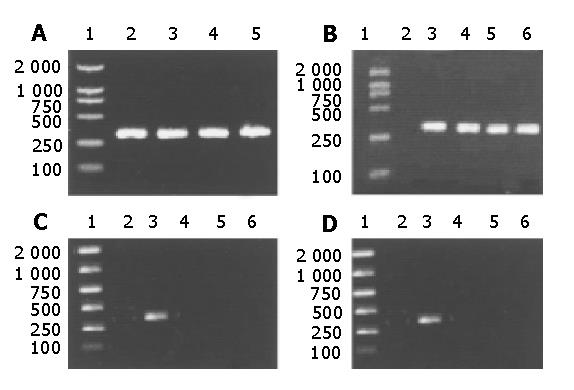
The HBV S DNA with A, G, C or T at nt551 was amplified respectively for control. The amplified DNA fragments were 314 bp in length. This result is shown in Figure 1.
One hundred and one serum samples positive for HBV S DNA were detected. After gel electrophoresis, 35 samples (positive for HBsAg and anti-HBs) were positive for P551A-PPS amplification, that is to say, each of these 35 samples was a wild genotype HBV genome with an A at nt551. Among the 66 samples negative for HBsAg and positive for anti-HBs, 3 samples were negative for amplification by P551A-PPS. They were samples of No.127, No.210 and No.216.Then, these 3 samples were amplified by using P551G-PPS, P551C-PPS and P551T-PPS. The electrophoresis results of them were as follows.
As positive and negative controls, No.2, No.8 and No.57 previously sequenced were known to have A, G and T at nt551 respectively. These results confirmed that No.127, No.210 and No.216 all had a G at nt551.
By using msPCR, 101 serum samples positive for HBV S DNA were detected and the other 16 samples were taken from the result we detected before. The results showed that 112 samples were positive for nt551A, 4 samples were positive for nt551G (one of them was taken from the previous DNA sequence). One sample was positive for nt551T. No nt551C of HBV DNA was found. Thus, the incidence of HBsAg nt551G mutant, nt551C mutants, nt551T mutants, and nt551A of wild genotype HBV DNAs was 3.42%, 0%, 0.85% and 95.73% respectively among the 117 samples. The statistical analysis results are summarized in Tables 1, 2 and 3.
| nt551 A | nt551 G | |
| Number of case | 112 | 4 |
| Incidence | 112/117 (95.37) | 4/117 (3.42)b |
| nt551 A | nt551 C | |
| Number of case | 112 | 0 |
| Incidence | 112/117 (95.37) | 0/117 (0)b |
| nt551 A | nt551 T | |
| Number of case | 112 | 1 |
| Incidence | 112/117 (95.37) | 1/117 (0.85)b |
It is well known that mutants of HBsAg are able to cause infection and horizontal transmission despite the presence of anti-HBs[18-22]. It has been found that increasing use of HBV vaccine has an overwhelming positive influence on the prevention of hepatitis B viral infection, but has no effective impact on those mutants[23]. Mutations in the coding region of “a” determinant of HBsAg could not be detected in some routine assays[24,25]. In our laboratory a specific and sensitive method for monitoring the HBsAg mutant with a mutation at nt551 has been established. The method of msPCR is different from immunoassays based on the antigen-antibody reaction[26,27]. To detect the mutant HBsAg, unique specific monoclonal antibodies are required. But these kinds of antibodies are limited or not available commercially. Because HBV is a double-stranded DNA virus, its genome is fairly stable in blood and tissues, PCR amplification of HBV DNA is relatively easy. msPCR is a practical method to detect the specific site mutation. This method was firstly developed to detect point mutation of allele-special genes of β-globin genome DNA for sickle cell anemia[28]. Then it is used in virological studies. The feasibility of msPCR has been confirmed by DNA sequence analysis, that is to say, the result of msPCR is accordant with that of DNA sequencing. After msPCR establishment, we have found that the annealing temperature is the key factor to msPCR method. Additionally, the concentration of primers is also an important factor for this msPCR. In short, different primers amplify HBV DNA specifically under different conditions.
In our study, all serum samples detected were taken from hepatitis B patients in Nanjing and its neighbourhood and four mutants of nt551G were found. Their detailed clinical data were as follows: No.57 taken from a man was negative for HBsAg and positive for anti-HBs. His ALT level was considered to be abnormal. No.127 taken from a 3-year-old boy was negative for HBsAg and positive for anti-HBs and HBeAg. His ALT level was higher than normal value. His mother was a HBsAg carrier and the boy was injected with HBV vaccine and HBIg when he was born. No. 210 taken from a 9-year-old boy, was negative for HBsAg and positive for anti-HBs and his ALT level was considered to be abnormal. No. 216 taken from an old male chronic hepatitis B patient, was negative for HBsAg and positive for anti-HBs and his ALT level was considered to be abnormal.
The reason why all these four samples were negative for HBsAg and positive for anti-HBs might be that the mutations of A→G at nt551 could cause a change from Met to Val at 133aa of HBsAg and the structure of HBsAg was altered accordingly. The affinity between HBsAg and anti-HBs might diminish subsequently. The alteration of Met to Val at 133aa of HbsAg can reduce the combination ability between HBsAg and mAb of anti-HBs.
The detection results of these four samples have shown that the anti-HBs loses its protection against HBsAg as usual. On the contrary, they may form immune pressure by recognizing and combining with wild genotype HBV strains, but those mutants having no epitopes can survive and multiply. Therefore, a specific and sensitive method should be used to find those mutants of HBsAg and provide theoretical data for clinical diagnosis and treatment of hepatitis B.
In conclusion, most HBV infections are caused by wild genotype HBV strains. Whether it is necessary to add the component of HBsAg mutants to HBV vaccine needs further study and investigation.
Edited by Wang XL and Zhu LH
| 1. | Koyanagi T, Nakamuta M, Sakai H, Sugimoto R, Enjoji M, Koto K, Iwamoto H, Kumazawa T, Mukaide M, Nawata H. Analysis of HBs antigen negative variant of hepatitis B virus: unique substitutions, Glu129 to Asp and Gly145 to Ala in the surface antigen gene. Med Sci Monit. 2000;6:1165-1169. [PubMed] |
| 2. | Brunetto MR, Rodriguez UA, Bonino F. Hepatitis B virus mutants. Intervirology. 1999;42:69-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 51] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 3. | Kfoury Baz EM, Zheng J, Mazuruk K, Van Le A, Peterson DL. Characterization of a novel hepatitis B virus mutant: demonstration of mutation-induced hepatitis B virus surface antigen group specific "a" determinant conformation change and its application in diagnostic assays. Transfus Med. 2001;11:355-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 4. | Dong J, Cheng J, Wang Q, Huangfu J, Shi S, Zhang G, Hong Y, Li L, Si C. The study on heterogeneity of hepatitis B virus DNA. Zhonghua YiXue ZaZhi. 2002;82:81-85. [PubMed] |
| 5. | Kreutz C. Molecular, immunological and clinical properties of mutated hepatitis B viruses. J Cell Mol Med. 2002;6:113-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 55] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 6. | Zhong S, Chan JY, Yeo W, Tam JS, Johnson PJ. Hepatitis B envelope protein mutants in human hepatocellular carcinoma tissues. J Viral Hepat. 1999;6:195-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 7. | Weinberger KM, Zoulek G, Bauer T, Böhm S, Jilg W. A novel deletion mutant of hepatitis B virus surface antigen. J Med Virol. 1999;58:105-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 8. | Yukimasa N, Ohkushi H, Fukasawa K, Fukuchi K, Takagi Y, Gomi K. Hepatitis B virus gene mutations in the sera of three patients with coexisting hepatitis B surface antigen and anti-surface antibody. Rinsho Byori. 2000;48:184-188. [PubMed] |
| 9. | Carman WF, Zanetti AR, Karayiannis P, Waters J, Manzillo G, Tanzi E, Zuckerman AJ, Thomas HC. Vaccine-induced escape mutant of hepatitis B virus. Lancet. 1990;336:325-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 817] [Cited by in RCA: 772] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 10. | Yang X, Lei J, Zhang Y, Luo H, Huang L, Zheng Y, Tang X, Li L. A novel stop codon mutation in S gene: the molecular basis of a patient with cryptogenic cirrhosis. Zhonghua YiXue ZaZhi. 2002;82:400-402. [PubMed] |
| 11. | Zhu Q, Lu Q, Xiong S, Yu H, Duan S. Hepatitis B virus S gene mutants in infants infected despite immunoprophylaxis. Chin Med J (Engl). 2001;114:352-354. [PubMed] |
| 12. | Santantonio T, Gunther S, Sterneck M, Rendina M, Messner M, Launois B, Francavilla A, Pastore G, Will H. Liver graft infection by HBV S-gene mutants in transplant patients receiving long-term HBIg prophylaxis. Hepatogastroenterology. 1999;46:1848-1854. [PubMed] |
| 13. | Rodriguez-Frias F, Buti M, Jardi R, Vargas V, Quer J, Cotrina M, Martell M, Esteban R, Guardia J. Genetic alterations in the S gene of hepatitis B virus in patients with acute hepatitis B, chronic hepatitis B and hepatitis B liver cirrhosis before and after liver transplantation. Liver. 1999;19:177-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 14. | He C, Nomura F, Itoga S, Isobe K, Nakai T. Prevalence of vaccine-induced escape mutants of hepatitis B virus in the adult population in China: a prospective study in 176 restaurant employees. J Gastroenterol Hepatol. 2001;16:1373-1377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 48] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 15. | Cooreman MP, Leroux-Roels G, Paulij WP. Vaccine- and hepatitis B immune globulin-induced escape mutations of hepatitis B virus surface antigen. J Biomed Sci. 2001;8:237-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 141] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 16. | Cooreman MP, Leroux-Roels G, Paulij WP. Vaccine- and hepatitis B immune globulin-induced escape mutations of hepatitis B virus surface antigen. J Biomed Sci. 2001;8:237-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 141] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 17. | Komatsu H, Fujisawa T, Sogo T, Isozaki A, Inui A, Sekine I, Kobata M, Ogawa Y. Acute self-limiting hepatitis B after immunoprophylaxis failure in an infant. J Med Virol. 2002;66:28-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 18. | Heijtink RA, van Bergen P, van Roosmalen MH, Sünnen CM, Paulij WP, Schalm SW, Osterhaus AD. Anti-HBs after hepatitis B immunization with plasma-derived and recombinant DNA-derived vaccines: binding to mutant HBsAg. Vaccine. 2001;19:3671-3680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 19. | Banerjee K, Guptan RC, Bisht R, Sarin SK, Khandekar P. Identification of a novel surface mutant of hepatitis B virus in a seronegative chronic liver disease patient. Virus Res. 1999;65:103-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 20. | Chen WN, Oon CJ. Hepatitis B virus surface antigen (HBsAg) mutants in Singapore adults and vaccinated children with high anti-hepatitis B virus antibody levels but negative for HBsAg. J Clin Microbiol. 2000;38:2793-2794. [PubMed] |
| 21. | Oon CJ, Chen WN, Goo KS, Goh KT. Intra-familial evidence of horizontal transmission of hepatitis B virus surface antigen mutant G145R. J Infect. 2000;41:260-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 42] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 22. | Chen WN, Oon CJ, Koh S. Horizontal transmission of a hepatitis B virus surface antigen mutant. J Clin Microbiol. 2000;38:938-939. [PubMed] |
| 23. | Schories M, Peters T, Rasenack J. Isolation, characterization and biological significance of hepatitis B virus mutants from serum of a patient with immunologically negative HBV infection. J Hepatol. 2000;33:799-811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 32] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 24. | Weinberger KM, Bauer T, Böhm S, Jilg W. High genetic variability of the group-specific a-determinant of hepatitis B virus surface antigen (HBsAg) and the corresponding fragment of the viral polymerase in chronic virus carriers lacking detectable HBsAg in serum. J Gen Virol. 2000;81:1165-1174. [PubMed] |
| 25. | Chen WN, Oon CJ, Moh MC. Detection of hepatitis B virus surface antigen mutants in paraffin-embedded hepatocellular carcinoma tissues. Virus Genes. 2000;20:263-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 26. | Ijaz S, Torre F, Tedder RS, Williams R, Naoumov NV. Novel immunoassay for the detection of hepatitis B surface 'escape' mutants and its application in liver transplant recipients. J Med Virol. 2001;63:210-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 27. | Jolivet-Reynaud C, Lésenéchal M, O'Donnell B, Becquart L, Foussadier A, Forge F, Battail-Poirot N, Lacoux X, Carman W, Jolivet M. Localization of hepatitis B surface antigen epitopes present on variants and specifically recognised by anti-hepatitis B surface antigen monoclonal antibodies. J Med Virol. 2001;65:241-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 26] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 28. | Wu DY, Ugozzoli L, Pal BK, Wallace RB. Allele-specific enzymatic amplification of beta-globin genomic DNA for diagnosis of sickle cell anemia. Proc Natl Acad Sci USA. 1989;86:2757-2760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 407] [Cited by in RCA: 402] [Article Influence: 11.2] [Reference Citation Analysis (0)] |