Published online Jan 15, 2004. doi: 10.3748/wjg.v10.i2.234
Revised: August 9, 2003
Accepted: August 16, 2003
Published online: January 15, 2004
AIM: Over 90% of drugs are metabolized by the cytochrome P-450 (CYP) family of liver isoenzymes. The most important enzymes are CYP1A2, 3A4, 2C9/19, 2D6 and 2E1. Although CYP2D6 accounts for < 2% of the total CYP liver enzyme content, it mediates metabolism in almost 25% of drugs. In order to study its enzymatic activity for drug metabolism, its cDNA was cloned and a HepG2 cell line stably expressing CYP2D6 was established.
METHODS: Human CYP2D6 cDNA was amplified with reverse transcription-polymerase chain reaction (RT-PCR) from total RNA extracted from human liver tissue and cloned into pGEM-T vector. cDNA segment was identified by DNA sequencing and subcloned into a mammalian expression vector pREP9. A cell line was established by transfecting the recombinant plasmid of pREP9-CYP2D6 to hepatoma HepG2 cells. Expression of mRNA was validated by RT-PCR. Enzyme activity of catalyzing dextromethorphan O-demethylation in postmitochondrial supernant (S9) fraction of the cells was determined by high performance liquid chromatography (HPLC).
RESULTS: The cloned cDNA had 4 base differences, e.g. 100 C→T, 336 T→C, 408 C→G and 1 457 G→C, which resulted in P34S, and S486T amino acid substitutions, and two samesense mutations were 112 F and 136 V compared with that reported by Kimura et al (GenBank accession number: M33388). P34S and S486T amino acid substitutions were the characteristics of CYP2D6*10 allele. The relative activity of S9 fraction of HepG2-CYP2D6*10 metabolized detromethorphan O-demethylation was found to be 2.31 ± 0. 19 nmol·min-1·mg-1 S9 protein (n = 3), but was undetectable in parental HepG2 cells.
CONCLUSION: cDNA of human CYP2D6*10 can be successfully cloned. A cell line, HepG2-CYP2D6*10, expressing CYP2D6*10 mRNA and having metabolic activity, has been established.
- Citation: Zhuge J, Yu YN, Wu XD. Stable expression of human cytochrome P450 2D6*10 in HepG2 cells. World J Gastroenterol 2004; 10(2): 234-237
- URL: https://www.wjgnet.com/1007-9327/full/v10/i2/234.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i2.234
Over 90% of drugs are metabolized by the cytochrome P-450 (CYP) family of liver isoenzymes[1]. The most important enzymes are CYP1A2, 3A4, 2C9/19, 2D6 and 2E1. Although CYP2D6 accounts for < 2% of the total CYP liver enzyme content, it mediates metabolism in almost 25% of drugs. Among these are many antipsychotics and antidepressants, beta-blockers, antiarrhythmic agents and opiates[2,3]. CYP2D6 exhibits extensive polymorphism. Over 40 CYP2D6 allelic variants have been discovered[4] (http://www.imm.ki.se/ CYPalleles/cyp2d6.htm).
Human CYP1A1[5], CYP2B6[5], CYP2A6[6], CYP3A4[7], CYP2C9[8], CYP2C18[9] and a phase II metabolism enzyme UDP-glucuronosyltransferase, UGT1A9[10] have been stably expressed in Chinese hamster lung CHL cells in our laboratory. Among the human hepatic cell lines, HepG2 is derived from a human liver tumor and characterized by many xenobiotic-metabolizing activities as compared to fibroblasts. Therefore, HepG2 cell is useful in the prediction of the metabolism and cytotoxicity of chemicals in human liver[11]. But it does not produce a significant amount of CYP[12,13]. Yoshitomi et al[14] have established stable expression of a series of human CYP subtypes, e.g. CYP1A1, CYP1A2, CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP2E1 and CYP3A4, respectively in the HepG2 cells.
In this study human CYP2D6*10 cDNA was amplified with reverse transcription-polymerase chain reaction (RT-PCR), and a cell line stably expressing CYP2D6.10 was established.
Restriction endonucleases and Moloney murine leukemia virus (M-MuLV) reverse transcriptase were supplied by MBI Fermentas AB, Lithuania. PCR primers, DNA sequence primers, random hexamer primers and dNTPs were synthesized or supplied by Shanghai Sangon Biotechnology Co. Expand fidelity PCR system and NADPH were from Roche Molecular Biochemicals. DNA sequencing kit was purchased from Perkin-Elmer Co. TRIzol reagent, G418, Dulbecco’s modified Eagle’s medium (DMEM) and newborn bovine calf sera were from Gibco. Diethyl pyrocarbonate (DEPC), dextromethorphan HBr and dextrophan D-tartrate were purchased from Sigma/RBI. T4 DNA ligase and pGEM-T vector system were from Promega. HPLC solvents and other chemicals were all of the highest grade from commercial sources.
Cloning of human CYP2D6 cDNA from human liver Total RNA was extracted from a surgical specimen of human liver with TRIzol reagent according to the manufacture’s instructions. RT-PCR amplifications using expand fidelity PCR system were described before. Two specific 28-mer oligonucleotide PCR primers were designed according to the cDNA sequence of CYP2D6 reported by Kimura et al[15] (GenBank accession no.M33388). The sense primer corresponding to base position -12 to 13 was CYP2D6 F1: 5’-CTCGAGGCAGGTATGGGGCTAGAAG-3’, with a restriction site of Xho I, and the anti-sense one, corresponding to the base position from 1 503 to 1 530, was CYP2D6 R1: 5’-GGATCCTGAGCAGGCTGGGGACTAGGTA-3’, with a restriction site of BamH I. The anticipated PCR products were 1.543 kb in length. PCR was performed at 94 °C for 5 min, then 35 cycles at 94 °C for 60 s, at 62 °C for 60 s, at 72 °C for 2 min, and a final extension at 72 °C for 10 min. An aliquot (10 μL) from PCR was subjected to electrophoresis in a 1% agarose gel stained with ethidium bromide.
Construction of recombinant pGEM-CYP2D6 and sequencing of CYP2D6 cDNA[8] The PCR products were ligated with pGEM-T vector, and transformed to E. coli DH5α. cDNA of CYP2D6 cloned in pGEM-T was sequenced by dideoxy chain-termination method marked with BigDye with primers of T7, SP6 promoters and two specific primers of 5’-ACCTCATGAATCACGGCAGT-3’ (1 088-1 069), and 5’-CCGTGTCCAACAGGAGA-3’ (987-1 003). The termination products were dissolved and detected using an automated DNA sequencer (Perkin-Elmer-ABI Prism 310).
Construction of pREP9 based expression plasmid forCYP2D6[8] Xho I/BamH I fragment having the total span of human CYP2D6 cDNA in pGEM-CYP2D6 was subcloned to a mammalian expression vector pREP9 (Invitrogen). The recombinant was transformed to E. coli Top 10, screened by ampicillin resistant and identified by restriction mapping.
Transfection and selection[8,16] HepG2 cells were maintained as monolayer cell cultures at 37 °C in DMEM supplemented with 10% new born calf sera. HepG2 cells were transfected with the resultant recombinant plasmid, pREP9-CYP2D6, using a modified calcium phosphate method. A cell line named HepG2-CYP2D6 was established by selecting in the culture medium containing G418.
RT-PCR assay of CYP2D6 mRNA expression in HepG2-CYP2D6 and HepG2 cells Total RNA was preprared from G-418-resistant clones by TRIzol reagent. RT-PCR was performed as described before[8], using 200 mmol·L-1 of CYP2D6F1 and CYP2D6R1 primers and 200 mmol·L-1 primers of beta-actin as internal control. The sense and anti-sense primers used for PCR amplification of beta-actin (GenBank accession no. NM_001101) are 5’-TCCCTGGAGAAGAGCTACGA-3’ (776-795) and 5’-CAAGAAAGGGTGTAACGCAAC-3’ (1 217-1 237), respectively. PCR was performed at 94 °C for 2 min, then 35 cycles at 94 °C for 30 s, at 62 °C for 30 s, at 72 °C for 90 s, and a final extension at 72 °C for 7 min. The anticipated beta-actin PCR products were 462 bp in length and that of CYP2D6 were 1 543 bp in length. An aliquot (10 μL) from PCR was subjected to electrophoresis in a 1.2% agarose gel stained with ethidium bromide.
Preparation of postmitochondrial supernant (S9) of HepG2-CYP2D6 The procedure for the preparation of S9 fraction was described before[8]. The protein in S9 was determined by Lowry’s method, with bovine serum albumin as standard.
Dextromethorphan O-demethylation assays[17-20] CYP2D6 dextromethorphan O-demethylation activity of S9 was determined by reversed phase high performance liquid chromatography (HPLC). Briefly, incubation reactions were performed in 50 mmol·L-1 potassium phosphate buffer (pH7.4), containing 3 mmol·L-1 MgCl2, 1 mmol·L-1 EDTA, 40 mmol·L-1 dextromethophan and 200 μg S9 protein in a final volume of 200 μL. Reactions were initiated by addition of 1 mmol·L-1 NADPH and terminated with 30% acetic acid after incubation for 10 min at 37 °C. Protein was precipitated by centrifugation at 10 000 g for 4 min, and the supernatant was stored at -20 °C for analysis. On HPLC analysis, 10 μL of supernatant was injected into a Water HPLC equipped with a Shimadzu RF-535 fluorescence detector. A CLC phenyl column (15 cm×4.5-mm i.d.) was used to separate the metabolites. The mobile phase consisted of a mixture of 30% acetonitrile, 1% acetic acid, and 0.05% triethylamine in water. The flow rate through the column at 25 °C was 0.75 ml·min-1. The excitation and emission wavelengths of the fluorescence detector were 285 nm and 310 nm, respectively. The rates of product formation were determined from standard curves prepared by adding varying amounts of dextrophan D-tartrate to incubations conducted without NADPH.
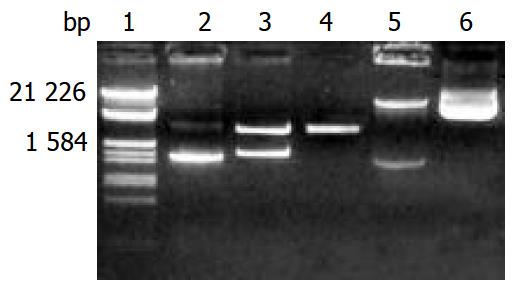
The pGEM-CYP2D6 recombinant was constructed by inserting human CYP2D6 cDNA into the pGEM-T vector. Selection and identification of the recombinant were carried out by Xho I/BamH I endonuclease digestion, agarose gel electrophoresis (Figure 1) and DNA sequencing. Compared with the cDNA sequence reported by Kimura et al[15] (GenBank accession no. M33388), differences were found in 100 C→T, 336 T→C, 408 C→G and 1457 G→C, that result in P34S and S486T amino acid substitutions, and two samesense mutations of 112 F and 136 V.
The Xho I/BamH I fragment (1.543 kb) containing the complete CYP2D6 cDNA was subcloned into the Xho I/BamH I site of mammalian expression vector pREP9. Selection and identification of the recombinant were carried out by Xho I/ BamH I endonuclease digestion and agarose gel electrophoresis (Figure 1). The resulting plasmid was designated as pREP9-CYP2D6 and contained the entire coding region, along with 11 bp of the 5’ and 35 bp of the 3’ untranslation region of CYP2D6 cDNA, respectively.
HepG2 cells were transfected with pREP9-CYP2D6, and selected with G418. The surviving clones were subcultured and the cell line termed HepG2-CYP2D6 was established.
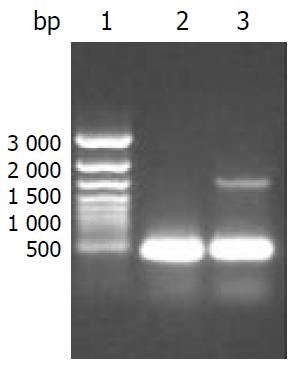
CYP2D6 mRNA expression in HepG2-CYP2D6 cells was detected by RT-PCR with CYP2D6F1 and CYP2D6R1 primers. It was easily to identify a 1.5 kb band from HepG2-CYP2D6 cells, but not from HepG2 cells (Figure 2).
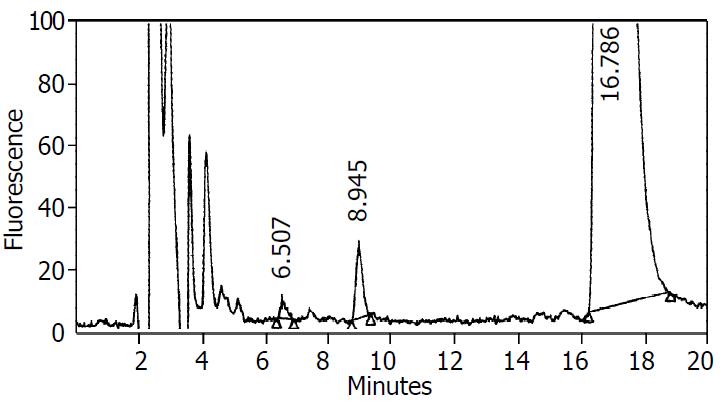
The dextromethorphan O-demethylation activity in S9 of HepG2-CYP2D6 cells was assayed by reverse HPLC. A typical elution profile of metabolites in supernatant was shown (Figure 3). The retention times for dextrophan and dextromethorphan were 6.5 min and 16.8 min, respectively. The CYP2D6 enzyme activity towards dextromethorphan O-demethylation was found to be 2.31 ± 0.19 nmol·min-1·mg-1 S9 protein (n = 3), but was undetectable in parent HepG2 cells.
The gene encoding CYP2D6 enzyme is localized on chromosome 22. Three major mutant alleles, termed CYP2D6*3, 4, and 5, associated with the poor metabolizer (PM) phenotype, were found early on in Caucasians[3]. CYP2D6 gene has turned out to be extremely polymorphic with 44 alleles described to 10-Nov-2003 (http://www.imm.ki.se/CYPalleles/cyp2d6.htm). Three fairly population specific alleles have been found with CYP2D6*4 in Caucasians, *10 in Asians and *17 in Africans[3]. The CYP2D6*10 allele with 100 C→T and 1457 G→C, can result in P34S and S486T amino acids substitute and an unstable enzyme with decreased catalytic activity. This allele occurred from 38% to 70% in Asian population[4]. The most frequent allele in Chinese was CYP2D6*10 allele with a frequency of about 51.3%[21], it was 57.2% in Guangdong Chinese population[22], 41.17% in Hong Kong Chinese population[23]. The CYP2D6 cDNA we cloned has the characteristics of CYP2D6*10 allele with 2 amino acid substitutions of P34S and S486T.
Ramamoorthy et al[24] have compared CYP2D6.10 with CYP2D6.1 in vitro in a baculovirus expression system using various substrates, such as dextromethorphan, P-methoxyamphetamine, 1-methyl-4-phenyl-1, 2, 3, 6-tetrahydropyridine and (+/-)3, 4-methylenedioxymethamphetamine, the ratio of intrinsic clearance (Vmax/Km) of CYP2D6.1 to CYP2D6.10 was 50, 34, 22 and 123, respectively.
Yu et al[25] reported that the purified CYP2D6.10 enzyme prepared from in vitro and to a high homogeneity was reconstituted with lipid and cytochrome P450 reductase, and exhibited an estimated enzyme efficiency (as Vmax/Km) 50-fold lower for dextromethophan O-demethylation and 100-fold lower for fluoxetine N-demethylation when compared with CYP2D6.1, whereas no measurable catalytic activity was observed for this variant toward codeine.
The intrinsic clearances (Vmax/Km) in reconstituted microsomes expressing CYP2D6.10 were reduced by 135-fold with (+/-)-3, 4-methylenedioxymethamphetamine and by 164-fold with dextromethorphan compared with that of wild-type CYP2D6.1[26].
Bufuralol 1’-hydroxylase activity in microsomes of yeast expressing CYP2D6.10 was rapidly decreased by heat treatment, supporting the idea that the thermal stability of the enzyme was reduced by amino acid replacement. Thermal instability together with the reduced intrinsic clearance of CYP2D6.10 is one of the causes responsible for the known fact that Orientals show lower metabolic activities than Caucasians for drugs metabolized mainly by CYP2D6[27].
Subjects homozygous for CYP2D6*10 had higher total areas under the plasma concentration-time curve, lower apparent oral clearances, and longer mean plasma half-life of nortriptyline than subjects in the CYP2D6*1/*1 and the heterozygous groups[28].
The plasma haloperidol concentration/dose ratio was significantly higher in older subjects (at least 50 years old) than in younger subjects with non-2D6*10 homozygous genotypes, but not for those with 2D6*10 homozygous genotype[29]. No significant differences in plasma concentration of fluvoxamine divided by daily dose of fluvoxamine per body weight ratio were found between subjects with no, one or two CYP2D6*10 alleles in Japanese subjects[30].
Cai et al[31] found that patients with homozygous mutant of CYP2D6*10 not only had a plasma concentration at peak (Cmax) of propafenone two times as high as those of wild-type genotype, but also showed a two-fold higher inhibitory rate of ventricular premature contractions compared with those with homozygous CYP2D6*1.
Venlafaxine, a new antidepressant, is metabolized mainly by CYP2D6 to an active metabolite, O-desmethylvenlafaxine. Cmax and areas under the plasma concentration-time curve of venlafaxine were 184% and 484% higher in the CYP2D6*10/*10 subjects than in the CYP2D6*1/*1 subjects[32].
Bufuralol 1’-hydroxylation has been commonly used by pharmaceutical industry to study in vitro drug interactions for CYP2D6[33]. Dextromethorphan has been a widely used probe drug for human CYP2D6 activity both in vitro and in vivo[34]. In humans, dextromethorphan is metabolized to dextrorphan, 3-methoxymorphinan and 3-hydroxymorphinan. CYP2D6 contributes at least 80% to the formation of dextrorphan, and CYP3A4 contributes more than 90% to the formation of 3-hydroxymorphinan. Dextromethorphan as a marker for monitoring both CYP2D6 and CYP3A activities has been found to be practical in human liver microsomal preparation[35].
The expression of CYP2D6*10 mRNA was validated by RT-PCR. The detromethorphan O-demethylation of HepG2-CYP2D6*10 was 2.31 ± 0.19 nmol·min-1·mg-1 S9 protein, which was higher than baculovirus expressed CYP2D6 (1.3420 ± 0.1466 nmol·min-1·mg-1 protein) and human liver microsome 0.17 to 0.30 nmol·min-1·mg -1 protein[36]. This cell line is a useful tool for further studies of the function and biochemical mechanism of CYP2D6.10 enzyme.
Edited by Zhang JZ and Wang XL
| 1. | Danielson PB. The cytochrome P450 superfamily: biochemistry, evolution and drug metabolism in humans. Curr Drug Metab. 2002;3:561-597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 520] [Cited by in RCA: 503] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 2. | Anzenbacher P, Anzenbacherová E. Cytochromes P450 and metabolism of xenobiotics. Cell Mol Life Sci. 2001;58:737-747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 686] [Cited by in RCA: 634] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 3. | Bertilsson L, Dahl ML, Dalen P, Al-Shurbaji A. Molecular genet-ics of CYP2D6: clinical relevance with focus on psychotropic drugs. Br J Clin Pharmacol. 2002;53:111-122. [RCA] [DOI] [Full Text] [Cited by in Crossref: 406] [Cited by in RCA: 384] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 4. | Bradford LD. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics. 2002;3:229-243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 593] [Cited by in RCA: 543] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 5. | Wu J, Dong H, Cai Z, Yu Y. Stable expression of human cytochrome CYP2B6 and CYP1A1 in Chinese hamster CHL cells: their use in micronucleus assays. Chin Med Sci J. 1997;12:148-155. [PubMed] |
| 6. | Yan L, Yu Y, Zhuge J, Xie HY. Cloning of human cytochrome P450 2A6 cDNA and its expression in mammalian cells. Zhongguo Yaolixue Yu Dulixue Zazhi. 2000;14:31-35. |
| 7. | Chen Q, Wu J, Yu Y. [Establishment of transgenic cell line CHL-3A4 and its metabolic activation]. Zhonghua Yu Fang Yi Xue Za Zhi. 1998;32:281-284. [PubMed] |
| 8. | Zhu GJ, Yu YN, Li X, Qian YL. Cloning of cytochrome P-450 2C9 cDNA from human liver and its expression in CHL cells. World J Gastroenterol. 2002;8:318-322. [PubMed] |
| 9. | Zhu-Ge J, Yu YN, Qian YL, Li X. Establishment of a transgenic cell line stably expressing human cytochrome P450 2C18 and identification of a CYP2C18 clone with exon 5 missing. World J Gastroenterol. 2002;8:888-892. [PubMed] |
| 10. | Li X, Yu YN, Zhu GJ, Qian YL. Cloning of UGT1A9 cDNA from liver tissues and its expression in CHL cells. World J Gastroenterol. 2001;7:841-845. [PubMed] |
| 11. | Rueff J, Chiapella C, Chipman JK, Darroudi F, Silva ID, Duverger-van Bogaert M, Fonti E, Glatt HR, Isern P, Laires A. Development and validation of alternative metabolic systems for mutagenicity test-ing in short-term assays. Mutat Res. 1996;353:151-176. [RCA] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 46] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 12. | Rodríguez-Antona C, Donato MT, Boobis A, Edwards RJ, Watts PS, Castell JV, Gómez-Lechón MJ. Cytochrome P450 expression in human hepatocytes and hepatoma cell lines: molecular mechanisms that determine lower expression in cultured cells. Xenobiotica. 2002;32:505-520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 294] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 13. | Jover R, Bort R, Gómez-Lechón MJ, Castell JV. Cytochrome P450 regulation by hepatocyte nuclear factor 4 in human hepatocytes: a study using adenovirus-mediated antisense targeting. Hepatology. 2001;33:668-675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 165] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 14. | Yoshitomi S, Ikemoto K, Takahashi J, Miki H, Namba M, Asahi S. Establishment of the transformants expressing human cytochrome P450 subtypes in HepG2, and their applications on drug metabolism and toxicology. Toxicol In Vitro. 2001;15:245-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 84] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 15. | Kimura S, Umeno M, Skoda RC, Meyer UA, Gonzalez FJ. The human debrisoquine 4-hydroxylase (CYP2D) locus: sequence and identification of the polymorphic CYP2D6 gene, a related gene, and a pseudogene. Am J Hum Genet. 1989;45:889-904. [PubMed] |
| 16. | Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning,A Labo-ratory Manual, 2nd ed. New York: Cold Spring Harbor Laboratory Press 1989; 6.28-6.29. |
| 17. | Kronbach T, Mathys D, Umeno M, Gonzalez FJ, Meyer UA. Oxidation of midazolam and triazolam by human liver cytochrome P450IIIA4. Mol Pharmacol. 1989;36:89-96. [PubMed] |
| 18. | Palamanda JR, Casciano CN, Norton LA, Clement RP, Favreau LV, Lin C, Nomeir AA. Mechanism-based inactivation of CYP2D6 by 5-fluoro-2-[4-[(2-phenyl-1H-imidazol-5-yl)methyl]-1-piperazinyl]pyrimidine. Drug Metab Dispos. 2001;29:863-867. [PubMed] |
| 19. | Yu A, Dong H, Lang D, Haining RL. Characterization of dextromethorphan O- and N-demethylation catalyzed by highly purified recombinant human CYP2D6. Drug Metab Dispos. 2001;29:1362-1365. [PubMed] |
| 20. | Barecki ME, Casciano CN, Johnson WW, Clement RP. In vitro characterization of the inhibition profile of loratadine, desloratadine, and 3-OH-desloratadine for five human cytochrome P-450 enzymes. Drug Metab Dispos. 2001;29:1173-1175. [PubMed] |
| 21. | Ji L, Pan S, Marti-Jaun J, Hänseler E, Rentsch K, Hersberger M. Single-step assays to analyze CYP2D6 gene polymorphisms in Asians: allele frequencies and a novel *14B allele in mainland Chinese. Clin Chem. 2002;48:983-988. [PubMed] |
| 22. | Gao Y, Zhang Q. Polymorphisms of the GSTM1 and CYP2D6 genes associated with susceptibility to lung cancer in Chinese. Mutat Res. 1999;444:441-449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 33] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 23. | Garcia-Barceló M, Chow LY, Chiu HF, Wing YK, Lee DT, Lam KL, Waye MM. Genetic analysis of the CYP2D6 locus in a Hong Kong Chinese population. Clin Chem. 2000;46:18-23. [PubMed] |
| 24. | Ramamoorthy Y, Tyndale RF, Sellers EM. Cytochrome P450 2D6.1 and cytochrome P450 2D6.10 differ in catalytic activity for multiple substrates. Pharmacogenetics. 2001;11:477-487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 68] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 25. | Yu A, Kneller BM, Rettie AE, Haining RL. Expression, purification, biochemical characterization, and comparative function of human cytochrome P450 2D6.1, 2D6.2, 2D6.10, and 2D6.17 allelic isoforms. J Pharmacol Exp Ther. 2002;303:1291-1300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 75] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 26. | Ramamoorthy Y, Yu AM, Suh N, Haining RL, Tyndale RF, Sellers EM. Reduced (+/-)-3,4-methylenedioxymethamphetamine ("Ecstasy") metabolism with cytochrome P450 2D6 inhibitors and pharmacogenetic variants in vitro. Biochem Pharmacol. 2002;63:2111-2119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 49] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 27. | Nakamura K, Ariyoshi N, Yokoi T, Ohgiya S, Chida M, Nagashima K, Inoue K, Kodama T, Shimada N, Kamataki T. CYP2D6.10 present in human liver microsomes shows low catalytic activity and thermal stability. Biochem Biophys Res Commun. 2002;293:969-973. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 36] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 28. | Yue QY, Zhong ZH, Tybring G, Dalén P, Dahl ML, Bertilsson L, Sjöqvist F. Pharmacokinetics of nortriptyline and its 10-hydroxy metabolite in Chinese subjects of different CYP2D6 genotypes. Clin Pharmacol Ther. 1998;64:384-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 54] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 29. | Ohara K, Tanabu S, Ishibashi K, Ikemoto K, Yoshida K, Shibuya H. Effects of age and the CYP2D6*10 allele on the plasma haloperidol concentration/dose ratio. Prog Neuropsychopharmacol Biol Psychiatry. 2003;27:347-350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 30. | Ohara K, Tanabu S, Ishibashi K, Ikemoto K, Yoshida K, Shibuya H. CYP2D6*10 alleles do not determine plasma fluvoxamine concentration/dose ratio in Japanese subjects. Eur J Clin Pharmacol. 2003;58:659-661. [PubMed] |
| 31. | Cai WM, Xu J, Chen B, Zhang FM, Huang YZ, Zhang YD. Effect of CYP2D6*10 genotype on propafenone pharmacodynamics in Chinese patients with ventricular arrhythmia. Acta Pharmacol Sin. 2002;23:1040-1044. [PubMed] |
| 32. | Fukuda T, Yamamoto I, Nishida Y, Zhou Q, Ohno M, Takada K, Azuma J. Effect of the CYP2D6 genotype on venlafaxine phar-macokinetics in healthy adult volunteers. Br J Clin Pharmacol. 1999;47:450-453. [RCA] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 64] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 33. | Yuan R, Madani S, Wei XX, Reynolds K, Huang SM. Evaluation of cytochrome P450 probe substrates commonly used by the pharmaceutical industry to study in vitro drug interactions. Drug Metab Dispos. 2002;30:1311-1319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 271] [Cited by in RCA: 251] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 34. | Streetman DS, Bertino JS, Nafziger AN. Phenotyping of drug-metabolizing enzymes in adults: a review of in-vivo cytochrome P450 phenotyping probes. Pharmacogenetics. 2000;10:187-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 314] [Cited by in RCA: 300] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 35. | Yu A, Haining RL. Comparative contribution to dextromethorphan metabolism by cytochrome P450 isoforms in vitro: can dextromethorphan be used as a dual probe for both CTP2D6 and CYP3A activities. Drug Metab Dispos. 2001;29:1514-1520. [PubMed] |
| 36. | Abdel-Rahman SM, Marcucci K, Boge T, Gotschall RR, Kearns GL, Leeder JS. Potent inhibition of cytochrome P-450 2D6-mediated dextromethorphan O-demethylation by terbinafine. Drug Metab Dispos. 1999;27:770-775. [PubMed] |