Published online Dec 1, 2004. doi: 10.3748/wjg.v10.i23.3518
Revised: March 22, 2004
Accepted: April 5, 2004
Published online: December 1, 2004
AIM: To investigate the expression of NDRG2 and mutation of the entire coding region of NDRG2 in human liver and pancreatic cancers, and to further discuss the possible causes of NDRG2 distinct expression patterns.
METHODS: Reverse transcription-polymerase chain reaction (RT-PCR) was used to analyze the expression of NDRG2 mRNA in 37 fresh cancer specimens (including 8 cases of pancreatic cancer and 29 cases of liver cancer) and adjacent normal tissues collected from clinical operation. In addition, mutation analysis of the whole coding region of NDRG2 in these cancers was examined by polymerase chain reaction-single strand conformational polymorphism (PCR-SSCP).
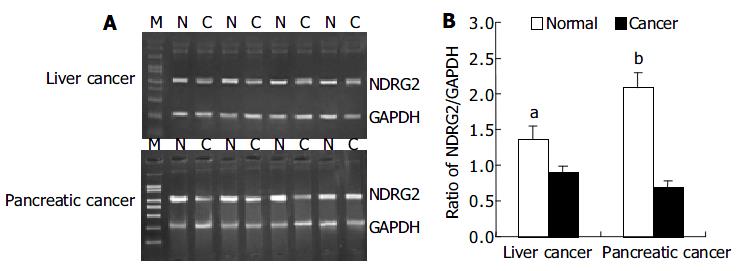
RESULTS: Compared with adjacent normal tissues, the expression levels of NDRG2 mRNA in corresponding cancer tissues reduced significantly (pancreatic cancer: 0.680 ± 0.112 vs 2.089 ± 0.214, P < 0.01) (liver cancer: 0.894 ± 0.098 vs 1.345 ± 0.177, P < 0.05). Using PCR-SSCP, the mutation of the whole coding region of NDRG2 was not found in those cancer tissues where the expression of NDRG2 mRNA reduced markedly.
CONCLUSION: NDRG2 gene might express differently between normal tissues and cancer tissues, and might play an important role in the development of pancreatic cancer and liver cancer. Low expression of NDRG2 might be unrelated to the mutation of coding region of NDRG2.
- Citation: Hu XL, Liu XP, Lin SX, Deng YC, Liu N, Li X, Yao LB. NDRG2 expression and mutation in human liver and pancreatic cancers. World J Gastroenterol 2004; 10(23): 3518-3521
- URL: https://www.wjgnet.com/1007-9327/full/v10/i23/3518.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i23.3518
NDRG2, a new member of the N-Myc downstream-regulated gene (NDRG) family including NDRG1-4, was first discovered and cloned from normal human whole brain cDNA library by subtractive hybridization in our laboratory[1]. NDRG2 is located on chromosome 14q11.2, including 16 exons and 15 introns, and its cDNA is 2024 bp in length[2]. Studies from our laboratory and others have shown that NDRG2 was highly expressed in many normal tissues, especially in the brain, heart, skeletal muscle, and kidney, whereas low expressed or not detected in various tumors and tumor cell lines[3,4]. This suggests that NDRG2 inactivation might play an important role in some tumor genesis or evolution, and NDRG2 might be a new cancer suppressor gene candidate. To date, no report on mutation analysis of NDRG2 in a variety of tumors has been presented. This study was conducted to determine whether the distinct expression of NDRG2 was related to gene mutation as most cancer suppressor genes. The expression of NDRG2 mRNA in 37 patients with liver cancer and pancreatic cancer was detected, and the mutation of the whole coding region of NDRG2 in normal and cancer specimens was analyzed by PCR-SSCP.
Thirty-seven fresh cancer specimens (including 8 cases of pancreatic cancer and 29 cases of liver cancer) and corresponding adjacent normal tissues were collected from patients who underwent surgery at First Affiliated Hospital of Xi’an Jiaotong University, Tangdu Hospital and Xijing Hospital between May 2003 and September 2003. All specimens were examined by microscope after haematoxylin and eosin (H&E) staining.
Total RNA was extracted from tissue specimens using TRIzol reagent (GIBCO BRL, USA) according to the manufacturer’s instructions. Reverse transcription reaction was set up according to Promega’s reverse transcription system protocol. cDNAs were then amplified by PCR using specific primers for human NDRG2: upstream primer 5’- GC GGA TCC ATG GCG GAG CTG CAG GAG GTC -3’; and downstream primer 5’- GC GAA TTC AAC AAG GGC CAT TCA ACA GGA GAC -3’, about 1200 bp. The primer sequences of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) which was used as an internal quantitative control for the amplification were as follows: upstream primer 5’ -GCC TCA AGA TCA GCA AT- 3’; downstream primer 5’ -AGG TCC ACC ACT GAC ACG TT- 3’, 310 bp. Cycling condition for NDRG2 was at 94 °C for 5 min, followed by 32 cycles of 94 °C for 40 s, 59 °C for 50 s, and 72 °C for 60 s, and the cycling condition for GAPDH was at 94 °C for 5 min, followed by 30 cycles of 94 °C for 30 s, 57 °C for 30 s, and 72 °C for 30 s, and finally for 10 min at 72 °C. The PCR products were then analyzed and visualized on 12 g/L agarose gel containing 5 g/L ethidium bromides. Results were normalized by the ratio of band density of NDRG2 mRNA to GAPDH mRNA.
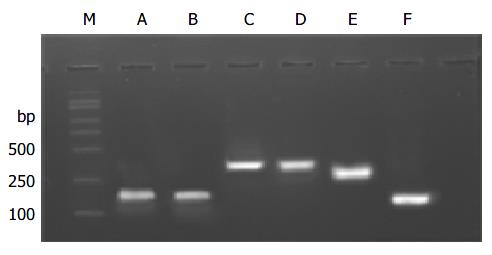
According to cDNA sequence of NDRG2 gene, we designed by DNAstar analysis software and synthesized 6 pairs of primers covering the whole coding region of NDRG2 gene, with length of each PCR product around 150 to 370 bp, and PCR reaction conditions are described in Table 1. The PCR products were separated by electrophoresis on 12 g/L agarose gel to check the specificity of each PCR reaction. The products were mixed with the same amount of formamide-loading buffer (950 g/L formamide, 10 mmol/L EDTA, and 0.5 g/L bromophenol blue, 0.5 g/L xylene cyanol) and heated at 100 °C for 5 min, and loaded on 80 g/L non-denaturing polyacrylamide gel in 1 × TBE buffer for 8-12 h at 40 V. After electrophoresis, the gel was fixed, silver-stained, and finally developed, photographed and analyzed. The sample was considered normal if the band position was the same as that of the normal tissue. Otherwise, it was abnormal.
| Primer | Sequence | Annealing temperature (°C) | PCR product size (bp) | |
| A | Upstream | 5’-ATGGCGGAGCTGCAGGAGGTC-3’ | 54 | 186 |
| Downstream | 5’-ATTTATAGTTGAGTCCCACATC-3’ | |||
| B | Upstream | 5’-CACCATACGGCTCTGTCACTTTCACT-3’ | 58 | 188 |
| Downstream | 5’-CTCTTCCATTCCAGGGGCATCCAC-3’ | |||
| C | Upstream | 5’-GCCAGCGATCCTTACCTACC-3’ | 58 | 365 |
| Downstream | 5’-GGCTGCCCAATCCATCCAACC-3’ | |||
| D | Upstream | 5’-CCGGACACTGTTGAAGGT-3’ | 55 | 364 |
| Downstream | 5’-GGGTCCAGTTTGAGTTACATT-3’ | |||
| E | Upstream | 5’-CTGTGATGCTGGTGGTAGGAGA-3’ | 58 | 286 |
| Downstream | 5’-TGGGACAGGTGCGAGAG-3’ | |||
| F | Upstream | 5’-GTCCGGTCTCGTACAGCCTCTC-3’ | 56 | 153 |
| Downstream | 5’-AACAAGGGCCATTCAACAGGAGAC-3’ |
Statistical analysis was performed by SPSS version 10.0. Data were expressed as mean±SD, and analyzed by Student’s t test. P < 0.05 was considered statistically significant.
We investigated the expression patterns of NDRG2 mRNA by RT-PCR in 37 cases of human cancers including 29 liver cancers and 8 pancreatic cancers. Figure 1A shows PCR products of representative agarose gels for NDRG2 and GAPDH in human liver cancer and pancreatic cancer. The size of the RT-PCR products was as predicted, 1200 bp for NDRG2 and 310 bp for GAPDH. The levels of NDRG2 mRNA expression were quantified by the ratio of band optical density of NDRG2/ GAPDH (Figure 1B). Compared with adjacent normal tissues, there were marked decreases in NDRG2 mRNA expression in liver cancer (0.894 ± 0.098 vs 1.345 ± 0.177, P < 0.05) and pancreatic cancer (0.680 ± 0.112 vs 2.089 ± 0.214, P < 0.01).
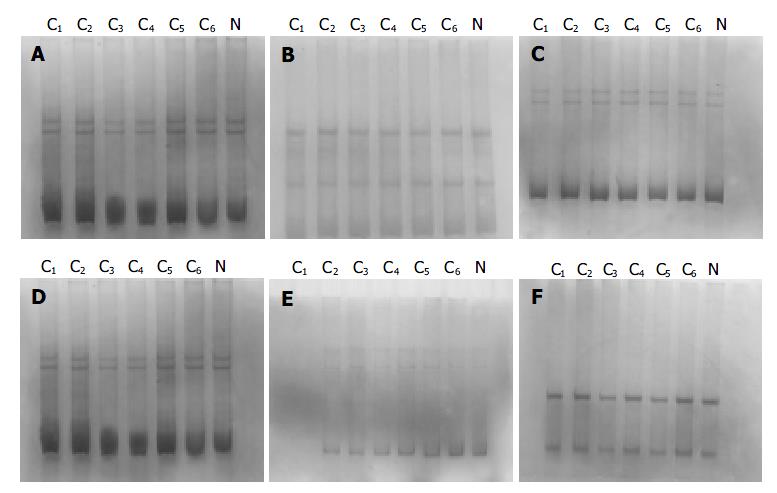
The PCR products, which were amplified by 6 pairs of primers covering the whole coding region of NDRG2 gene, were separated by electrophoresis on 12 g/L agarose gel to check the specificity of each PCR reaction (Figure 2). PCR products with only one specific band of right size in the gel can further be used for SSCP assay. Figure 3 shows representative results of SSCP silver staining of the PCR products amplified by 6 pairs of primers covering the whole coding region of NDRG2. By SSCP, all the PCR products from the cancer tissues of 27 patients of NDRG2 low-expression confirmed by RT-PCR, did not show any mutation.
NDRG2, together with NDRG1, NDRG3 and NDRG4, constitute NDRG gene family. The four members, which share 53%-65% amino acid identity, belong to the alpha/beta hydrolase super family[5]. Phylogenetic analysis of the family demonstrated that human NDRG1 and NDRG3 belong to a subfamily, and NDRG2 and NDRG4 to another. Expression of the fusion proteins showed that all of them were cytosolic proteins. Northern and dot blot analysis showed that NDRG gene family was evidently expressed in adult brain and almost not detected in some human cancer lines[6-8]. In addition, NDRG2 is highly expressed in adult skeletal muscle and brain, NDRG3 remarkably expresses in brain and testis, and NDRG4 evidently expresses in brain and heart, suggesting that they might display different specific functions in distinct tissues[9,10]. Although NDRG gene family is believed to be involved in cell differentiation, the exact molecular and cellular function of members of this family is still unknown.
In 2000, our laboratory first discovered and successfully cloned human NDRG2 cDNA from normal human whole brain cDNA library by subtractive hybridization. Our previous studies showed that the expression level of NDRG2 mRNA was very high in brain, salivary gland, skeletal muscle and mammary gland, and low in bone marrow, testis, peripheral blood and placenta, and not detectable in leukocyte, colon and some tumor cell lines. Other studies have also shown similar result[11,12], suggesting that NDRG2 inactivation might play an important role in some cancer genesis or evolution, and NDRG2 might be a new cancer suppressor gene candidate.
A recent study showed that over expression of NDRG1 in tumor cells decreased the proliferation rate, enhanced differentiation, and suppressed metastasis potency of cancer cells. More interestingly, a nonsense mutation in NDRG1 gene may be the cause of hereditary motor and sensory neuropathy-Lom (HMSNL)[13].
The activation of oncogene and inactivation of cancer suppressor gene can lead to uncontrolled cell proliferation and occurrence of cancer. The inactivation of most cancer suppressor genes has been confirmed to relate to gene mutation such as p53, p16 and Rb gene[14]. To the authors’ knowledge, we first investigated the expression patterns of NDRG2 mRNA by RT- PCR in 37 cases of human cancers including 29 liver cancers and 8 pancreatic cancers. The results showed that there was marked decrease in NDRG2 mRNA expression in human liver cancer (P < 0.05) and pancreatic cancer (P < 0.01), compared with adjacent normal tissue. We speculated that different expression pattern of NDRG2 might relate to gene mutation. The designed 6 pairs of primers covering the whole coding region of NDRG2 gene to amplify the cancer tissue of 27 patients of NDRG2 low-expression were confirmed by RT-PCR, but no mutation was found.
In conclusion, our results indicate that NDRG2 gene expresses differently between normal tissues and cancer tissues, and it might play an important role in the development of pancreatic cancer and liver cancer. The low expression of NDRG2 might be unrelated to mutation of the coding region of NDRG2. In addition, whether a mutation exists in the upstream sequence of transcription initial site of NDRG2, deserves further investigation.
We are grateful to Dai-Hua Yu, Bang-Yong Sun, Wei Lin, and Jiang-Wei Liu for the collection of tissue specimens and Jiang-Tian Yu, Li-Hu Wu, and Li-Feng Wang for digital photographs.
Edited by Chen WW Proofread by Zhu LH and Xu FM
| 1. | Deng YC, Yao LB, Liu XP, Nie XY, Wang JC, Zhang XG, Su CZ. Exploring a new gene containing ACP like domain in human brain and expression it in E.coli. Prog Bichem Biophys. 2001;28:72-76. |
| 2. | Okuda T, Kondoh H. Identification of new genes ndr2 and ndr3 which are related to Ndr1/RTP/Drg1 but show distinct tissue specificity and response to N-myc. Biochem Biophys Res Commun. 1999;266:208-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 101] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 3. | Qu X, Zhai Y, Wei H, Zhang C, Xing G, Yu Y, He F. Characterization and expression of three novel differentiation-related genes belong to the human NDRG gene family. Mol Cell Biochem. 2002;229:35-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 204] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 4. | Deng Y, Yao L, Chau L, Ng SS, Peng Y, Liu X, Au WS, Wang J, Li F, Ji S. N-Myc downstream-regulated gene 2 (NDRG2) inhibits glioblastoma cell proliferation. Int J Cancer. 2003;106:342-347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 188] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 5. | Shaw E, McCue LA, Lawrence CE, Dordick JS. Identification of a novel class in the alpha/beta hydrolase fold superfamily: the N-myc differentiation-related proteins. Proteins. 2002;47:163-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 74] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 6. | Zhou RH, Kokame K, Tsukamoto Y, Yutani C, Kato H, Miyata T. Characterization of the human NDRG gene family: a newly identified member, NDRG4, is specifically expressed in brain and heart. Genomics. 2001;73:86-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 172] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 7. | van Belzen N, Dinjens WN, Diesveld MP, Groen NA, van der Made AC, Nozawa Y, Vlietstra R, Trapman J, Bosman FT. A novel gene which is up-regulated during colon epithelial cell differentiation and down-regulated in colorectal neoplasms. Lab Invest. 1997;77:85-92. [PubMed] |
| 8. | Piquemal D, Joulia D, Balaguer P, Basset A, Marti J, Commes T. Differential expression of the RTP/Drg1/Ndr1 gene product in proliferating and growth arrested cells. Biochim Biophys Acta. 1999;1450:364-373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 117] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 9. | Boulkroun S, Fay M, Zennaro MC, Escoubet B, Jaisser F, Blot-Chabaud M, Farman N, Courtois-Coutry N. Characterization of rat NDRG2 (N-Myc downstream regulated gene 2), a novel early mineralocorticoid-specific induced gene. J Biol Chem. 2002;277:31506-31515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 112] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 10. | Zhao W, Tang R, Huang Y, Wang W, Zhou Z, Gu S, Dai J, Ying K, Xie Y, Mao Y. Cloning and expression pattern of the human NDRG3 gene. Biochim Biophys Acta. 2001;1519:134-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 38] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 11. | Nichols NR. Ndrg2, a novel gene regulated by adrenal steroids and antidepressants, is highly expressed in astrocytes. Ann N Y Acad Sci. 2003;1007:349-356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 58] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 12. | Choi SC, Kim KD, Kim JT, Kim JW, Yoon DY, Choe YK, Chang YS, Paik SG, Lim JS. Expression and regulation of NDRG2 (N-myc downstream regulated gene 2) during the differentiation of dendritic cells. FEBS Lett. 2003;553:413-418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 82] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 13. | Kalaydjieva L, Gresham D, Gooding R, Heather L, Baas F, de Jonge R, Blechschmidt K, Angelicheva D, Chandler D, Worsley P. N-myc downstream-regulated gene 1 is mutated in hereditary motor and sensory neuropathy-Lom. Am J Hum Genet. 2000;67:47-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 262] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 14. | Wunderlich V, Rajewsky MF. "Tumour suppressor gene" concept of carcinogenesis. Lancet. 1995;345:1570-1571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.1] [Reference Citation Analysis (0)] |