Published online Dec 1, 2004. doi: 10.3748/wjg.v10.i23.3511
Revised: November 22, 2003
Accepted: December 8, 2003
Published online: December 1, 2004
AIM: To clone and sequence the cagA gene fragment of Helicobacter pylori (H pylori) with coccoid form.
METHODS: H pylori strain NCTC11637 were transformed to coccoid form by exposure to antibiotics in subinhibitory concentrations. The coccoid H pylori was collected. cagA gene of the coccoid H pylori strain was amplified by PCR. After purified, the target fragment was cloned into plasmid pMD-18T. The recombinant plasmid pMD-18T-cagA was transformed into E.coli JM109. Positive clones were screened and identified by PCR and digestion with restriction endonucleases. The sequence of inserted fragment was then analysed.
RESULTS: cagA gene of 3 444 bp was obtained from the coccoid H pylori genome DNA. The recombinant plasmid pMD-18T-cagA was constructed, then it was digested by BamH I+Sac I, and the product of digestion was identical with the predicted one. Sequence analysis showed that the homology of coccoid and the reported original sequence H pylori was 99.7%.
CONCLUSION: The recombinant plasmid containing cagA gene from coccoid H pylori has been constructed successfully. The coccoid H pylori contain completed cagA gene, which may be related to pathogenicity of them.
-
Citation: Wang KX, Wang XF. Cloning and sequencing of cagA gene fragment of
Helicobacter pylori with coccoid form. World J Gastroenterol 2004; 10(23): 3511-3513 - URL: https://www.wjgnet.com/1007-9327/full/v10/i23/3511.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i23.3511
Helicobacter pylori (H pylori) is one of the common bacteria causing chronic infection, which infects more than 50% of the human population, causes chronic gastritis and plays an important role in the pathogenesis of gastroduodenal ulceration. H pylori has also been suggested to be involved in the genesis of adenocarcinoma and MALT lymphoma of the stomach.[1-5] H pylori cells growing actively in vitro are curved rods, which evolve into metabolically active but nonculturable coccoid cells after prolonged incubation[6-8]. In the stomach mostly spiral-shaped bacteria are found, but coccoid cells have been observed in the more severely damaged regions of the gastric mucosa[9,10]. Many scholars believed that coccoid H pylori might lead to difficult recovery, easy relapse and epidemical transmission[11]. But the pathogenicity of coccoid H pylori is unclear at present. The cytotoxin-associated protein encoded by cagA (cytotoxin-associated gene A) is important virulence determinated by H pylori. cagA+H pylori strains have been linked to more severe gastric inflammation, peptic ulcer disease, and gastric cancer in adults[12-15]. CagA gene is a mono-replicon and locates 3’ region of cag-PAI (cag pathogenity island), whose length varies from 3400 bp to 4000 bp. In order to probe into possible pathogenesis of coccoid H pylori, the recombinant plasmid encoding cagA gene of coccoid H pylori was constructed and detected for the sequence in this study.
The strain NCTC11637 of H pylori was afforded by Chinese center for disease control and prevention. JM109 E.coli strains were preserved by our laboratory, pMD-18T (T-Vector), restriction endonuclease enzymes (BamH I, Sac I), T4 DNA ligase, LA Taq DNA polymerase, DNA Extraction Kit and DNA purification reagent kit were provided by TaKaRa Company.
Bacterial culture and induction of coccoid formsH pylori strains were grown on Columbia agar with 50 m/L frozen-melting sheep blood, 100 mL/L fetal bovine serum, and Skirrow’s antibiotic supplement in a microaerophilic atmosphere for 3 d at 37 °C, then the bacteria were suspended in brucella broth and supplemented with 0.02 mg/L of amphotericin, still were incubated at 37 °C for 3, 5, 7, and 10 d. Baterial morphology was determined by light microscopy after Gram staining. The coccoid forms were collected and stored at -20 °C.
Extraction of genomic DNAH pylori of coccoid form were added to a 1.5 mL microcentrifuge tube, rinsed once with phosphate-buffered saline (PH 7.2), and pelleted by centrifugation at 11000 g. Genomic DNA was extracted by TaKaRa MiniBEST Bacterial Genomic DNA Extraction Kit, the DNA pellet was suspended in TE (10 mmol/L Tris-HCL, 1 mmol/L PH 8.0 EDTA), and stored at -20 °C.
Synthetic primers A single primer pair was used to amplify coccoid H pylori cagA gene based on GenBank. The primers had a BamH I site incorporated into the 5’ end and a Sac I site at the 3’ end and their sequences as follows (5’-3’): AAGGATCC ACTAACGAAACCATTGACCA (forward) and AAGAGCTCA GATTTTTGGAAACCACCTT (reverse). The 5’ region initiator and 3’ end stop codon were banned.
PCR amplification PCR was performed in a 100 μL reaction mixture in 0.6-mL tube in an automatic thermal cycler (TP3000; TaKaRa BIO INC). The PCR mixture contained 10 μL of 10 × PCR buffer, 1 μL of sample DNA, 10 μL of 2.5 mmol/L deoxynucleoside triphoshpate, 4 μL of 10 μmol/L oligonucleotide primers, 0.5 μL of LA Taq polymerase, 74.5 μL of molecular-biology-grade distilled water. The mixtures were incubated for 1 min at 94 °C for initial denaturation of the target DNA and then subjected to 30 cycles of denaturation at 98 °C for 10 s, annealing at 55 °C for 30 s, and extension at 72 °C for 210 s. The amplified products (5 μL) were analyzed by electrophoresis on 10 g/L agarose gel containing 0.1 μg of ethidium bromide per ml in TBE buffer. The PCR product was visualized under UV light and photographed.
Construction of recombinant plamids The PCR product was purified by TaKaRa PCR Fragment Recovery Kit. The purified product was cloned into the compatible sites of the T-vector pMD-18T by using T4 DNA ligase at a molar ratio of 6:1 at 16 °C for 3 h. After the above product was transformed into E.coli JM109, pMD-18T/cagA was selected and identified by PCR and enzyme digestion.
Extraction of recombinant plasmid The single bacterial colony (JM109/pMD-18T/cagA) was picked, and cultivated in 3 mL LB broth containing 100 mg/L of ampicillin, at 300 r/min at 37 °C overnight, then recombinant plasmids were extracted according to manufacturer’s instructions (TaKaRa MiniBest DNA Purification Kit), in the meantime, identified by PCR and restriction endomuclease enzyme digestion.
Sequence determination and homology analysis The sequence determination of cagA gene of recombinant plasmid was carried out by Takara Company, in the meantime, the sequence of gene and amino acid were analyzed by software sequence 3.0, and compared the homology based on the GenBank.
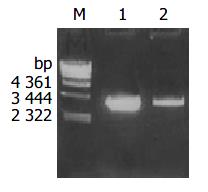
H pylori with coccoid form cagA was amplified by PCR from the above primers and The PCR product was electrophoresed and visualized by 10 g/L agarose gel (Figure 1). It revealed that the size of cagA DNA fragment amplified by PCR was 3444 bp, and was compatible with the expectant size.
The plasmid was extracted from recombinant bacteria and conducted as template to amplify by PCR under the condition mentioned above. The PCR products were visualized by 10 g/L agarose gel electrophoresis (Figure 2). It indicated that recombinant plasmid contained the objective gene. At the same time, it was successful in transforming recombinant plasmid into JM109 E.coli.
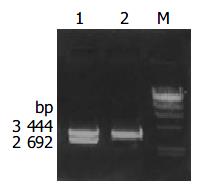
Recombinant plasmid pMD-18T/cagA was digested by bienzyme digestion with BamH I and Sac I, then digestive product was visualized on 10 g/L agarose gel (Figure 3). It demonstrated that recombinant plasmid was digested to 3444 bp and 2692 bp DNA fragment, which contained the objective gene.
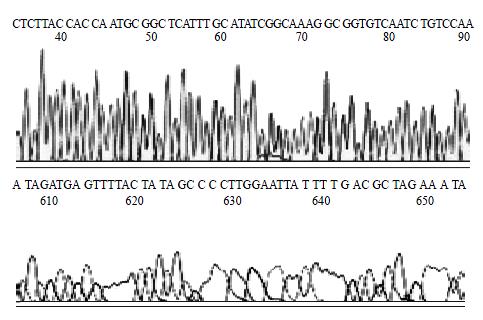
H pylori Sequence of inserted DNA was analyzed with BcaBEST Primer M13-47/BcaBEST Primer RV-M using automatic sequence analyzer by Sanger dideoxy chain termination method. The result of analysis showed that the size of inserted DNA was about 3444 bp and 99.7% affinity in comparison with DNA sequence published on GenBank (locus: AB015416). The sequence of partial vacA gene was showed in Figure 4.
Morphological conversion from spiral H pylori to coccoid forms has been described under several suboptinal conditions. These conditions include aerobiosis, alkaline pH, high temperature, extended incubation, or treatment with proton pump inhibitor or antibiontics[16-18]. This coccoid form conversion phenomenon, which has been thought to result in a viable but nonculturable form of the bacterium, is not exclusive to H pylori, as it is common for other enteric pathogens. Controversy remains about the pathogenicity of coccoid H pylori. Many investigators have suggested that the coccoid form of H pylori represents a degenerative form with no infectious capability, but others believed that the coccoid form retains a weak metabolic activety, important structural components, and patogenicity[19-21]. Recently, successful infection with coccoid forms of H pylori in animal models has been reported[22-24]. These findings have highlighted the possible role of the coccoid forms in transmission of infection and morphological conversion of coccoids to the spiral form. It is well known that cagA is an important virulence factor of H pylori and related to severe gastrointestinal diseases. However, the research of cagA gene of coccoidal H pylori is few at present. In order to observe cagA and vacA expression during conversion to the coccoid form, Sisto et al[25] analyzed the expression of ureA, cagA, vacA genes after prolonged incubation in a liquid medium in 2000, the results showed that although the coccoid forms had decreased DNA and RNA levels after 31 d, they were not degraded and still expressed the urease, cytotoxic island and vacuolating toxin genes. So in conclusion, coccoid forms are therefore viable and may act as a transmissible agent that plays a crucial role in disease relapses after antibiotic therapy. She et al explored the virulence and the potential pathogenicity of coccoid H pylori transformed from spiral form by exposure to antibiotic in 2001, and found that the content of the protein with the molecular weight over Mr 74000 decreased, but vacA, cagA, urea, ureB, hpaA gene remained to be preserved, so they concluded that the virulence and the proteins with molecular weight over Mr 74000 in coccoid H pylori decrease, but no deletion exists in amplification fragments from ureA, ureB, hpaA, vacA and cagA genes, and suggested that coccoid H pylori may have potential pathogenicity. Furthermore, Monstein et al[26] confirmed that the transcription and translation of cagA and vacA gene might actively take place in coccoid H pylori cells.
In order to research the cagA gene existence, and explore the possible mechanism of pathogenicity in coccoid H pylori, we designed the specific primers based on cagA gene sequence reported in GenBank, and successfully amplified the cagA gene of coccoid H pylori by PCR, then inserted into pMD-18T vector. The recombinant plasmids were successfully identified containing cagA gene fragment by PCR and enzyme digestion, and sequence determination confirmed that cagA gene existed in coccoid H pylori, though existence of 0.3% difference with the reported sequence in GenBank. The reason for the discrepancy might be as follows: (1) the mutant base could come from the process of PCR amplification and sequencing, (2) H pylori provided have the transformation ability, which could lead to H pylori variation and genome reset. Our results also support the assumption that virulence-gene expression is differently regulated among H pylori strains[26].
In this study, the NCTC11637 strain with amphotericin for 3 d generated a high proportion of coccoid forms. Forms obtained after removal of bacterial clumps and amorphous debris by centrifugation at 600 r/min for 5 min were nearly 100% coccoid. Due to similar to the condition in vivo by antibiotic induction, the collected coccoid H pylori still remained complete celluar structure, and their genome DNA lost was less, so some important virulent gene, such as vacA, cagA and so on, could exist in their cells. Once coccoid H pylori live in suitable condition, they can recover their virulence and lead to the occurrence of diseases. Or coccoid H pylori may revert helical form, and result in the transmission and/or relapse of diseases with the complete cagA gene.
Edited by Xia HHX Proofread by Xu FM
| 1. | Nguyen TN, Barkun AN, Fallone CA. Host determinants of Helicobacter pylori infection and its clinical outcome. Helicobacter. 1999;4:185-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 2. | Vandenplas Y. Helicobacter pylori infection. World J Gastroenterol. 2000;6:20-31. [PubMed] |
| 3. | Sakai T, Ogura Y, Narita J, Suto T, Kimura D, Ainai S, Fujita H, Kamada M. Simultaneous early adenocarcinoma and mucosa-associated lymphoid tissue (MALT) lymphoma of the stomach associated with Helicobacter pylori infection. Gastric Cancer. 2003;6:191-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 18] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 4. | Satoh K, Sugano K. [Causal relationship between Helicobacter pylori infection and upper gastroduodenal diseases]. Nihon Rinsho. 2001;59:239-245. [PubMed] |
| 5. | Müller S, Seifert E, Stolte M. Simultaneous MALT-type lymphoma and early adenocarcinoma of the stomach associated with Helicobacter pylori gastritis. Z Gastroenterol. 1999;37:153-157. [PubMed] |
| 6. | Ren Z, Pang G, Musicka M, Dunkley M, Batey R, Beagley K, Clancy R. Coccoid forms of Helicobacter pylori can be viable. Microbios. 1999;97:153-163. [PubMed] |
| 7. | Willén R, Carlén B, Wang X, Papadogiannakis N, Odselius R, Wadström T. Morphologic conversion of Helicobacter pylori from spiral to coccoid form. Scanning (SEM) and transmission electron microscopy (TEM) suggest viability. Ups J Med Sci. 2000;105:31-40. [PubMed] |
| 8. | Mizoguchi H, Fujioka T, Nasu M. Evidence for viability of coccoid forms of Helicobacter pylori. J Gastroenterol. 1999;34 Suppl 11:32-36. [PubMed] |
| 9. | Saito N, Konishi K, Sato F, Kato M, Takeda H, Sugiyama T, Asaka M. Plural transformation-processes from spiral to coccoid Helicobacter pylori and its viability. J Infect. 2003;46:49-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 52] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 10. | Janas B, Czkwianianc E, Bak-Romaniszyn L, Bartel H, Tosik D, Płaneta-Małecka I. Electron microscopic study of association between coccoid forms of Helicobacter pylori and gastric epithelial cells. Am J Gastroenterol. 1995;90:1829-1833. [PubMed] |
| 11. | Mizoguchi H, Fujioka T, Kishi K, Nishizono A, Kodama R, Nasu M. Diversity in protein synthesis and viability of Helicobacter pylori coccoid forms in response to various stimuli. Infect Immun. 1998;66:5555-5560. [PubMed] |
| 12. | Mao HY, Fang PC, Ye SJ. [Analysis of babA2 cagA and vacA genotypes of Helicobacter pylori in chronic gastritis and peptic ulcer]. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2003;32:29-32. [PubMed] |
| 13. | Oleastro M, Gerhard M, Lopes AI, Ramalho P, Cabral J, Sousa Guerreiro A, Monteiro L. Helicobacter pylori virulence genotypes in Portuguese children and adults with gastroduodenal pathology. Eur J Clin Microbiol Infect Dis. 2003;22:85-91. [PubMed] |
| 14. | Debets-Ossenkopp YJ, Reyes G, Mulder J, aan de Stegge BM, Peters JT, Savelkoul PH, Tanca J, Peña AS, Vandenbroucke-Grauls CM. Characteristics of clinical Helicobacter pylori strains from Ecuador. J Antimicrob Chemother. 2003;51:141-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 15. | Bravo LE, van Doom LJ, Realpe JL, Correa P. Virulence-associated genotypes of Helicobacter pylori: do they explain the African enigma? Am J Gastroenterol. 2002;97:2839-2842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 81] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 16. | Kusters JG, Gerrits MM, Van Strijp JA, Vandenbroucke-Grauls CM. Coccoid forms of Helicobacter pylori are the morphologic manifestation of cell death. Infect Immun. 1997;65:3672-3679. [PubMed] |
| 17. | Narikawa S, Kawai S, Aoshima H, Kawamata O, Kawaguchi R, Hikiji K, Kato M, Iino S, Mizushima Y. Comparison of the nucleic acids of helical and coccoid forms of Helicobacter pylori. Clin Diagn Lab Immunol. 1997;4:285-290. [PubMed] |
| 18. | Costa K, Bacher G, Allmaier G, Dominguez-Bello MG, Engstrand L, Falk P, de Pedro MA, García-del Portillo F. The morphological transition of Helicobacter pylori cells from spiral to coccoid is preceded by a substantial modification of the cell wall. J Bacteriol. 1999;181:3710-3715. [PubMed] |
| 19. | Sato F. Helicobacter pylori in culture: an ultrastructural study. Hokkaido Igaku Zasshi. 2000;75:187-196. [PubMed] |
| 20. | Nakamura A, Park A, Nagata K, Sato EF, Kashiba M, Tamura T, Inoue M. Oxidative cellular damage associated with transformation of Helicobacter pylori from a bacillary to a coccoid form. Free Radic Biol Med. 2000;28:1611-1618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 21. | Monstein HJ, de la Cour CD, Jonasson J. Probing 23S ribosomal RNA cleavage sites in coccoid Helicobacter pylori. Helicobacter. 2001;6:100-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 22. | Rabelo-Gonçalves EM, Nishimura NF, Zeitune JM. Acute inflammatory response in the stomach of BALB/c mice challenged with coccoidal Helicobacter pylori. Mem Inst Oswaldo Cruz. 2002;97:1201-1206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 23. | Wang X, Sturegård E, Rupar R, Nilsson HO, Aleljung PA, Carlén B, Willén R, Wadström T. Infection of BALB/c A mice by spiral and coccoid forms of Helicobacter pylori. J Med Microbiol. 1997;46:657-663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 57] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 24. | Cao J, Li ZQ, Borch K, Petersson F, Mårdh S. Detection of spiral and coccoid forms of Helicobacter pylori using a murine monoclonal antibody. Clin Chim Acta. 1997;267:183-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 15] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 25. | Sisto F, Brenciaglia MI, Scaltrito MM, Dubini F. Helicobacter pylori: ureA, cagA and vacA expression during conversion to the coccoid form. Int J Antimicrob Agents. 2000;15:277-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 31] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 26. | Monstein HJ, Jonasson J. Differential virulence-gene mRNA expression in coccoid forms of Helicobacter pylori. Biochem Biophys Res Commun. 2001;285:530-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |