INTRODUCTION
Transcriptional regulation is controlled through interactions between DNA and protein complex, the latter contains transcription factors with highly conserved protein motifs. The most well known motifs are helix-turn-helix, helix-loop-helix, and zinc finger. During cell differentiation and development, each of these domains is involved in the binding of transcription factors to their cognate DNA recognition sites, resulting in specific activation or repression of gene expression[1].
Zinc finger gene family belongs to one of the largest human gene families and plays an important role in the regulation of transcription. This large family may be divided into many subfamilies such as Cys2/His2 type, glucocorticoid receptor, ring finger, GATA-1 type, GAL4 type, and LIM family[2-4]. In the Cys2/His2 type of zinc finger genes, there is a highly conserved consensus sequence TGEKPYX (X representing any amino acid) between adjacent zinc finger motifs. The zinc finger proteins containing this specific structure are named Krüppel-like zinc finger proteins because the structure was first found in Drosophila Krüppel protein[1].
ZNF191 is a putative transcription factor belonging to Krüppel-like zinc finger gene family. It contains four Cys2/His2 zinc fingers in its C-terminus, and one SCAN box element (also known as LeR domain for leucine-rich) in its N-terminus[5-7]. Biochemical binding studies showed SCAN as a selective heterologous and homologous oligomerization domain[7,8]. Tissue mRNA analysis showed that ZNF191 gene was ubiquitously expressed[5,9]. ZNF191 can specifically bind to the TCAT repeats (HUMTH01) in the first intron of the human tyrosine hydroxylase (TH) gene. HUMTH01 may regulate transcription of TH gene, which encodes the rate-limiting enzyme in the synthesis of catecholamines[9-11]. The disturbances of catecholaminergic neurotransmission have been implicated in neuropsychiatric and cardiovascular diseases[12-17]. These studies suggested that ZNF191 might be relevant to these diseases. Analysis of amino acid sequence of ZNF191 showed 94% identity with the murine sequence of ZF-12[18]. Mouse ZF-12 gene likely represents the murine counterpart of human ZNF191. ZF-12 also contains four zinc finger motifs of the Cys2/His2 type and one SCAN box. ZF-12 mRNA is expressed during embryonic development and in different organs in adult[19]. ZF-12 may play an important role in cartilage differentiation and basic cellular processes[19].
To facilitate the functional studies of ZNF191, we established transgenic mice carrying ZNF191 gene. Four transgenic mice were identified by PCR and Southern blot, and used as founders to establish transgenic mouse lineages. The results of F1 identification with PCR showed that ZNF191 gene could be transmitted stably. RT-PCR analysis demonstrated that ZNF191 gene was expressed in multiple tissues of transgenic mice. Pathological analysis results demonstrated that over-expression of ZNF191 did not cause obvious pathological changes in multiple tissues of the transgenic mice.
MATERIALS AND METHODS
Plasmid
pcDNA3-ZNF191 containing full length ZNF191 cDNA under control of CMV promoter.
Animals
C57, CBA mice were maintained by Shanghai Nanfang Research Center for Biomodel Organism. Transgenic mice were raised and bred in the Laboratory Animal Centre of Second Military Medical University, Shanghai.
Generation of transgenic mice
The 6.5 kb linearized pcDNA3-ZNF191 was purified from agarose gel with QIAGEN gel extraction kit (Qiagen), adjusted to a final concentration of 2 μg/ml in TE buffer and used as DNA solution in microinjection. The first generation (F1) female hybrids of C57 and CBA mice were hormonally superovulated and mated with F1 male hybrids. The next morning fertilized one-cell eggs were collected from the oviduct. The eggs were microinjected with the DNA solution under a microscope. The injected fertilized eggs were transplanted into the oviduct of pseudo-pregnant F1 hybrids of C57 and CBA mice.
Identification of transgenic mice
Founder (G0) mice were identified by PCR and Southern blotting analysis. For PCR analysis, genomic DNA was extracted from tails, and amplified using pcDNA3-ZNF191 primers (P1: 5’-ATGCGGTGGGCTCTATG-3’; P2: 5’-CGGCTTCCATCCGAGTA-3’) which produced a 1 353 bp fragment from mice carrying the transgene. For Southern blotting analysis, genomic DNA was digested overnight with HindIII and subjected to electrophoresis in a 0.7% agarose gel and transferred to nylon membrane (Millipore Co., Ltd). Hybridization was performed under stringent conditions with a random-primed (α-32P)-labeled ZNF191 cDNA probe.
Expression of transgene
One of the transgenic mouse lineages was used to study the expression of transgene. Total RNA was isolated from tissues with TRIzol reagent (Invitrogen), according to the manufacturer’s instructions. After digestion of the RNA with DNase I (RNase free), first strand cDNA was synthesized by reverse transcription (Promega). RT-PCR reactions were performed using primers (P3:5’-GTACTAGTAGAAGACATGGT-3’, P4:5’-CGCACACAAAAGAACAAATCT-3’) for ZNF191 cDNA, which produced a 394 bp fragment. PCR reactions were performed 35 cycles at 95 °C for 45 s, at 55 °C for 1 min, and at 72 °C for 1 min. The PCR products were electrophoresed on 1.2% agarose gel.
Transmission of transgene
To study transmission of the transgene in mice, transgenic founder mice were mated with normal C57 mice to produce the first generation (F1) which was identified by PCR analysis using primers P1/P2. F1 mice of the same founder carrying the transgene were mated between brother and sister mice to produce the second generation (F2). F2 mice were also identified by PCR analysis.
Histological examination
Various organs were dissected from the 10 weeks old mice and fixed in 10% formalin. Sections were obtained from paraffin-embedded tissue samples, stained with H&E, and examined under a microscope and photographed. Pathological analysis was carried out in the Department of Pathology, Changhai Hospital of Second Military Medical University, Shanghai.
RESULTS
Establishment of ZNF191 transgenic mice
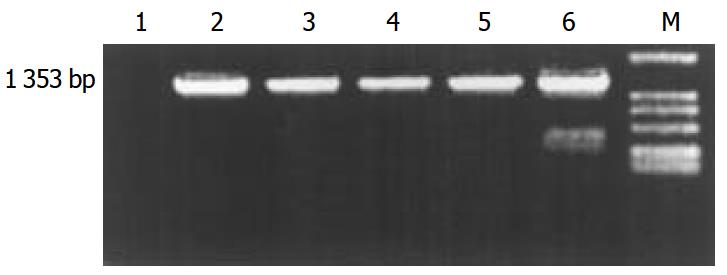
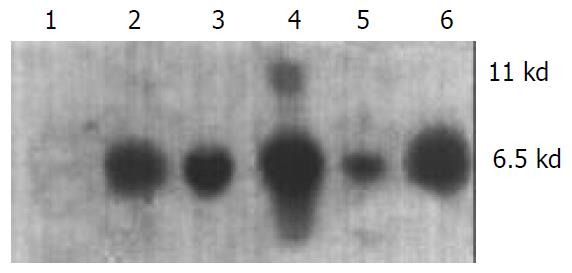
The transgene fragment containing full length ZNF191 cDNA was microinjected into the male pronuclei of 582 fertilized oocytes of F1 hybrids between C57 and CBA mice. The injected eggs were implanted into the oviducts of 27 pseudo-pregnant foster mothers, of which 15 mice became pregnant and gave birth to 81 offsprings. Four offsprings were identified to carry ZNF191 cDNA by PCR and Southern blotting analysis (Figures 1 and 2). The ratio of transgene integration was 4.9% by PCR and Southern blotting analysis.
Figure 1 PCR results in transgenic mice.
M: DL-2000 Marker, 1: Negative control (wild type mouse), 2-5: Transgenic founder mice, 6: Positive control (pcDNA3-ZNF191).
Figure 2 Results of genomic DNA Southern blotting analysis in transgenic mice.
1: Negative control (wild type mouse), 2-5: Transgenic founder mice, 6: Positive control (pcDNA3-ZNF191).
ZNF191 transgene expression in multiple tissues of transgenic mice
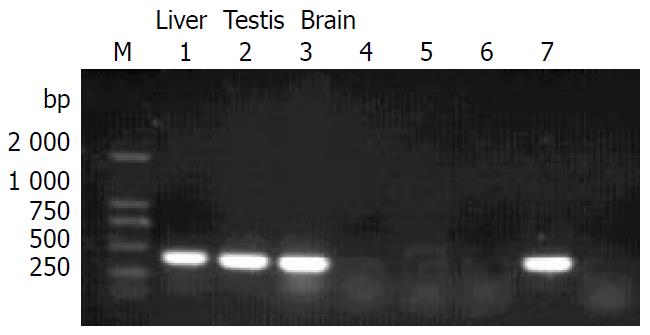
The transgene was driven by CMV promoter. To study whether it could be expressed in multiple tissues of transgenic mice, we analysed the tissue expression profile of ZNF191 transgene by RT-PCR. The results (Figure 3) showed that ZNF191 transgene was expressed in the liver, testis and brain.
Figure 3 RT-PCR results in transgene expression.
M: DL-2000 Marker, 1-3: Transgenic mice tissues, 4-5: Negative control (non-reverse transcripted RNA), 6:C57 mouse liver, 7: Positive control (pcDNA3-ZNF191).
Genetics of transgenic mice
To establish transgenic mouse lineages, the founder mice were mated to C57 mice to produce F1 mice. Among the 43 mice of first generation, 19 were identified as carrying ZNF191 cDNA transgene by PCR analysis. The ratio of transgene transmission was 44.2%. F1 mice from the same founder were mated with each other to produce F2 mice. Sixty-one out of 86 F2 mice were ZNF191 transgenic mice, with a transgene transmission ratio of 71%. These results showed that the inheritance of ZNF191 transgene was in accordance with Mendel’s laws, and the transgene was integrated into the chromosome at a single site and could be transmitted stably.
Pathological analysis of tissues
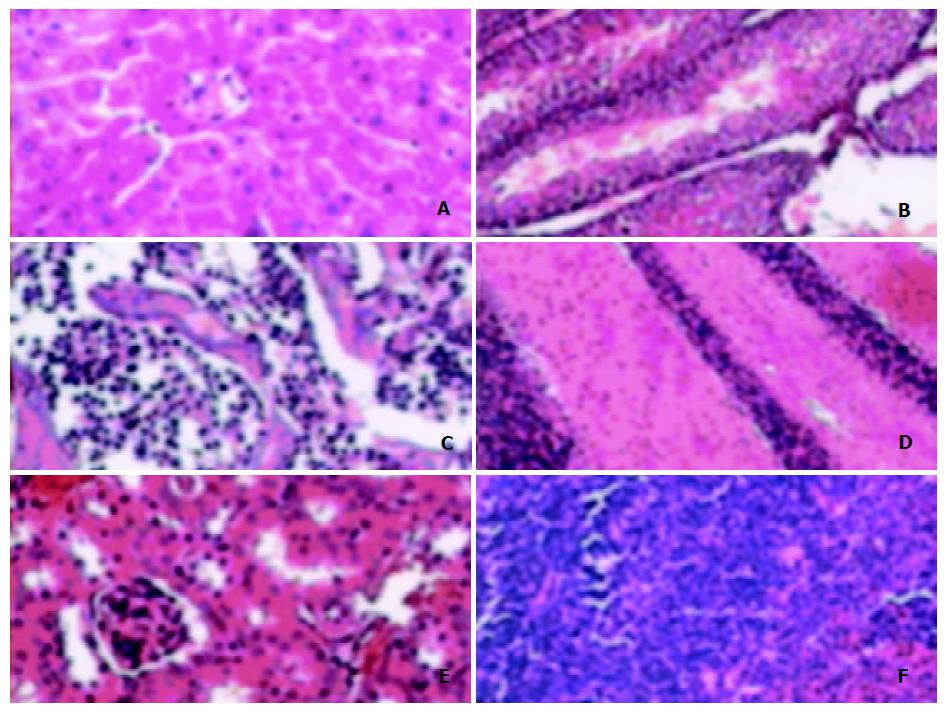
Pathological analysis was carried out to see whether the expression of ZNF191 transgene would cause any pathological changes in tissues of the transgenic mice. The results (Figure 4) showed there were no obvious pathological changes in the tissues examined. Therefore, so far as the tissues examined, the expression of exogenous ZNF191 did not cause any pathological consequences in the transgenic mice.
Figure 4 Morphology of tissues of transgenic mouse.
A: Liver, B: Testis, C: Marrow, D: Brain, E: Kidney, F: Spleen.
DISCUSSION
Cys2/His2 type zinc finger gene family is one of the largest gene families, and each member has repeated zinc finger motifs containing a finger-like structure, in which two cysteines and two histidines could covalently bind to one zinc ion[20]. It is estimated that in this huge family, about one third of the members are Krüppel-like genes as characterized by the presence of highly conserved connecting sequences “TGEKPYX” between adjacent zinc finger motifs. Substantial evidences indicate that Krüppel proteins are important players in many physiological processes as transcriptional regulators. Krüppel-like zinc finger genes are key transcriptional repressors in the development of Drosophila. In mammalian animals and human, these genes have been found involved in embryo development and carcinogenesis. For example, KLF6 was reported as a tumor suppressor gene in prostate cancer[21-23]. Gut-enriched Krüppel-like factor (GKLF) was expressed abundantly in epithelial cells of gastrointestinal tract, and deregulation of GKLF was linked to several types of cancer[24-27].
ZNF191 gene is a novel member of the Krüppel-like zinc finger gene family. It was previously cloned from bone marrow and promyelocytic leukemia cell line NB4 using homologous PCR amplification with primers based on conserved sequences of the Krüppel-like family of transcription factors[5]. This first identification suggested that ZNF191 played a role in hematopoiesis. ZNF191 has been found to contain a SCAN domain mediating selective protein oligomerization[6-8]. ZNF191 specifically binds to HUMTH01 in vitro. The microsatellite HUMTH01, located at the first intron of tyrosine hydroxylase (TH) gene, is characterized by a TCAT repeated motif and has been used in genetic studies of neuropsychiatric and cardiovascular diseases, in which disturbances of catecholaminergic neurotransmission were implicated[10-17]. Allelic variations of HUMTH01 had a quantitative silencing effect on TH gene expression in vitro, and correlated with quantitative and qualitative changes in the binding by ZNF191[9]. Since TCAT repeated sequence is widespread throughout the genome, this phenomenon might be relevant to the quantitative expression of several genes implicated in complex genetic traits, both normal and pathological[28-31]. These studies suggested that ZNF191 might be relevant to neuropsychiatric and cardiovascular diseases. It is vital to have a transgenic or knock-out animal model for the study of the biological role of ZNF191. So far, the attempt to produce ZNF191 transgenic and knock-out mice has not been reported. To our knowledge, we have generated the first transgenic mice carrying ZNF191 transgene and ZF-12+/- mice (to be reported in an other paper).
In in vivo situation, ZNF191 transcript was found in various organs[5,9]. Moreover, ZNF191 expression was significantly up-regulated in liver cancer (personal communication), suggesting that ZNF191 might be relevant to hepatocellular carcinogenesis. In this study, ZNF191 transgene was expressed in multiple tissues, which are capable of expressing endogenous ZF-12 in vivo. Therefore, the ZNF191 transgenic mouse we generated would be an invaluable animal model not only for the investigation of ZNF191 functions in hepatic tissues, but also in other tissues, which may ultimately reveal the undefined biological functions of ZNF191.
Given that ZNF191 has important biological functions and is relevant to many diseases, it is somewhat unexpected that no pathological changes occurred in the ten-week transgenic mice with ZNF191 over-expression. It is possible that overexpression of exogenous ZNF191 may trigger a negative feedback response to the expression of endogenous ZF-12. It remains to be determined whether the expression of endogenous ZF-12 would change in cells overexpressing exogenous ZNF191. On the other hand, liver cancer, neuropsychiatric and cardiovascular diseases involve many factors and usually have a long incubation time before any pathological phenotypes can be observed. So, it is necessary to continue the investigation into possible pathological changes in a long term follow-up.
In conclusion, we reported here the successful generation of a transgenic mouse model expressing ZNF191 gene. Future studies should focus on the physiological and pathological changes in this mouse model using powerful analytic methods such as microarray comparisons of gene expression profiles among normal, transgenic, ZF-12+/- and ZF-12-/- mice.