Published online Dec 1, 2004. doi: 10.3748/wjg.v10.i23.3394
Revised: April 3, 2004
Accepted: April 16, 2004
Published online: December 1, 2004
AIM: To explore the effect of DNA methyltransferase, demethylase and methyl-CpG binding protein MeCP2 on the expressions and methylation of hMSH2 and proto-oncogene in human gastric cancer.
METHODS: Paired samples of primary gastric cancer and corresponding para-cancerous, non-cancerous gastric mucosae were obtained from surgically resected specimens of 28 patients. Transcription levels of Dnmt1, mbd2, MeCP2, p16INK4A, hMSH2 and c-myc were detected by using real-time PCR or RT-PCR. Promoter methylation of p16INK4A, c-myc and hMSH2 genes was assayed by methylation-specific PCR (MSP) and sequencing (mapping). Their relationships were analyzed by Fisher’s exact test using the software SPSS.
RESULTS: The average mRNA level of Dnmt1 gene from cancerous tissue was higher and that of mbd2 gene from cancerous tissue was lower than that from non-cancerous tissue, respectively. mbd2 was lower in cancerous tissue than in non-cancerous tissue in 14 (50.0%) of patients but higher in 3 cases (10.7%) of non-cancerous gastric tissue (P < 0.001). c-myc expression was up-regulated in cancer tissues (P < 0.05). The up-regulation of mbd2 was found in all patients with hypomethylated c-myc. The transcriptional levels of p16INK4A and MeCP2 genes did not display any difference between gastric cancerous and matched non-cancerous tissues. There were down-regulation and hypermethylation of hMSH2 in cancer tissues, and the hypermethylation of hMSH2 coexisted with down-regulated transcription. However, the transcription level of the above genes was not associated with biological behaviours of gastric cancers.
CONCLUSION: The up-regulation of proto-oncogene may be the consequence of epigenetic control of gene expression by demethylase, and mbd2 is involved in the regulation of hMSH2 expression in human gastric cancer.
- Citation: Fang JY, Cheng ZH, Chen YX, Lu R, Yang L, Zhu HY, Lu LG. Expression of Dnmt1, demethylase, MeCP2 and methylation of tumor-related genes in human gastric cancer. World J Gastroenterol 2004; 10(23): 3394-3398
- URL: https://www.wjgnet.com/1007-9327/full/v10/i23/3394.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i23.3394
Methylation of gene regulatory elements, a well-known epigenetic change, acts as an important alternative to genetic alteration for gene inactivation. DNA methyltransferase (Dnmt) and DNA demethylase are the enzymes potentially affecting promoter methylation status. Human DNA demethylase has been cloned[1]. The inactivity of demethylase may play a role in production and maintenance of regional DNA hypermethylation, which frequently results in reduced expression of tumor suppressor genes in human cancers.
Gastric cancer is the second most common malignant tumor in Asia, with a much higher incidence than in Western countries. During gastric carcinogenesis, hypomethylation of c-myc and c-Ha-ras proto-oncogenes was frequently seen[2,3], however the average level of mRNA for Dnmt1 was significantly higher in gastric cancers than in corresponding non-cancerous mucosa[4]. mbd2 has been proposed to be a DNA demethylase, and the average level of mbd2 mRNA was significantly lower in gastric cancer than in non-cancerous mucosa. But there was no significant association between DNA demethylase mRNA level and malignant potential in gastric cancers[5]. The mRNA expression levels for pro-methylation of Dnmt1, Dnmt3a and Dnmt3b and mbd2 are not the critical determinant of tumor-specific promoter hypermethylation of hMLH1, p16INK4A or CDH1 in gastric cancer[6]. However, little is known about the relationship between mbd2 or Dnmt1 and the methylation status of proto-oncogene or hMSH2, another important mismatch repair (MMR) gene. Furthermore, up-to date, there has not been any report about methyl-CpG binding protein MeCP2 associated with tumor-related genes in gastric cancer.
In the present study, we examined the methylation and transcriptional level of tumor-related genes including p16INK4A, hMSH2 and c-myc, and detected the expression of Dnmt1, mbd2 and MeCP2. We therefore analyzed their relationship in normal and cancerous stomach tissues.
Paired samples of histologically verified primary gastric cancer and corresponding para-cancerous (3-5 cm from cancer margin) and non-cancerous gastric mucosae were obtained immediately from surgically resected specimens of 28 patients (C1-C28) treated at Renji Hospital, Shanghai, China. A complete written informed consent was obtained from all patients. They were all diagnosed pathologically by HE-stained sections and classified according to WHO’s histological classifications of gastric carcinoma. The histological characteristics in para-cancerous area were chronic gastritis or intestinal metaplasia, and dysplasia was present in ten cases. The clinico-pathological features of each patient were reviewed and recorded (Table 1). The mean age of patients (19 men and 9 women) at resection was 58 (range 45-79) years. A portion of the tissues (approximately 1-3 g) was snap-frozen on dry ice and kept in liquid nitrogen until used for DNA or RNA extraction. Other portions were used for histological examination.
| Subtotal | Dnmt1 transcription | P | |||
| Up-regulated | Normal | Down-regulated | |||
| Total or subtotal | 28 (100.0) | 9 (32.14) | 9 (32.14) | 10 (35.72) | |
| Age (yr) > 50 | 18 (64.30) | 7 (38.89) | 4 (22.22) | 7 (38.89) | 0.30 |
| < 50 | 10 (35.70) | 2 (20.00) | 5 (50.00) | 3 (30.00) | |
| Size of tumor >5 cm | 14 (50.00) | 4 (28.57) | 5 (35.71) | 5 (35.71) | 0.88 |
| <5 cm | 14 (50.00) | 4 (28.57) | 5 (35.71) | 5 (35.71) | |
| Borrmann II | 6 (21.43) | 2 (33.33) | 0 (0.00) | 4 (66.67) | 0.34 |
| III | 15 (53.57) | 5 (33.33) | 6 (40.00) | 4 (26.67) | |
| IV | 7 (25.00) | 2 (28.57) | 3 (42.86) | 2 (28.57) | |
| Lymph node Positive | 21 (75.00) | 6 (28.57) | 8 (38.10) | 7 (33.33) | 0.49 |
| invasion Negative | 7 (25.00) | 3 (42.86) | 1 (14.29) | 3 (42.86) | |
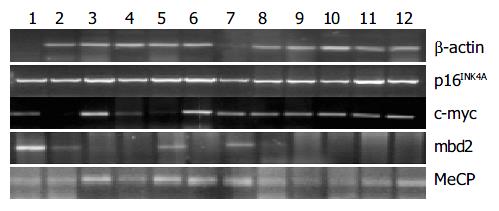
First, mRNA expression of tumor-related genes in cancerous, para-cancerous and non-cancerous tissues from each patient was detected by using RT-PCR and real-time PCR. Total RNA was extracted using a commercial kit (Trizol) according to the manufacturer’s instructions (Gibco BRL). RT reactions using 5 μg of total RNA in 20 μL of reaction buffer were performed with Superscript II reverse transcriptase (Life Technologies, Inc). mRNA transcription levels of mbd2, MeCP2, p16INK4A and c-myc genes were evaluated by using RT-PCR. The sequence and PCR reaction for each primer are shown in Table 2. For control of RT-PCR, a 612 bp fragment of β -actin cDNA was also amplified. At the end of 40 cycles, reaction products were separated electrophoretically on a 3% agarose gel and stained with ethidium bromide for visual confirmation of PCR products. The density of bands in RT-PCR in each lane was normalized to the amount of total RNA as determined by the density of the band in RT-PCR for β -actin[7]. RT-PCR analysis was repeated at least thrice.
| Primers | Sense (5’→3’) | Antisense (5’→3’) | Product size (bp) PCR program | GenBank accession number |
| β -Actin | GGA GTC CTG TGG CAT CCA CG | CTA GAA GCA TTT GCG GTG GA | 322 bp 94 °C 30 s, 60 °C 1 min, 72 °C 1 min, 27 cycles | BC023204 |
| p16INK4A RT-PCR | CCC GCT TTC GTA GTT TTC AT | TTA TTT GAG CTT TGG TTC TG | 355 bp 94 °C 1 min, 58 °C 1 min, 72 °C, 1 min, 35 cycles | L27211 |
| c-myc RT-PCR | CCA ACA GGA GCT ATG ACC TC | CTC GGT CAC CAT CTC CAG CT | 290 bp 94 °C 1 min, 52 °C 1 min, 72 °C, 1 min, 35 cycles | V00568 |
| MeCP2 RT-PCR | ACT CCT CAG AAT ACA CCT TGC TT | TGA GGC CCT GGA GGT CCT | 112 bp 95 °C 1 min, 50 °C 1 min, 72 °C, 1 min, 35 cycles | MCB, 2000; 20: 3316 |
| mbd2 RT-PCR | AAC CCT GCT GTT TGC TTA AC | CGT ACT TGC TGT ACT CGC TCT TC | 101 bp 94 °C 1 min, 60 °C 1 min, 72 °C, 1 min, 40 cycles | AF072242 |
| p16INK4A M-MSP | TTA TTA GAG GGT GGG GCG GAT CGC | GAC CCC GAA CCG CGA CCG TAA | 150 bp 95 °C 30 s, 65 °C 30 s, 72 °C, 1 min, 40 cycles | X94154 |
| p16INK4A UM-MSP | TTA TTA GAG GGT GGG GTG GAT TGT | CAA CCC CAA ACC ACA ACC ATA A | 151 bp 95 °C 30 s, 60 °C 30 s, 72 °C, 1 min, 40 cycles | X94154 |
| c-myc M-MSP | TAG AAT TGG ATC GGG GTA AA | CGA CCG AAA ATC AAC GCG AAT | 131 bp 95 °C 30 s, 56 °C 30 s, 72 °C, 1 min, 40 cycles | AF002859 |
| c-myc UM-MSP | TAG AAT TGG ATT GGG GTA AA | CCA ACC AAA AAT CAA CAT GAA T | 132 bp 95 °C 30 s, 56 °C 30 s, 72 °C, 1 min, 40 cycles | AF002859 |
| hMSH2 sequencing 1st round | TGT TTA GAA AGA AAA AGG GA | AAA CCT CCT CAC CTC CT | 94 °C 1 min, 55 °C 1 min, 72 °C, 1 min, 35 cycles | AB006445 |
| hMSH2 sequencing 2nd round | AAA TAT TGG GAG GAG GAG GA | ACC CAC TAA ACT ATT TCC CA | 327 bp 94 °C1 min,, 55 °C 1 min, 72 °C, 1 min, 35 cycles | AB006445 |
mRNA levels of Dnmt1 and hMSH2 were measured using a real-time quantitative PCR system. Relative quantitation using the comparative Ct method with the data from ABI PRISM 7700 sequence detection system (version 1.6 software) was performed according to the manufacturer’s protocol.
Real-time PCR was also performed with Taqman β -actin to normalize each of the extracts for amplifiable human DNA. The results were expressed as the ratio of copies of each gene to β -actin, respectively. The Ct values were measured, and the average Ct value of triplicate samples was calculated. Alteration of mRNA expression was defined as a 3-fold difference in the expression level[8]. The primers and the Taqman fluorogenic probes for Dnmt1, hMSH2 and β -actin were provided by Jikang Company, Shanghai, China.
The sequences of all primers and probes, as well as PCR programs are shown in Table 3.
| Gene | Sense (5’→3’) | Antisense (5’→3’) | Probe | GenBank No |
| Dnmt1 | GCA CCT CAT TTG CCG AAT ACA | TCT CCT GCA TCA GCC CAA ATA | AGT CCC GAG TAT GCG C | XM_017218 |
| hMSH2 | ATC CAA GGA GAA TGA TTG GTA TTT G | CAA AGA GAA TGT CTT CAA ACT GAG AGA | CAT ATA AGG CTT CTC CTG GC | HSU04045 |
| β -actin | CTG GCA CCC AGC ACA ATG | GGA CAG CGA GGC CAG GAT | ATC ATT GCT CCT CCT GAG | BC016045 |
To amplify the promoters of p16INK4A and c-myc genes, we carried out bisulfite modification[9] and MSP.
Bisulfite could convert unmethylated cytosine residues to uracil, but methylated cytosines remained non-reactive. PCR amplified uracil as thymine while methylated cytosines were only amplified as cytosines. Two micrograms of total genomic DNA (from at least two independent treatments corresponding to RT-PCR experiments) were isolated by using QIAamp DNA blood mini kit (QIAGEN Inc.), then denatured by NaOH and modified by sodium bisulfite solution (2.35 mol/L) containing hydroquinone (40 mmol/L) freshly prepared. The bisulfite-treated DNA was desalted using Wizard DNA clean up kit (Promega) and amplified by PCR using primers specific for methylated and unmethylated p16INK4A and c-myc promoters. PCR reaction buffer contained 0.1 mmol/L dNTP, 2.0 mmol/L MgCl2, and 0.5 μmol/L primers. PCR products were directly loaded onto 3% agarose gels and electrophoresed. The gel was stained with ethidium bromide and directly visualized under UV illumination. Furthermore, the primers for wild-type p16INK4A and c-myc were used to monitor complete conversion of DNA obtained in the bisulfite reaction.
Primers were designed in the region without CpG dinucleotides to amplify both methylated and unmethylated alleles in hMSH2. The PCR products were recovered using QIA quick gel extraction kit (Gibco). Direct sequencing was performed by using ABI PRISM BigDye terminator kit (PE Biosystems, Foster City, CA). The products of sequencing PCR included 22 CpG sites in promoter of hMSH2 gene.
The sequences of all primers and PCR programs are shown in Table 2.
Results were representative of at least three independent experiments performed in triplicate and presented as mean±SD. Comparisons between groups were made using Student’s paired t test. Their relationship was analyzed by Fisher’s exact test using the software SPSS.
After quantitative PCR, the specific products visually confirmed on agarose gels were about 101- and 96 bp, and no non-specific products were obtained upon amplification of Dnmt1 and β -actin, respectively. In all cancerous, para-cancerous and non-cancerous tissues, their mRNA expressions were detected, and 32.2% (9/28) of cancerous tissues over-expressed Dnmt1, and the copies of Dnmt1 were higher than in non-cancerous tissues (5.08 vs 1).
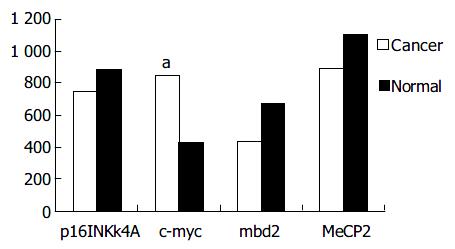
mbd2 was expressed in all samples. However, mbd2 was down-regulated in 50% (14/28) of gastric cancer tissues, up-regulated in 10.7% (3/28) cases, and no significant change in 3.39% (11/28), respectively. Average value of mbd2 mRNA expression was lower in cancerous tissues than in non-cancerous tissue (Figure 1, Figure 2). In 11 patients, mbd2 mRNA level in gastric cancer was particularly low, being reduced by 50% or more when compared with that in the corresponding non-cancerous mucosae. However, we did not find association between expressions of MeCP2 and Dnmt1 and mbd2.
In addition, we did not find the association of overall aberrant methylation and expressions of Dnmt1, MeCP2, mbd2 with the age of patients.
Figure 1 shows the results from RT-PCR and real-time PCR, and Figure 2 shows the relative mRNA levels of each gene after normalization with the signal of β -actin. These data demonstrated that the expressions of p16INK4A and hMSH2 (data not shown) were down-regulated in cancerous tissues from 11 cases (39.3%) and 10 cases (35.7%) respectively, but there was no significant difference between cancerous and non-cancerous tissues. As shown in Figure 2, c-myc transcription level was up-regulated in 14 patients with gastric cancer, and the mean value of expression in cancerous tissues was greater than that in non-cancerous tissues (P < 0.05).
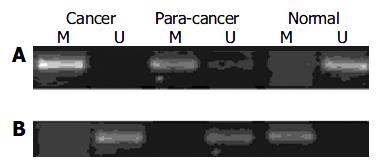
To determine whether the expression of p16INK4A, hMSH2 and c-myc genes in gastric cancer was associated with methylation, we selected the samples from cancerous, para-cancerous and non-cancerous tissues in ten patients for MSP or genomic bisulfite sequencing. Each tissue showed a positive 150 bp and a 151 bp band for methylated and unmethylated specific primer sets for p16INK4A, indicating that the p16INK4A gene was partially methylated in human gastric cancer. The methylated bands for the p16INK4A gene in the cancerous and para-cancerous tissues were consistently stronger than the products of non-cancerous tissues (Figure 3A). Regarding c-myc gene, the methylated product (131 bp) was significantly lower and the unmethylated product (132 bp) was higher in cancerous and para-cancerous tissues than in non-cancerous tissues. Figure 3B displays representative examples of MSP products analyzed by electrophoresis on an agarose gel.
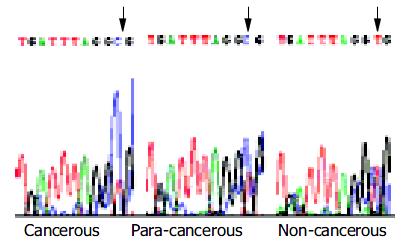
Bisulfite genomic sequencing of the representative PCR products of hMSH2 showed that all cytosines at non-CpG sites were converted to thymine. This excluded the possibility that successful amplification could be attributable to incomplete bisulfite conversion. Interestingly, methylation occurred at the -166 CpG site only (Figure 4).
Taken together, the results from MSP, sequencing, RT-PCR and real-time PCR indicated that hypermethylation of hMSH2 might be induced, inactivation and activation of c-myc were associated with its hypomethylation in human gastric cancer.
Dnmt1 was not associated with the methylation status of p16INK4A or c-myc gene.
Among Dnmt1 down-regulated cases, 6 (60%) had over-expression of c-myc in cancerous tissues. In the cancer cases with up-regulated mbd2, all 3 had an expression of c-myc. We also found that there was a positive correlation in the expression of c-myc and mbd2 (r = 0.59, P = 0.026), MeCP2 (r = 0.64, P = 0.02), but c-myc had no positive correlation with Dnmt1.
In contrast to cancer tissues, all normal tissues had a positive association of p16INK4A with MeCP2 (r = 0.483, P = 0.02) or mbd2 (r = 0.483, P = 0.027).
Among the 10 cases with down-regulated hMSH2, 6 cases had a decreased mbd2 mRNA, but there was no significant association between expressions of Dnmt1 and hMSH2.
There was no significant association between Dnmt1, mbd2 mRNA level and clinico-pathological parameters of the tumors, such as histological differentiation, size of tumor and lymph node metastasis. These data failed to indicate their aberrant expression as an early event in gastric cancer.
Generally, the global DNA methylation level is lower in cancer cells than in normal cells, and some loci tend to show hypomethylation of proto-oncogene in human gastrointestinal cancers[10]. Recent studies demonstrated that DNA methylation could contribute to inactivation of tumor suppressor genes, a key event in tumorigenesis of a wide spectrum of human tumors[11,12].
Three enzymes are responsible for DNA methylation. The carboxy-terminal domain of Dnmt1 could catalyse the methylation of DNA containing hemi-methylated CpG dinucleotides more efficiently than unmethylated DNA in vitro[13]. DNA demethylase was first identified by Szyf’s group, and they demethylated both fully methylated and hemimethylated DNA, showing dinucleotide specificity, and could demethylate mCpG in different sequence contexts[5]. The shortest form of methyl- CpG binding domain (MBD)2, mbd2b, has been proposed to be a DNA demethylase which has been cloned and characterized[14]. MeCP2 is the first true member of the family of proteins that could selectively recognize methylated CpG[15]. It is a single polypeptide characterized by an MBD and a transcriptional repression domain[16,17]. Aberrant methylation in tumor-related genes was frequently detected in gastric intestinal metaplasia of both cancer and non-cancer patients, suggesting their early involvement in the multi-step progression of gastric carcinogenesis[18]. However, little is known about the relationship of Dnmt1, demethylase, MeCP2 with tumor-related genes.
We found that the average mRNA level of Dnmt1 gene in cancerous tissue was higher and that of mbd2 gene was lower than that in non-cancerous tissue. Regarding Dnmt1 expression, these results are consistent with a study by Kanai[4]. On the other hand, Patra found that MBD2 protein expression was significantly higher in benign prostatic hyperplasia BPH-1 cells and deficient in prostate cancer cell lines and in BPH tissues[19]. For mbd2, our result is consistent with another experiment of gastrointestinal cancer[3]. They suggested that the average levels of demethylase mRNA expression normalized to GAPDH mRNA were significantly lower in colorectal (0.81 ± 0.55) and gastric (2.88 ± 0.23) cancers than in the non-cancerous mucosae (1.90 ± 0.16 and 5.11 ± 0.34, respectively, P < 0.0001). However, up to date, we have not found any report about the expression of MeCP2 in gastric cancer. Darwanto found that colonic mucinous adenocarcinomas showed a strong MeCP2 expression[20], and Muller observed a higher expression level of MeCP2 mRNA in breast cancer tissues than in non-cancerous tissues[21].
Previous studies indicated that the mRNA expression level for Dnmt1 and mbd2 was not a critical determinant of promoter hypermethylation of tumor-suppressor gene p16INK4A in gastric cancer[6,22]. Regarding the hMLH1, the result was different in different studies[6,23]. During gastric carcinogenesis, hypomethylation of c-myc proto-oncogene is common, and it has not been linked to Dnmt1, mbd2 or MeCP2, or hMSH2. In the present study, we demonstrated that there was a positive association of mbd2 and c-myc proto-oncogene in gastric cancerous tissues. The up-regulation of mbd2 was found in all patients with hypomethylated c-myc. The correlation of mbd2 expression with unmethylation of c-myc promoter suggests that c-myc may be a target for demethylation by the enzymes.
Another interesting gene we tested was hMSH2. The second important finding in this study was the decrease of mbd2 mRNA in most patients with down-regulated hMSH2, but we did not find any correlation between hMSH2 and Dnmt1 or MeCP2. It suggests that mbd2 is also involved in the regulation of hMSH2 expression in human gastric cancer. We did not test the promoter region of hMLH1 gene, because the promoter was not associated with Dnmt1[6,23] and mbd2[6] in human gastric cancer.
In addition, the data from our study demonstrated that the transcription level of Dnmt1, mbd2 and MeCP2 might not be associated with biological behaviours in human gastric cancer. Kanai also suggested there was no significant association between DNA demethylase mRNA level and malignant potential in gastric cancers[5]. Although the mbd2 expression was lower both in early and in advanced cancers, we could not demonstrate a definitive relationship between the histology and mbd2 expression due to the small number of patients studied.
In conclusion, the up-regulation of proto-oncogene may be the consequence of epigenetic control of gene expression by demethylase, and mbd2 is also involved in the regulation of hMSH2 expression in human gastric cancer.
Thanks are given to Dr. Yao Shi and Xiao-Yu Chen, Ms. Hong-Yin Zhu and Ms. Wei-Qi Gu for performing the diagnosis of pathology, MSP and real-time PCR.
Edited by Wang XL Proofread by Zhu LH and Xu FM
| 1. | Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579-583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 464] [Cited by in RCA: 432] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 2. | Fang JY, Zhu SS, Xiao SD, Jiang SJ, Shi Y, Chen XY, Zhou XM, Qian LF. Studies on the hypomethylation of c-myc, c-Ha-ras oncogenes and histopathological changes in human gastric carcinoma. J Gastroenterol Hepatol. 1996;11:1079-1082. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 3. | Fang J, Zhu S, Xiao S, Shi Y, Jiang S, Zhou X, Qian L. Alterations of level of total genomic DNA methylation and pattern of c-myc, c-Ha-ras oncogene methylation in human gastric carcinogenesis. Chin Med J (Engl). 1996;109:787-791. [PubMed] |
| 4. | Kanai Y, Ushijima S, Kondo Y, Nakanishi Y, Hirohashi S. DNA methyltransferase expression and DNA methylation of CPG islands and peri-centromeric satellite regions in human colorectal and stomach cancers. Int J Cancer. 2001;91:205-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 5. | Kanai Y, Ushijima S, Saito Y, Nakanishi Y, Sakamoto M, Hirohashi S. MRNA expression of genes altered by 5-azacytidine treatment in cancer cell lines is associated with clinicopathological parameters of human cancers. J Cancer Res Clin Oncol. 2001;127:697-706. [PubMed] |
| 6. | Oue N, Shigeishi H, Kuniyasu H, Yokozaki H, Kuraoka K, Ito R, Yasui W. Promoter hypermethylation of MGMT is associated with protein loss in gastric carcinoma. Int J Cancer. 2001;93:805-809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 66] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 7. | Fang JY, Mikovits JA, Bagni R, Petrow-Sadowski CL, Ruscetti FW. Infection of lymphoid cells by integration-defective human immunodeficiency virus type 1 increases de novo methylation. J Virol. 2001;75:9753-9761. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 69] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 8. | Scanlan MJ, Welt S, Gordon CM, Chen YT, Gure AO, Stockert E, Jungbluth AA, Ritter G, Jäger D, Jäger E. Cancer-related serological recognition of human colon cancer: identification of potential diagnostic and immunotherapeutic targets. Cancer Res. 2002;62:4041-4047. [PubMed] |
| 9. | Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532-2534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 913] [Cited by in RCA: 884] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 10. | Fang JY, Xiao SD. Alteration of DNA methylation in gastrointestinal carcinogenesis. J Gastroenterol Hepatol. 2001;16:960-968. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 37] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 11. | Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068-1070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1432] [Cited by in RCA: 1408] [Article Influence: 58.7] [Reference Citation Analysis (0)] |
| 12. | Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1168] [Cited by in RCA: 1165] [Article Influence: 46.6] [Reference Citation Analysis (0)] |
| 13. | Bestor TH. Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. EMBO J. 1992;11:2611-2617. [PubMed] |
| 14. | Ramchandani S, Bhattacharya SK, Cervoni N, Szyf M. DNA methylation is a reversible biological signal. Proc Natl Acad Sci U S A. 1999;96:6107-6112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 239] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 15. | Nan X, Meehan RR, Bird A. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;21:4886-4892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 412] [Cited by in RCA: 428] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 16. | Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 636] [Cited by in RCA: 646] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 17. | Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471-481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 908] [Cited by in RCA: 955] [Article Influence: 34.1] [Reference Citation Analysis (0)] |
| 18. | To KF, Leung WK, Lee TL, Yu J, Tong JH, Chan MW, Ng EK, Chung SC, Sung JJ. Promoter hypermethylation of tumor-related genes in gastric intestinal metaplasia of patients with and without gastric cancer. Int J Cancer. 2002;102:623-628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 99] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 19. | Patra SK, Patra A, Zhao H, Dahiya R. DNA methyltransferase and demethylase in human prostate cancer. Mol Carcinog. 2002;33:163-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 156] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 20. | Darwanto A, Kitazawa R, Maeda S, Kitazawa S. MeCP2 and promoter methylation cooperatively regulate E-cadherin gene expression in colorectal carcinoma. Cancer Sci. 2003;94:442-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 41] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 21. | Müller HM, Fiegl H, Goebel G, Hubalek MM, Widschwendter A, Müller-Holzner E, Marth C, Widschwendter M. MeCP2 and MBD2 expression in human neoplastic and non-neoplastic breast tissue and its association with oestrogen receptor status. Br J Cancer. 2003;89:1934-1939. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 60] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 22. | Sato M, Horio Y, Sekido Y, Minna JD, Shimokata K, Hasegawa Y. The expression of DNA methyltransferases and methyl-CpG-binding proteins is not associated with the methylation status of p14(ARF), p16(INK4a) and RASSF1A in human lung cancer cell lines. Oncogene. 2002;21:4822-4829. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 68] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 23. | Etoh T, Kanai Y, Ushijima S, Nakagawa T, Nakanishi Y, Sasako M, Kitano S, Hirohashi S. Increased DNA methyltransferase 1 (DNMT1) protein expression correlates significantly with poorer tumor differentiation and frequent DNA hypermethylation of multiple CpG islands in gastric cancers. Am J Pathol. 2004;164:689-699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 223] [Article Influence: 10.6] [Reference Citation Analysis (0)] |