INTRODUCTION
Retinoids, the important factors in modulating cell growth, differentiation, apoptosis and suppressing carcinogenesis in vitro and in vivo, are a group of natural and synthetic vitamin A analogs. In many animal models, such as cancers of skin, lung, prostate and esophagus, retinoids could suppress or reverse epithelial carcinogenesis[1]. The effects of retinoids are mainly mediated through two classes of nuclear receptors: the retinoic acid receptors (RARs) and retinoid X receptors (RXRs) which belong to a steroid/thyroid hormone-receptor superfamily. Each of them is composed of 3 subtypes (α, β and γ), of which RAR β is expressed as three isoforms: β1, β2 and β4[2]. Most human cells express RAR β2 as the major isoform. Many studies have confirmed that altered expression or function of the retinoid receptors may be related to malignant transformation of human cells[3,4].
RAR β2 was decreased in many tumors, including lung carcinoma, breast cancer and esophageal cancers[5-7]. A study of esophageal cancer demonstrated that loss of RAR β expression was an early event associated with esophageal carcinogenesis and the status of squamous differentiation[6], however the mechanisms underlying the inactivation of RAR β2 expression in cancer, especially the esophageal cancer, have not been well known yet. In breast cancer, no mutation or polymorphism was detected within the βRARE promoter, which was located in the promoter regions of target genes, and the lack of correlation between RAR β2 expression and LOH at chromosome 3p24 was demonstrated in esophageal cancer[8,9]. The RAR β2 promoter was characterized by an island, which was located in the 5’-untranslated region, and increasing data showed that aberrant methylation of the CpG islands in tumors was associated with transcriptional repression of tumor suppressor genes, such as p16 in esophageal cancer[10]. To clarify the epigenetic mechanism of RAR β2 loss or reduction in ESCC cell lines, we analyzed the methylation status of the RAR β2 promoter region by MSP assay and bisulfite sequencing. Our results indicated that downregulation of RAR β2 expression was associated with the aberrant methylation of the CpG islands in some ESCC cell lines, but not in others.
MATERIALS AND METHODS
Reagents and chemicals
5-aza-dc (Sigma, St. Louis, MO) was dissolved in pure ethanol at a stock concentration of 2 mM, and stored in aliquots at -70 °C.
ESCC cell cultures and treatments
Six cell lines were used in this study: KYSE180, KYSE410, KYSE450, KYSE510, COLO680N, and TE12 were purchased from DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH). Each of the cell lines was cultured in either 90% RPMI 1640 medium (Invitrogen, Carlsbad, CA) or 45% RPMI 1640/45% Ham’s F-12 medium (Invitrogen) with 100 mL/L fetal bovine serum at 37 °C containing 50 mL/LCO2. In the demethylation experiment, the cells with lower RAR β2 expression were treated with 5-aza-dc at a final concentration of 5 µmol/L for 96 h. The growth medium with or without the drug was changed every 24 h.
DNA preparation
The esophageal cell cultures were digested with trypsin-EDTA before collection. Genomic DNA from cell lines was extracted by proteinase-K digestion and phenol/chloroform extraction as described previously[11]. DNA was dissolved in TE buffer and stored at -20 °C.
RNA isolation and semi-quantitative RT-PCR
Total RNAs were isolated from ESCC cell lines with Trizol reagent (Invitrogen) according to the manufactures’ instructions. RNA quality was assessed with agarose gel electrophoresis and spectrophotometric analysis. Ten µg of total RNA of each sample was treated with 2 µL DNase I (10 units/µL, Promega, Madison, WI), 1 µL LRNasin (40 units/µL, Promega) at 37 °C for 15 min to remove contaminated DNA. First strand cDNA was reversely transcribed from 5 µg of total RNAs using SuperScriptTM first-strand synthesis system for RT-PCR II kit (Invitrogen) at 42 °C for 80 min and 0.5-1 µL aliquots of the cDNA was then subjected to RT-PCR. The primers used for the amplification of RAR β2 were as follows: sense 5’-ATCGATGCCAATACTGTCGA - 3 ’, antisen se 5 ’ - GACTCGATGGTCAGCACTG-3’[8]. As, GAPDH was used as an internal control to ensure quality and quantity of cDNA for each RT-PCR. A product with 241 bp was generated, and PCR products were analyzed on 20g/L agarose gels. The PCR assay for each sample was repeated at least twice.
Methylation-specific PCR
Bisulfite modification of genomic DNA from primary esophageal cancer cell lines was carried out essentially as described previously[12]. Briefly, 2 µg of genomic DNA was denatured with freshly prepared NaOH (final concentration 0.3 mol/L) for 15 min in a 37 °C water bath, 30 µL of freshly prepared 10 mmol/L hydroquinone (Sigma) and 520 µL of freshly prepared 3.6 mol/Lsodium bisulfite (Sigma) at pH 5 were added and mixed. The samples were then incubated under mineral oil for 16 h at 50°C. The modified DNA was purified with a Wizard DNA clean-up system (Promega). After purification, the samples were treated with NaOH (0.3 M) for 15 min at 37 °C. The DNA was ethanol-precipitated and re-suspended in water for MSP analysis.
The bisulfite-induced changes affecting unmethylated (U) and methylated (M) alleles were detected by nested PCR. The primers for the first round PCR were as follows: sense 5’ - AAGTGAGTTGTTTAGAGGTAGGAGGG3’, and antisense 5’- CCTATAATTAATCCAAATAATCATTTACC-3’[13]. The amplification was performed under the following modified conditions: pre-denaturation at 95 °C for 5 min; 35 amplification cycles (denature at 95 °C for 15 s; annealing at 53 °C for 15 s; and extension at 72 °C for 30 s) and final extension at 72 °C for 5 min for the first round of PCR. The PCR products were diluted (1:10 dilution) and used in the second round of PCR with the primers: forward 5’-TTGAGAATGTGAGTGATTTGA-3’, reverse 5’-AACCAATCCAACCAAAACAA-3’ for the unmethylated sequence; and forward 5’-TCGAGAACGCG-AGCGATTCG-3’, reverse 5’-GACCAATCCAACCGAAA-CGA-3’ for the methylated sequences[14]. The amplification was performed under following conditions: pre-denaturation at 95 °C for 5 min; 35 amplification cycles (denature at 95 °C for 15 s; annealing at 52 °C for 15 s; and extension at 72 °C for 15 s for U-primers) and pre-denaturation at 95 °C for 5 min; 20 amplification cycles (denature at 95 °C for 15 s; annealing at 62 °C for 15 s; and extension at 72 °C for 15 s M-primers). DNA from normal lymphocytes was used as negative control for methylated alleles, and placental DNA treated in vitro with SssI methyltransferase (New England biolabs, Beverly, MA) was used as positive control for methylated genes. The product of 146 bp was generated, and the PCR products were analyzed on 25g/L agarose gels and photographed. The controls without DNA were performed in each set of PCR, and MSP assay for each sample was repeated at least twice.
Bisulfite sequencing
The PCR products amplified with primers specific either for the methylated or for the unmethylated DNA were purified and cloned into the pMD-T vector (Takara, Dalian, China) according to the manufacture’s protocol. Recombinants were transformed into Escherichia coli. Plasmid DNA was isolated and integrated PCR fragments were verified by enzyme digestion and PCR. The cloned PCR fragments were further analyzed by automated sequencing (Bioasia, Shanghai, China).
RESULTS
Expression of RAR β2 in ESCC cell lines
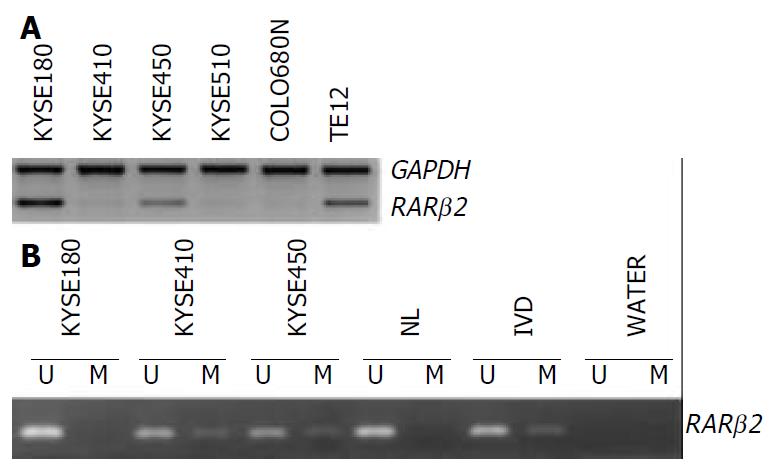
The expression of the RAR β2 mRNA was detected in the six ESCC cell lines by RT-PCR. Figure 1 shows representative examples of RT-PCR and MSP results. Markedly downregulation of RAR β2 expression was observed in the cell lines KYSE410, KYSE510 and COLO680N as shown in Figure 1A.
Figure 1 RARβ2 expression and methylation in ESCC cell lines.
A: RT-PCR analysis of RAR β2 expression, GAPDH was used as internal control. B: MSP analysis of RAR β2 promoter methyla-tion status, (U) lanes and (M) lanes represent amplification of unmethylated and methylated alleles, respectively. In vitro methylated DNA (IVD) and normal human peripheral lym-phocytes (NL) serve as positive and negative methylation controls, respectively.
Methylation analysis of the RAR β2 promoter region
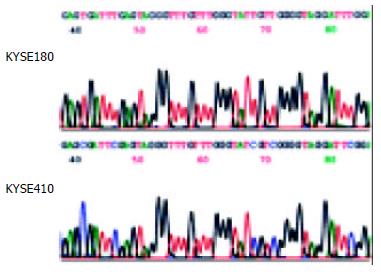
Using MSP method, both the methylated and unmethylated alleles were found in the four cell lines except KYSE180 and TE12 in which only the unmethylated alleles were present (Figure 1B). The quantitation of the unmethylated or the methylated products varied in different cell lines. To confirm the reliability of the nested MSP method, PCR products amplified with primers specific either for the methylated (cell lines KYSE410) or for the unmethylated (cell line KYSE180) DNA were cloned and sequenced. Bisulfite sequencing of 4 individual clones of PCR products from cell line KYSE180 revealed no methylation of CpGs within the promoter region, whereas the methylated products showed dense methylation within the CpG islands in cell line KYSE410 (Figure 2).
Figure 2 Sodium bisulfite sequencing of RAR β2 in the cell lines that were found to include only unmethylated alleles (cell line KYSE180) or partial methylated alleles (cell line KYSE410) by MSP.
Association between RAR β2 promoter methylation and downregulation of its expression in ESCC cell lines
The reduction of RAR β2 mRNA expression was observed in three of the four cell lines that had partially methylated alleles, however the another one cell line showed strong RAR β2 gene expression, which suggesting that the aberrant methylation might play other roles except leading to gene silencing.
Restoration of RAR β2 expression after 5-aza-dc treatment
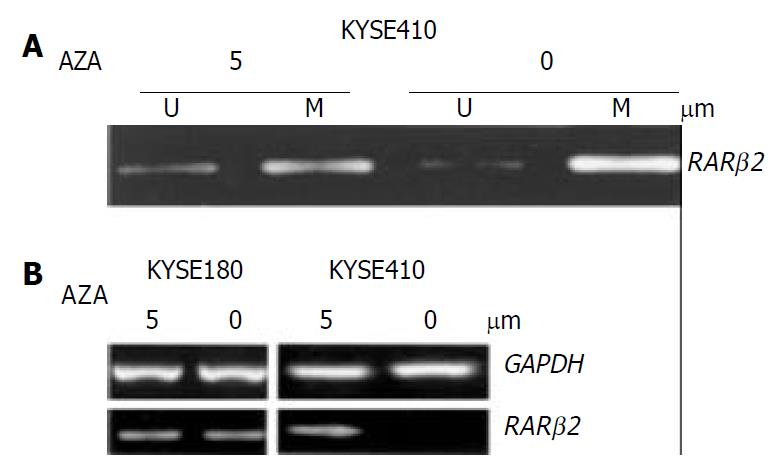
To confirm whether RAR β2 promoter methylation could be further linked to downregulation of its expression, the cancer cell lines which showed evidently decreased expression of RAR β2 and cell line KYSE180 without methylated alleles were treated with the demethylating agent (5-aza-dc). To determine the effects of demethylating agent, we analyzed the methylation status in KYSE410 cells after 5-aza-dc treatment, and partial demethylation was detected by MSP as shown in Figure 3A. In addition, the MSP product was still observed after treatment because 5-aza-dc only inhibited methylation of newly synthesized DNA. The level of RAR β2 mRNA was not changed in cell line KYSE180 before and after demethylating treament. Among the three cell lines where RAR β2 genes were methylated, the treatment led to up-regulation of RAR β2 mRNA expression only in cell line KYSE410 (Figure 3B). The other two cell lines showed no detectable re-expression of RAR β2 mRNA after 5-aza-dc treatment (data not shown). These data indicated that RAR β2 promoter methylation was closely associated with downregulation of RAR β2 expression in some ESCC cell lines.
Figure 3 Treatment with the indicated concentrations of 5-aza-dc induced demethylation and RARβ2 mRNA expression in ESCC cell lines.
A: MSP analysis shows partial demethylation of the RARβ2 promoter region after 5-aza-dc treatment. B: RT-PCR analysis of RARβ2 mRNA expression before and af-ter 5-aza-dc treatment in cell lines that were found to have either positive (cell line KYSE180) or reduced (cell line KYSE410) baseline RARβ2 expression, GAPDH was used as internal control
DISCUSSION
There were two major RAR β2 antineoplastic mechanisms in breast cancer cell lines, which were RA-dependent and RA-independent apoptosis[15,16], however little was known how RAR β2 performed its anticancer activity. A growing evidence indicated that RAR β2 was required for the growth inhibitory action of RA. In RAR β2-negative cancer cells, RA-induced growth inhibition and decreased tumorigenicity could be restored by expression of RAR β2; on the contrary, in RAR β2-positive cancer cells, RA effects could be abolished by inhibition of RAR β2 expression[17-19]. Another study indicated that not only expression but also up-regulation of RAR β was linked to retinoid sensitivity in esophageal cancer cell lines[20], and our previous study also suggested that RA resistance in EC109 cells paralleled with the loss of RAR β2 inducibility [21]. A clinical trial using N-4-(ethoxycarbophenyl) retinamide, a synthetic retinoid, demonstrated that cancer incidence among the treatment groups with severe esophageal dysplasia was decreased by 43.2% compared with the placebo group[22], whereas this was not the case for recent clinical trials of 13-cis retinoic acid in the treatment of advanced esophageal cancer patients, for which an explanation was that the loss of RAR β expression could lead to the resistance to treatment with 13-cis RA in esophageal cancer[23,24]. Recent study suggested that hypermethylation of RAR β might play an important role in the early stage of esophageal squamous cell carcinogenesis[25]. However, the mechanisms of RAR β2 suppression remained largely unknown in esophageal cancer.
Genetic and epigenetic mechanisms may be involved in progressive decrease in RAR β2 mRNA expression during esophageal carcinogenesis. RAR β was localized on chromosome 3p24, and LOH occurred frequently on chromosome arm 3p in many cancers, including esophageal cancer[2]. However, no correlation was observed between the expression of RAR β and the LOH on chromosome 3p24 in ESCC; moreover, no mutations or other genetic alterations were found in different cell lines without RAR β2 mRNA expression[9,26,27]. Thus, other mechanisms for RAR β2 suppression should be considered. DNA methylation, which led to the inactivation of all kinds of essential genes, such as HLA class I, E-cadherin, FHIT and p16, was an important mechanism for gene silencing or suppressing in ESCC[10,28-30]. In addition, Arapshian et al[31] concluded that methylation of the RARE region might be particularly important in RAR β2 suppression. Data presented in this report demonstrated that epigenetic silencing of the RAR β2 gene promoter might be an important event, and that downregulation of RAR β2 expression was closely associated with RAR β2 gene promoter methylation in some ESCC cell lines. In our study, of the three RAR β2-downregulated cell lines, only the one poorly differentiated cell line showed evidently re-expression of RAR β2 with the partial demethylation of its promoter region, indicating that repression was, at least in part, mediated by methylation in the poorly differentiated cell lines, and multiple mechanisms were involved in the gene silencing in the other two cell lines. Our results suggested that a relationship between methylation status and decreased RAR β2 expression only existed in some differentiated ESCC cell lines or ESCC. Previous study has demonstrated that for breast carcinoma a relationship between methylation status and decreased RAR β2 expression in breast cancer was found only in invasive grade II lesions[13]. There was mounting evidence that suggested epigenetic modifications might be a mechanism responsible for the lack of RAR β2 expression in many epithelial cancers or cancer cell lines, such as breast cancer, colon cancer and prostate cancer[8,13,32,33]. Taken together the further analysis of the cytogenetic data of the cell lines, we found that there was no deletion in 3p in the cell line exhibiting re-expression of RAR β2. We therefore speculate that methylation may be at least one of the major mechanisms resulting in loss of RAR β2 expression in such cell lines. It has been reported that in ESCC cell lines without any structural alteration, hypermethylation at CpG sites was the important mechanism for transcription inactivation of the FHIT gene[30]. Given that the RAR β2 gene is recognized as a putative tumor suppressor gene, restoration of its function may have implications for both cancer therapy and prevention.
Notely, demethylation could only lead to the restoration of RAR β2 expression in 1/3 ESCC cell lines without 3p deletion, but not in the other cell lines with 3p deletion, indicating that there were other mechanisms, such as deletion, involved in the loss of RAR β2 expression in ESCC cell lines. It was reported that differential function of coactivators and corepressors might determine the level of RAR β induction that might mediate retinoid action in colon cancer cells[34]. In lung cancer cell lines, loss of RAR β gene expression was linked to aberrant histone H3 acetylation, so the aberrant acetylation status might have effects on RAR β2 expression[35]. In addition, orphan receptor COUP-TF have been identified to play a key role in modulating RAR β expression and retinoid sensitivity in cancer cells[36]. Thus, multiple mechanisms may contribute to the loss of RAR β2.
In conclusion, this study not only shows a high frequency of methylation of RAR β2 promoter region but also provides the first evidence that downregulation of RAR β2 is closely associated with DNA methylation in ESCC cell lines. These findings will be helpful to further understand the mechanisms underlying tumor progression in ESCC, and subsequently improve the prevention and treatment.